Abstract
Maize was genetically engineered for the biosynthesis of the high value carotenoid astaxanthin in the kernel endosperm. Introduction of a β-carotene hydroxylase and a β-carotene ketolase into a white maize genetic background extended the carotenoid pathway to astaxanthin. Simultaneously, phytoene synthase, the controlling enzyme of carotenogenesis, was over-expressed for enhanced carotenoid production and lycopene ε-cyclase was knocked-down to direct more precursors into the β-branch of the extended ketocarotenoid pathway which ends with astaxanthin. This astaxanthin-accumulating transgenic line was crossed into a high oil- maize genotype in order to increase the storage capacity for lipophilic astaxanthin. The high oil astaxanthin hybrid was compared to its astaxanthin producing parent. We report an in depth metabolomic and proteomic analysis which revealed major up- or down- regulation of genes involved in primary metabolism. Specifically, amino acid biosynthesis and the citric acid cycle which compete with the synthesis or utilization of pyruvate and glyceraldehyde 3-phosphate, the precursors for carotenogenesis, were down-regulated. Nevertheless, principal component analysis demonstrated that this compositional change is within the range of the two wild type parents used to generate the high oil producing astaxanthin hybrid.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are found in all groups of organisms with the exception of animals (Goodwin 1980) which have to supply their carotenoid demand from their diet. Biosynthesis of carotenoids other than β-carotene can be specific for certain families or even species. Carotenoids are present in all photosynthetic organisms. They protect against peroxidative processes (Krinsky 1989) and in animals they represent an essential source for vitamin A. Furthermore, color conferred by carotenoids is an aesthetic attractant (Vershinin 1999).
Carotenoids for animal feed or food coloring have the biggest share of the global carotenoid market (Tyczkowski and Hamilton 1986). Although natural sources of carotenoids are available and are used especially in poultry feeding, the market is dominated by chemically synthesized carotenoids (Sandmann 2015; Berman et al. 2015). Attempts have been made to develop and establish biological systems for carotenoid production which can commercially compete with chemical synthesis. This is especially attractive for astaxanthin which is the highest-priced carotenoids on the market and is used as a feed additive in salmon farming (Tyczkowski and Hamilton 1986). The only known natural sources for astaxanthin are a few bacteria, some green algae, one fungal species and plants belonging to the genus Adonis (Goodwin 1980). Currently, the Haematococcus and Paracoccus carotinifaciens are the only exploited natural astaxanthin sources (Ambati et al. 2014) but production costs are high and capacity low. Genetic engineering of an astaxanthin-producing organism for higher yields or the extension of carotenoid biosynthesis to astaxanthin are two promising strategies (Sandmann 2001). The first approach has been followed recently with the fungus Xanthophyllomyces dendrorhous (Gassel et al. 2014). Starting from a chemically induced mutant with already increased astaxanthin synthesis, limiting genes for carotenogenic reactions were over-expressed stepwise to generate a highly astaxanthin producing strain. Genetic engineering of astaxanthin high-level accumulation in a crop plant was most successful with a β-carotene forming tomato variety by extension of the pathway mediated by hydroxylase and ketolase transgenes (Huang et al. 2013).
In this study, our engineering approach involved the establishment of high carotenoid biosynthesis in a pathway ending with astaxanthin in maize seed endosperm by multigene transformation. The maize seeds can be used directly as feed additives. Engineering of high β-carotene maize was already described (Zhu et al. 2008)). Feeding chickens on this maize as sole pigment source resulted in superior pigmentation due to accumulation of carotenoids in muscle and skin including a higher vitamin A content as well as much improved tolerance to a pathogen which constraints poultry production if not treated with antibiotics (Nogareda et al. 2015). For better storage and extraction of astaxanthin, we chose a high-oil maize line to generate a prototype astaxanthin production line for further development. Here we report the generation and characterization of this line at the metabolome and proteome levels and conclude that even though perturbations in a number of primary metabolism pathways were identified, these were within the natural variation range in the parental lines.
Materials and methods
Maize material
Corn (Zea mays) variety M37W (white endosperm) was obtained from CSIR, Pretoria, South Africa. High-oil variety NSL 30876 referred to as NSL76 in this manuscript was kindly provided by USDA, ARS, NCRPIS, Iowa State University, Regional Plant Introduction Station, Ames, Iowa, USA. Both varieties, the selected transgenic line and the crosses were grown in the greenhouse at 28/20 °C day/night temperature with a 10 h photoperiod and 60–90 % relative humidity during the first 50 days, followed by maintenance at 21/18 °C day/night temperature with a 16 h photoperiod thereafter. Transgenic line M37W-bkt with the modified carotenoid pathway to synthesize astaxanthin was out-crossed with NSL76 resulting in line NSL76-bkt. Homozygous T2 and T3 generations were derived by selfing. Endosperm samples from immature seeds collected at 30 days after pollination (DAP) were frozen in liquid nitrogen and stored at −80 °C prior to use.
Vector construction and transformation
The phytoene synthase 1 cDNA (PSY1) was cloned from inbred maize line B73 by RT-PCR with forward primer 5′-AGGATCCATGGCCATCATACTCGTACGAG-3′ with BamHI site (underlined) and reverse primer 5′-AGAATTCTAGGTCTGGCCATTTCTCAATG-3′ with EcoRI site (underlined). Plasmid p326-ZmPSY1 (Zhu et al. 2008) is under the control of the LMW glutenin promoter (Colot et al. 1987). A maize lycopene ε-cyclase (LYCE) cDNA fragment was amplified by RT-PCR with ZmLYCE cDNA as template using forward (5′-GGAATTCTCTAGACGATCTCGGCGCCGCTCGGCTGCT-3′) with EcoRI and XbaI sites (underlined) and reverse primers (5′-gactagtggatcccaatgagacctacagtgagacct-3′) with SpeI and BamHI sites based on sequence information in GenBank (accession numbers EF622043). This sense DNA was cloned into the pHorP vector (Sørensen et al. 1996) containing the barley D-hordein promoter with a 300 bp gusA gene fragment and the ADP-glucose pyrophosphorylase terminator via restriction with XbaI and BamHI and the antisense LYCE fragment into the SpeI and EcoRI sites. The chemically synthesized truncated Chlamydomonas reinhardtii β-carotene ketolase gene (sCrBKT) (Zhong et al. 2011) fused to the pea small subunit of Rubisco (SSU) (Schreier et al. 1985) and 5´-untranslated region (5´UTR) of the rice alcohol dehydrogenase gene (Sugio et al. 2008) was inserted into pGZ63 vector (Zhu et al. 2008) between the maize γ-zein promoter and nos terminator using the BamHI and SacI restriction sites. The CrBKT and SSU codon usage was modified to a monocotyledonous plant preference. The CRTZ gene encoding beta-carotene hydroxylase from Brevundimonas sp. Strain SD212 (MBIC 03018) (Nishida et al. 2005) was chemically synthesized according to the codon usage of Brassica napus (accession number AB377272) and kindly provided by Dr. Norihiko Misawa, Ishikawa Prefectural University, Japan. The sBrCRTZ gene fused with the pea small subunit of Rubisco (SSU) and 5′-untranslated region (5´UTR) of the rice alcohol dehydrogenase gene was digested with BamHI and SacI. The digested fragments were cloned into the BamHI and SacI site of pGZ63 with the maize γ-zein gene promoter to generate pGZ63-sBrCRTZ. The bar gene encoding phosphinothricin N-acetyltransferase (Thompson et al. 1987) was used as a selectable marker. Details of the final plasmids can be found in Fig. 1a. The procedure used for transformation of M37W maize was described in Zhu et al. (2008). The highest-accumulating astaxanthin M37W-bkt lines were crossed with a high oil line (NSL76) to generate NSL76-bkt, which is the subject of the in depth metabolomic and proteomic analysis reported in this manuscript.
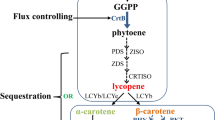
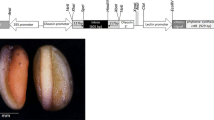
A Transgene expression vectors for ZmPSY1, RNAi-lyce, sBcrtZ and sCrbkt. Abbreviations: NOS nos terminator, ADGPP ADP-glucose pyrophosphorylase terminator; sBrcrtZ truncated β-carotene ketolase (BKT), UTR 5′-untranslated region of the rice alcohol dehydrogenase gene, TPS transit peptide sequence from the Phaseolus vulgaris small subunit of ribulose bisphosphate carboxylase, sCrbkt CRTZ gene encoding β-carotene hydroxylase. B Scheme describing the transformation of maize variety M37W yielding M37-bkt followed by crossing astaxanthin synthesis into the oil accumulating variety NSL76 and selection of high astaxanthin lines
Transcript analysis
Total RNA (30 µg) was fractionated on a denaturing 1.2 % (w/v) agarose gel containing formaldehyde prior to blotting. The membrane was probed with digoxigenin-labeled partial cDNAs prepared as above using the PCR-DIG Probe Synthesis Kit (Roche, Mannheim, Germany), with hybridization carried out at 50 °C overnight using DIG Easy Hyb. The membrane was washed twice for 5 min in 2 × SSC, 0.1 % SDS at room temperature, twice for 20 min in 0.2 × SSC, 0.1 % SDS at 68 °C, and then twice for 10 min in 0.1 × SSC, 0.1 % SDS at 68 °C. After immunological detection with anti-DIG-AP (Fab-Fragments Diagnostics GmbH, Germany) chemo luminescence generated by disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7] decan}-4-yl) phenyl phosphate (CSPD) (Roche, Mannheim, Germany) was detected on Kodak BioMax light film (Sigma-Aldrich, USA) according to the manufacturer’s instructions.
Primers for the probes were Zmpsy1-forward 5′-GTGTAGGAGGACAGATGAGCTTGT-3′, Zmpsy1-reverse 5′-CATCTGCTAGCCTGTGAGAGCTCA-3′, CrBKT-forward 5′-GGATCCTCAGCCAGGAGCCAGTGCAGCGCCTCT-3′, CrBKT-reverse 5′-GAATTCCATGGGGCCAGGCATTCAGCCCACTTCCG-3′, sBrCRTZ-forward 5′-ACGAATTCGATGGCCTGGCTGACGT-3′, sBrCRTZ-reverse 5′-TAGAGGATCCTCAGGCGCCGCTGCTGG-3′, GUS-forward 5-GGTCTAGAGGATCCGCACCTCTGGCAACCGGGTGAAGGT-3′, GUS-reverse 5-GTGAATTCACTAGTCGAGCATCTCTTCAGCGTAAGGGTAA-3′.
Quantitative real time PCR
Real-time PCR was performed on a BIO-RAD CFX96™ system using 25 µl mixture containing 10 ng of synthesized cDNA, 1X iQ SYBR green supermix (BIO-RAD) and 0.2 mM forward and reverse primer concentrations. Primers for maize endogenous lycopene ε-cyclase gene are 5′- AGTCCATCAATGCTTGCATGG-3′ (forward primer) and 5′-CATCTCGGCACCCTGAAAAAG-3′ (reverse primer). Primers for the internal control actin gene are 5′-CGATTGAGCATGGCATTGTCA-3′ (forward primer) and 5′-CCCACTAGCGTACAACGAA-3′ (reverse primer). To enable calculation of relative expression levels, serial dilutions (125–0.2 ng) were used for the generation of standard curves for each gene separately. PCR reactions were performed in triplicate using 96-well optical reaction plates. Cycling conditions consisted of a single incubation step at 95 °C for 5 min, followed by 40 cycles of 95 °C for 0:15 min, 58 °C for 1 min, 72 °C for 0:20 min. Specificity of amplification was confirmed by melt curve analysis of final PCR products with a temperature range of 50–90 °C with fluorescence acquired after every 0.5 °C increase. The fluorescence threshold value and gene expression data were calculated using the CFX96™ system software. Values represent the mean of three RT-PCR replicates ± SD.
Carotenoid analysis
The powdered seeds and endosperm samples were extracted with tetrahydrofurane/methanol (50:50, v/v) by heating for 20 min at 60 °C and then partitioned into 30 % ether in petrol. The collected upper phase was evaporated and re-dissolved in acetone for high-performance liquid chromatography (HPLC) analysis on a 15 cm Nucleosil C18, 3 µm column with a mobile phase of acetonitrile/2-propanol/methanol/(85:5:10, v/v/v). The flow was 1 ml/min, at 20 °C column temperature. Spectra were recorded online with a photodiode array detector 440 (Kontron, Straubenhard, Germany). Identification and quantification was performed by co-chromatography and comparison of spectral properties with authentic standards and reference spectra (Britton et al. 2004).
Metabolome analysis
General metabolite profiling of polar and non-polar metabolites from freeze-dried maize endosperm powder was performed as described recently (Decourcelle et al. 2015). After methanol extraction and addition of the internal standard ribitol, samples were derivatized with methoxyamine-HCl (Sigma-Aldrich) and N-methyltrimethylsilytrifluoroacetamide (Macherey–Nagel). Gas chromatography–mass spectrometry analysis was performed on an Agilent 7890A gas chromatograph with a 5975 MSD. For components identification, an in-house mass spectral library constructed from standards as well as the NIST 98MS library (Perez-Fons et al. 2014) were used. Quantification and identification were achieved using AMDIS (v 2.71) software facilitating integrated peak areas for specific compound targets (qualifier ions) relative to the ribitol internal standard peak. Data matrices were transformed using the pareto-scaled method (van den Berg et al. 2006) and multivariate analysis performed using SIMCA-P + 12.0 (Umetrics AB, Sweden). Pathway diagrams were created using the in-house developed software BioSynlab© (Royal Holloway, University of London) or Powerpoint. Means, standard deviation, p-values and q-values were calculated in Excel.
Proteome analysis
Proteome analysis included protein fractionation, tryptic digestion, peptide separation, and analysis by tandem mass spectrometry. Proteins of ground maize endosperms were extracted and proteins separated as recently described (Decourcelle et al. 2015). Mass spectrometry was carried out with a Q-TOF mass spectrometer (Maxis Impact, Bruker Daltonics) with a Captive Spray ion source interfaced with a nano-LC Ultimate 3000 (Thermo Scientific) at a flow rate of 20 µL/min using 0.1 % formic acid and separated with a reversed-phase capillary column (C18 PepMan10, 75 µm × 250 mm, 3 µm, 100A, Thermo Scientific) at a flow rate of 0.3 µl/min using a two steps gradient (8–28 % ACN with 0.1 % formic acid in 40 min then 28–42 % in 10 min), and eluted directly into the mass spectrometer.
Proteins were identified by MS/MS by data-dependent acquisition of fragmentation spectra of multiple charged peptides using Data Analysis software (Bruker Daltonics GmbH, Bremen, Germany) to generate peak lists. Protein identification was obtained by searching with X!Tandem (version 2013.09.01; http://www.thegpm.org/tandem/) against the Maizesequence.org (release AGPv3.21) and UniProtKB (maize taxonomy) (release 2014_03) combined database, using carbamidomethylation of cysteine as fixed modification, and N-terminus acetylation, deamidation of asparagine and glutamine, oxidation of methionine, and phosphorylation of serine, threonine and tyrosine as variable modifications. The functional annotations of proteins (GO) were established with MSDA (Mass Spectrometry Data Analysis, https://msda.unistra.fr/, Carapito et al. 2014).
Label-free quantification was carried out with the MassChroQ software (version 2.1) (Valot et al. 2011) based on extracted ion chromatograms. The detection threshold on min and max were set at 3000 and 5000, respectively. Data were filtered to remove (1) unreliable peptides for which standard deviation of retention time was superior to 60 s, (2) peptides shared by several proteins and (3) quantified peptides in less than 3 biological replicates. Normalization was performed to take into account possible global quantitative variations between LC–MS runs. Normalized peptide areas were calculated by dividing the area value of each peptide by the sum of all peptide area values for each LC–MS. Since a peptide can be detected in several SDS-PAGE bands, peptide abundance in one sample is calculated by summing the normalized areas of this peptide in each of these bands. And protein abundance was calculated by summing the normalized peptide area. Univariate differential analysis was performed with the more appropriate statistical test, Wilcoxon test (control of the normality and homoscedasticity hypotheses) with the “multi-test” package (‘R/Bioconductor’ statistical open source software. Multiple testing corrections enabled to adjust the p value of each marker to control the false discovery rate with the “multi-test” package (‘R/Bioconductor’ statistical open source software). Protein fold change was calculated after averaging protein areas between the biological replicates.
Results
Generation of astaxanthin maize lines
Our strategy for engineering a high-oil astaxanthin producing maize transformant is outlined in Fig. 1. It started with M37W as a suitable and efficient transformation host and the breeding of the engineered astaxanthin pathway into the high-oil NSL76 line. Starting with the wild type M37W with white endosperm, genetic transformation was carried out with additional copies of the endogenous phytoene synthase 1 gene, together with the bacterial β-carotene hydroxylase (crtZ) and algal β-carotene ketolase genes codon optimized for maize. Since astaxanthin is directly derived from zeaxanthin via β-carotene, and maize possess an active alternative route to lutein, an antisense construct of lycopene ε-cyclase was additionally included in the transformation (Fig. 1A) in order to divert lycopene preferentially into the β-branch of the carotenoid pathway. M37-bkt was selfed to generate lines which were used to breed with the high oil NSL76 line. The experimental strategy for the introgression of the modified carotenoid pathway into NSL76 yielding NSL76-bkt is shown in Fig. 1B.
Expression of carotenogenic genes
Transcript analysis for the expressed carotenogenic genes was carried out by mRNA blot analysis of the transgenes (Fig. 2). The ZmPSY1 transcript was not detectable in wild-type M37W endosperm, in agreement with previous investigations (Zhu et al. 2008), but it was expressed in the rest of the lines. The highest ZmPSY1 expression levels were observed in NSL76-bkt. sBrCRTZ, and CrBKT were expressed in the transgenic line and the crosses, although at different levels. Each transgene was expressed at similar levels in the transgenic line and the crosses and no expression was detected in the two wild-types M37W and NSL76. Amounts of the endogenous Zmlyce gene which was down-regulated by transformation with RNAi-LYCE were determined by quantitative real-time PCR in all four maize lines (Fig. 3). In both transformants M37-bkt and NSL76-bkt, the levels were only about one-sixth compared to the corresponding non-transformed lines, M37 and NSL76.
Carotenoid content and metabolite profiling
Due to the use of endosperm specific promoters, carotenoids were analysed from endosperm and whole seeds of the initial line M37 W, the M37-bkt transformant, the high oil line NSL76 and the NSL76-bkt hybrid (Table 1). The results demonstrate that M37W is low in carotenoids and only violaxanthin, lutein and zeaxanthin were detectable. The same carotenoids were found in the seeds of the other wild type NSL76 but with much higher concentrations of lutein. In NSL76 trace amounts of the lutein and zeaxanthin precursors α-cryptoxanthin and β-cryptoxanthin were detectable. In the transformant M37-bkt and the cross NSL76-bkt the two ketocarotenoids astaxanthin and 4-ketozeaxanthin were present in addition to β-carotene. Astaxanthin was the predominant carotenoid in M37-bkt as well as in NSL76-bkt. Astaxanthin concentrations in endosperm and whole seeds were in the same range in the two lines.
For the analysis of global metabolite changes of the final line NSL76-bkt versus its parent line NSL76, metabolite profiling of the intermediary metabolism was carried out with a GC-MS platform (Enfissi et al. 2010; Perez-Fons et al. 2014) which enables detection of most of the compound of maize intermediary metabolism (Decourcelle et al. 2015). Only the concentrations of sucrose and lactic acid were significantly increased (by more than 10-fold) (Table 2) whilst other metabolite pools were significantly decreased. These included mainly amino acids of the pyruvate family that is leucine, alanine and valine as well as serine and glycine and metabolically related glyceric acid. Furthermore, metabolites connected to the citric acid cycle were lower in NSL76-bkt. These comprise fumaric, malic and succinic acid together with amino acids of the aspartate family (aspartic acid and threonine) and of the glutamate familie (glutamic acid and proline).
Principal component analysis (PCA) of intermediary metabolism of the four lines described in this study was carried out in order to assess the effect of keto carotenoid accumulation on intermediary metabolism. PCA components 1 and 2 explain 61.8 and 17.7 % of the variability, respectively, and their corresponding score and loadings plots are shown in Fig. 4. The four lines separate out in the PCA score plot (Fig. 4A) indicating differences in the intermediary metabolism between groups of samples. The homozygous red endosperm line NSL76-bkt clusters closer to the M37W group (wild type) and away from its parental lines NSL76 and M37-bkt. The results in the loadings plot (Fig. 4B) indicate that sucrose, glucose and methyl galactose are responsible for the clustering of NSL76-bkt and M37W in the PCA score plot.
Proteomic profiling
Proteomic profiling was carried out with endosperm from seeds harvested 30 DAP and from mature seeds (MS) from NSL76 and NSL76-bkt lines. Using the decoy database and the tolerated presence of at least 2 peptides with an e-value smaller than 0.05 and a protein e-value smaller than 0.0001, the false discovery rate was 0.13 and 0.23 %, respectively, for peptide and protein identification for 30 DAP samples, and 0.12 and 0.17 %, respectively, for the MS samples. Our approach allowed the identification of 1688 non-redundant and grouped proteins at 30 DAP and 1220 at the MS stage. For data quantification, MassChroQ software was used and proteins with significant quantitative change, NSL76-bkt versus NSL-76, [Wilcoxon p value < 0.05 AUC (area under curve) >0.9 and with fold-change >1.5] were considered. In MS, 14 proteins varied in their abundance of which 3 were significantly increased and the others decreased. In endosperm of 30 DAP seeds, there were 17 variant proteins of which 4 were increased (Table 3).
In MS, an uncharacterized protein, stress-related peroxidase and acid phosphatase were found higher in NSL76-bkt maize compared to its parent line (Table 3). Of the characterized proteins related to glycolysis, the amounts of sugars and amino acid metabolism, malate dehydrogenase, endochitinase and glutamine synthase were decreased. At 30 DAP, no metabolic pathway-related protein with higher concentrations in the astaxanthin accumulating transformant was detectable (Table 4). Metabolic enzymes with lower abundance were 1,3-diphosphoglycerate phosphatase, trehalose-6-phosphate synthase and sucrose synthase. In addition, changes in storage proteins were observed with increased concentrations of a leguminin-like protein and a decrease of prolamin (Table 4).
Discussion
Carotenoid synthesis in seeds of engineered and high oil hybrid maize lines
We had reported earlier a library of maize transformants constructed by transfer of carotenogenic genes under endosperm-specific promoters for increased carotenoid production in seed endosperm (Zhu et al. 2008). Among these a particular line with low astaxanthin content (about 3 % of total carotenoids) was recovered. This proof of concept experiment encouraged us to select more suitable genes for a more efficient astaxanthin synthesis in maize. To overcome the problem of ketolase and hydroxylase competition during astaxanthin biosynthesis from β-carotene (Zhong et al. 2011), we transformed maize line M37W with β-carotene hydroxylase (crtZ) from Brevundimonas and β-carotene ketolase from Chlamydomonas reinhardtii which were shown to be highly efficient in tobacco (Hasunuma et al. 2008) and tomato (Huang et al. 2013). In order to enhance the pool of β-carotene as precursor of astaxanthin, we over-expressed phytoene synthase 1 (resulting in a seven- to eight-fold increase of total seed carotenoids) and knocked-down lycopene ε-cyclase. The latter modification decreased the transcript levels (Fig. 3) which in turn shifted metabolite distribution into the β branch of the carotenoid pathway from which astaxanthin originates. This is indicated by the changing ratios of β-branch carotenoids (all except lutein and α-carotene) versus the ε-branch carotenoids (lutein plus α-carotene) from 0.3 in M37W to 25 in the final NSL76-bkt line (Table 1). All modifications were monitored by mRNA blot analysis in M37-bkt and in NSL76-bkt (Fig. 2). After crossing of the astaxanthin pathway into the high-oil NSL76 line, astaxanthin accumulation in the seeds increased by about 50 % (Table 1). This may be due to higher storage capacity for lipophilc compound in an oil environment, although only about 15 % of the oil is stored in the endosperm (Hartings et al. 2012). NSL76-bkt accumulated 60 % of whole seed carotenoids as astaxanthin and 45 % of the endosperm carotenoids. This resembles a highly efficient astaxanthin biosynthesis pathway near the biosynthesis efficiency in transgenic tobacco (Hasunuma et al. 2008) and tomato (Huang et al. 2013). Due to the high starch content of the seeds and the endosperm, the astaxanthin content in maize was lower compared to tomato. However, the combination of astaxanthin and starch content makes NSL76-bkt a useful feed component, e.g. for fish farming and for raising poultry.
Metabolism in seeds of NSL76-bkt
The increase of total carotenoid content in M37 W-bkt and NSL76-bkt (Table 1) is due to the over-expression of the phytoene synthase 1 gene which has been shown in different plant species to limit carotenoid synthesis (Sandman et al. 2006). The enhanced metabolite flow into the carotenoid pathway affects primary metabolism as a source for terpenoid pathway precursors in the transformants as already demonstrated for maize high in zeaxanthin and other carotenoids (Decourcelle et al. 2015). For NSL76 and NSL76-bkt, metabolic changes were analysed by comparing pool sizes of compounds and amounts of enzymes. Most evident was the increase of sucrose and lactate pools in the endosperm (Table 2). Maize kernels are supplied from the green parts of the plant with different sugars including sucrose (Alfonso et al. 2011). Sucrose can be further metabolized to glucose and fructose by invertase or to fructose and UDP-glucose by sucrose synthase as the initial reaction for starch synthesis (Spielbauer et al. 2006). The concentration of the latter enzyme was decreased providing less UDP-glucose for the synthesis of other sugars, including trehalose which is additionally supported by lower trehalose synthase concentrations in NSL76-bkt (Table 4). These changes in enzyme amounts preferentially support glycolytic metabolism to the precursors for carotenoid biosynthesis. Lactate can be considered as a key compound for the provision of pyruvate together with glyceraldehyde 3-phosphate, the major substrates for the deoxyxylulose 5-phosphate pathway leading to terpenoids (Eisenreich et al. 2001). Lactate accumulation (Table 2) coincides with lower activities of pools of the citric acid cycle components and lower concentrations of malate dehydrogenase. This indicates a reduced flow of pyruvate into the citric acid cycle (Table 3). It appears that pathways competing with glycolytic pyruvate formation or competing with the deoxyxylulose 5-phosphate pathway for pyruvate are down-regulated. This is also the case for decreased formation of glycerate out of the glycolytic pathway (Table 2). A similar down-regulation of starch biosynthesis, with a concurrent increase of the sucrose pool, also competing with glycolysis was found in an engineered high-carotenoid maize line (Decourcelle et al. 2015).
A very strong decrease of amino acid pools from most amino acid families was measured in NSL76-bkt (Table 2). This includes all amino acids derived from pyruvate which is needed for higher carotenoid synthesis. The concentration of a legumin-like protein was increased in the endosperm at 30 DAP (Table 4). In this maize storage protein, the predominant amino acids are histidine and glutamine (Yamagata et al. 2003) which in contrast to several other amino acids remained unchanged in NSL76-bkt compared to NSL76. Unlike legumin, concentrations of prolamin, another storage protein were decreased in NSL76-bkt. This may be due to lack of leucine, alanine and proline in NSL76-bkt (Table 2) which are the major amino acids in maize prolamins (Kim and Okita 1988).
No characterized enzyme related to primary metabolism in endosperm either at 30 DAP or in mature seeds was up-regulated in NSL76-bkt. The only enzyme present in higher concentration in NSL76-bkt was a peroxidase (Table 3). In maize kernels, this enzyme is part of an antioxidative system to cope with oxidative stress (Corona-Carrillo et al. 2014) which seems to be enhanced in NSL76-bkt. Despite the variability in the metabolic profile of NSL76-bkt compared to NSL76, the principal component analysis (Fig. 4) indicated that the changes are within the natural variation of the two wild type varieties M37W and NSL76.
Conclusion
High astaxanthin accumulation in maize endosperm can be achieved by over-expression of the gene encoding phytoene synthase 1, the limiting enzyme of the pathway, knock-down of the competing ε-branch and selection of interactive hydroxylase and ketolase genes. Metabolome and proteome analysis provided basic insights into how maize primary metabolism supports the synthesis of high levels of astaxanthin from pyruvate and glyceraldehyde 3-phosphate in the NSL76-bkt line by favouring the flow of these precursors towards carotenoid synthesis. With this genetic engineering approach, we were successful in generating a staple crop as a source of corn oil containing the highly antioxidative carotenoid astaxanthin. This corn line can also be used directly as a feed to supply astaxanthin together with oil and carbohydrate e.g. in fish farming. Alternatively, NSL76-bkt can be used as a raw material for the extraction of an oily astaxanthin preparation which will form the basis for other industrial processes.
References
Alfonso AP, Val DL, Shachar-Hill T (2011) Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metab Eng 13:96–107
Ambati RR, Moi PS, Ravi S, Aswathanarayana RG (2014) Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar Drugs 12:128–152
Berman J, Zorrilla-López U, Farré G, Zhu C, Sandmann G, Twyman RM, Capell T, Christou C (2015) Nutritionally important carotenoids as consumer products. Phytochem Rev 14:727–743
Britton G, Liaaen-Jensen S, Pfander H (2004) Carotenoids Handbook. Birkhäuser Verlag, Basel
Carapito C, Burel A, Guterl P, Walter A, Varrier F, Bertile F, Van Dorsselaer A (2014) MSDA, a proteomics software suite for in-depth mass spectrometry data analysis using grid computing. Proteomics 14:1014–1019
Colot V, Roberts LS, Kavanagh TA, Bevan MW, Thomson RD (1987) Localization of sequences in wheat endosperm protein genes which confer tissue-specific expression in tobacco. EMBO J 6:3559–3564
Corona-Carrillo JI, Flores-Ponce M, Chávez-Nájera G, Díaz-Pontones DM (2014) Peroxidase activity in scutella of maize in association with anatomical changes during germination and grain storage. Springer Plus 3:399
Decourcelle M, Perez-Fons L, Baulande S, Steiger S, Couvelard L, Hem S, Zhu C, Capell T, Christou P, Fraser P, Sandmann G (2015) Combined transcript, proteome, and metabolite analysis of transgenic maize seeds engineered for enhanced carotenoid synthesis reveals pleotropic effects in core metabolism. J Exp Bot 66:3141–3150
Eisenreich W, Rojdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6:78–84
Enfissi EM, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, Fraser PD (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22:1190–1215
Gassel S, Breitenbach J, Sandmann G (2014) Genetic engineering of the complete carotenoid pathway towards enhanced astaxanthin formation in Xanthophyllomyces dendrorhous starting from a high-yield mutant. Appl Microbiol Biotechnol 98:345–350
Goodwin TW (1980) The biochemistry of the carotenoids: volume I plants. Chapman and Hall, London
Hartings H, Fracassetti M, Motto M (2012) Access genetic enhancement of grain quality- related traits in maize. In: Çiftçi Y (ed) Transgenic plants—advances and limitations. InTech Publisher, Rijeka, pp 191–218
Hasunuma T, Miyazawa SI, Yoshimura S, Shinzaki Y, Tomizawa KI, Shindo K, Choi SK, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55:857–868
Huang JC, Zhong YJ, Liu J, Sandmann G, Chen F (2013) Metabolic engineering of tomato for high-yield production of astaxanthin. Metab Eng 17:59–67
Kim WT, Okita TW (1988) Structure, expression, and heterogeneity of the rice seed prolamines. Plant Physiol 88:649–655
Krinsky NI (1989) Antioxidant functions of carotenoids. Free Radic Biol Med 7:617–635
Nishida Y, Adachi K, Kasai H, Shizuri Y, Shindo K, Sawabe A, Komemushi S, Miki W, Misawa N (2005) Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-beta-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol 71:4286–4296
Nogareda C, Moreno JA, Angulo E, Sandmann G, Portero M, Capell T, Zhu C, Christou P (2015) Carotenoid-enriched transgenic corn delivers bioavailable carotenoids to poultry and protects them against coccidiosis. Plant Biotechnol J [Epub ahead of print] PubMed
Perez-Fons L, Bramley PM, Fraser PD (2014) The optimisation and application of a metabolite profiling procedure for the metabolic phenotyping of Bacillus species. Metabolomics 10:77–90
Sandmann G (2001) Genetic manipulation of carotenoid biosynthesis: strategies, problems and achievements. Trends Plant Sci 6:14–17
Sandmann G (2015) Carotenoids of biotechnological importance. In: Schrader J, Bohlmann J (eds) Biotechnology of isoprenoids. Springer, Berlin, Heidelberg, pp 449–467
Sandmann G, Römer S, Fraser PD (2006) Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab Eng 8:291–302
Schreier PH, Seftor EA, Schell J, Bohnert HJ (1985) The use of nuclear-encoded sequences to direct the light-regulated synthesis and transport of a foreign protein into plant chloroplasts. EMBO J 4:25–32
Sørensen MB, Müller M, Skerritt J, Simpson D (1996) Hordein promoter methylation and transcriptional activity in wild-type and mutant barley endosperm. Mol Genet Genom 250:750–760
Spielbauer G, Margl L, Hannah LC, Römisch W, Ettenhuber C, Bacher A, Gierl A, Eisenreich W, Genschel U (2006) Robustness of central carbohydrate metabolism in developing maize kernels. Phytochem 67:1460–1475
Sugio T, Satoh J, Matsuura H, Shinmyo A, Kato K (2008) The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J Biosci Bioeng 105:300–302
Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J (1987) Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J 6:2513–2518
Tyczkowski JK, Hamilton PB (1986) Absorption, transport, and deposition in chickens of lutein diester, a carotenoid extracted from Marigold (Tagetes erecta) petals. Poult Sci 65:1526–1531
Valot B, Langella O, Nano E, Zivy M (2011) Mass ChroQ: a versatile tool for mass spectrometry quantification. Proteomics 11:3572–3577
Vershinin A (1999) Biological functions of carotenoids—diversity and evolution. Biofactors 10:99–104
Yamagata T, Kato H, Kuroda S, Abe S, Davies E (2003) Uncleaved legumin in developing maize endosperm: identification, accumulation and putative subcellular localization. J Exp Bot 54:913–922
Zhong YJ, Huang JC, Liu J, Li Y, Jiang Y, Xu ZF, Sandmann G, Chen F (2011) Functional characterization of various algal carotenoid ketolases reveals that ketolating zeaxanthin efficiently is essential for high production of astaxanthin in transgenic Arabidopsis. J Exp Bot 62:3659–3669
Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci USA 105:18232–18237
Acknowledgments
Funding through the Plant KBBE project CaroMaize is gratefully acknowledged. Further support to PC was by the Ministerio de Economia y Competitividad, Spain (BIO2014-54441-P, BIO2011-22525) and a European Research Council Advanced Grant (BIOFORCE); PROGRAMA ESTATAL DE INVESTIGACIÓN CIENTÍFICA Y TÉCNICA DE EXCELENCIA, Spain (BIO2015-71703-REDT). PDF and LP are grateful for funding from the EU FP7 project DISCO grant number 613513. We thank Sys2Diag team (CNRS, France) for statistical analyses of proteomic data and particularly Nicolas Salvetat and Franck Molina.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Farré, G., Perez-Fons, L., Decourcelle, M. et al. Metabolic engineering of astaxanthin biosynthesis in maize endosperm and characterization of a prototype high oil hybrid. Transgenic Res 25, 477–489 (2016). https://doi.org/10.1007/s11248-016-9943-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-016-9943-7