Abstract
Nitrogen is a primary macronutrient in plants, and nitrogen fertilizers play a critical role in crop production and yield. In this study, we investigated the effects of overexpressing a glutamine synthetase (GS) gene on nitrogen metabolism, and plant growth and development in sorghum (Sorghum bicolor L., Moench). GS catalyzes the ATP dependent reaction between ammonia and glutamate to produce glutamine. A 1,071 bp long coding sequence of a sorghum cytosolic GS gene (Gln1) under the control of the maize ubiquitin (Ubq) promoter was introduced into sorghum immature embryos by Agrobacterium-mediated transformation. Progeny of the transformants exhibited higher accumulation of the Gln1 transcripts and up to 2.2-fold higher GS activity compared to the non-transgenic controls. When grown under optimal nitrogen conditions, these Gln1 transgenic lines showed greater tillering and up to 2.1-fold increase in shoot vegetative biomass. Interestingly, even under greenhouse conditions, we observed a seasonal component to both these parameters and the grain yield. Our results, showing that the growth and development of sorghum Gln1 transformants are also affected by N availability and other environmental factors, suggest complexity of the relationship between GS activity and plant growth and development. A better understanding of other control points and the ability to manipulate these will be needed to utilize the transgenic technology to improve nitrogen use efficiency of crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen (N) is a primary macronutrient for plants. Plant growth and crop productivity highly depend on the application of N fertilizers. Due to the critical role that N fertilization plays in increasing yields in agriculture, world demand for N is predicted to grow by 1.3 % per year from 2012 to 2016, according to the Food and Agriculture Organization of the United Nations. However, while N fertilizers can contribute to enhanced agricultural productivity, they can also lead to environmental degradation. In fact, it is estimated that only 30–50 % of applied N is taken up by plants (McAllister et al. 2012), with the remainder causing soil, water and air pollution. In the US for example, fertilizer run-off into the Mississippi river, which drains into the Gulf of Mexico, causes excessive algal growth leading to a dead zone in the gulf every year. For this reason, there has been an increasing interest in improving N use efficiency (NUE) in crop plants, which is their efficiency to take-up N from the soil and utilize it in the production of biomass. Genetic engineering is not only an important tool for the characterization of candidate genes that are thought to improve NUE, but it will also be necessary to generate crop plants in the future that are better able to grow under low N conditions and efficiently utilize the available N.
Among the different biotechnological strategies used to date to enhance NUE in plants, multiple studies have been conducted on the overexpression of genes encoding glutamine synthetases (GS, E.C. 6.3.1.2) (Good et al. 2004; Foyer and Zhang 2011; McAllister et al. 2012; Xu et al. 2012). GS is known to catalyze the ATP dependent reaction between ammonia and glutamate to produce glutamine. This enzyme is likely to be an important checkpoint controlling NUE, since it is involved in the assimilation of all of the N, derived either from nitrate or ammonium, into organic forms (Miflin and Habash 2002). GSs, present either as cytosolic (GS1) or plastidic (GS2) isoforms (Hirel and Gadal 1982; Foyer and Zhang 2011), are differentially expressed in various plant tissues and have specific functions in N assimilation (Bernard and Habash 2009).
Several studies have demonstrated that the overexpression of GS genes (Gln) resulted in improved growth or biomass accumulation in different plant species. It has been shown that the overexpression of a cytosolic Gln gene (Gln1) from alfalfa (Medicago sativa L.) or pea (Pisum sativum L.) directed by the 35S cauliflower mosaic virus (CaMV 35S) promoter led to enhanced growth in tobacco (Nicotiana tabacum L.) (Fuentes et al. 2001; Oliveira et al. 2002). Likewise, constitutive overexpression of a Gln1 gene from pine (Pinus silvestris L.) resulted in increased dry weight (DW) in poplar (Populus tremula L.) (Gallardo et al. 1999; Fu et al. 2003). Habash et al. (2001) found that wheat (Triticum aestivum L.) plants overexpressing a Gln1 gene from bean (Phaseolus vulgaris L.), driven by the light-induced rubisco small subunit (rbcS) promoter, exhibited higher root and grain DW than the non-transgenic controls. In rice (Oryza sativa L.), ectopic expression of a Gln1 gene under the control of a ubiquitin (Ubq) promoter resulted in increased spikelet yield in plants grown under high N conditions (Brauer et al. 2011); however, no positive effect on NUE was observed when the CaMV 35S promoter was used (Cai et al. 2009). Migge et al. (2000) demonstrated that the overexpression of a plastidic Gln gene (Gln2) under the control of the rbcS promoter enhanced the growth of tobacco plants. Moreover, overexpression of a maize (Zea mays L.) gln1-3 gene driven constitutively by the cassava vein mosaic virus (CsVMV) promoter increased grain number in this crop (Martin et al. 2006).
In the current study, we investigated the effects of the overexpression of a Gln1 gene from sorghum (Sorghum bicolor L.) on its growth and development. The two sorghum GS isoforms, cytosolic and plastidic, were first identified by Hirel and Gadal (1982). Later on, El Omari et al. (2010) found that the cytosolic GS isoforms in sorghum were encoded by three different genes, SbGln1.1, SbGln1.2 and SbGln1.3, with the SbGln1.1 gene being more active in sorghum leaves, while the other two were expressed mainly in the roots. However, in maize, which belongs to the same family, cytosolic GS isoenzymes are known to be encoded by five distinct genes, all of them being differentially expressed in various tissues of the plant (Martin et al. 2006). In the study performed by Martin et al. (2006), the authors presented a model describing the expression and putative function of the five GS isoforms in maize and also reported that the constitutive overexpression of ZmGln1.3 (GenBank accession number: x65928) led to a 30 % increase in the grain number of transgenic plants. We conducted bioinformatic analyses (http://www.phytozome.net/) and identified a sorghum gene sequence that had the highest degree of homology to the ZmGln1.3. Based on EST sequences (http://www.ncbi.nlm.nih.gov/; accession numbers: CD462839, CD205511 and AW671987), the sequence identified in Phytozome seems to correspond to the SbGln1.2 gene reported by El Omari et al. (2010) and was chosen in this study for overexpression in sorghum. To this end, the 1,071 bp long coding sequence (CDS, Online resource 5) from sorghum was placed downstream of the maize Ubq promoter or the CaMV 35S promoter in two separate binary vectors, and each construct was introduced into sorghum by means of Agrobacterium-mediated transformation. Transgenic lines showing higher transgene expression and enzyme activity compared to the wild-type plants were subjected to biomass and yield evaluations as well as to nitrogen and metabolite analyses.
Materials and methods
Assembly of binary vectors and plant transformation
The CDS of the sorghum Gln1 gene was obtained by RT-PCR from mRNA isolated from sorghum leaf tissue. Primers Gln1-F1: 5′-GGATCCATGGCCTCCCTCACCGA-3′ and Gln1-R1: 5′-GAGCTCTCAGGGCTTCCAGAGGATG-3′ were used to amplify the 1,071 bp long sequence of the gene and to incorporate BamHI and SstI cloning sites at the 5′ and 3′ ends, respectively. A binary vector was assembled by ligating this fragment at the 3′-end of the maize Ubq promoter in a pCAMBIA 1200-based vector. The laboratory designation for this vector was LCT93 (Online resource 6). In addition, primers Gln1-F2: 5′-CCATGGATGGCCTCCCTCACCGA-3′ and Gln1-R2: 5′-GGATCCTCAGGGCTTCCAGAGGATG-3′ were used to amplify the same sequence, but to incorporate NcoI and BamHI cloning sites at the 5′ and 3′ ends, respectively. This fragment was ligated at the 3′-end of the tobacco etch virus (TEV) translation enhancer under the control of the CaMV 35S promoter in a pCAMBIA 1300-based vector. The laboratory designation for this second vector was LCT94 (Online resource 7). Each plasmid was mobilized into Agrobacterium tumefaciens strain NTL4 harboring the disarmed Chry5 Ti plasmid designated pTiKPSF2 (Palanichelvam et al. 2000). The transgene expression cassettes were introduced into S. bicolor L., genotype P898012, by Agrobacterium-mediated transformation of immature embryos, collected 14 days post-anthesis (dpa), as described by Kumar et al. (2011). Plants were regenerated following selection of transgenic events on hygromycin (20 mg/l). Plantlets with well-developed roots were transferred to three-gallon pots containing MetroMix 200 soil medium (SUN GRO Horticulture, Bellevue, WA, USA) and grown in a greenhouse. Plants were watered daily and, starting at the six-leaf stage, were fertilized every week.
Plant material for molecular and physiological studies
Homozygous or azygous (null segregant) status of the plants was determined by germinating T2 seeds from 10 different T1 plants on hygromycin medium. Their status was further confirmed by PCR analysis. T2 generation plants from a homozygous T1 plant and the corresponding null segregants were used in various analyses. One week-old seedlings were planted in 500 ml size plastic cups (with holes in the bottom covered with wire mesh) containing sand. The experiment was conducted in a greenhouse and plants were irrigated daily with a nutrient solution containing either 4 mM NO3 − (low N concentration) or 12 mM NO3 − and 2 mM NH4 + (optimal N concentration) (Online resource 1 and 2). The pH of each solution was adjusted to 5.8 before use. At each irrigation, solution was allowed to drain out from the bottom of the plastic cups. At the five-leaf stage, roots and the three youngest, fully expanded leaves were harvested for subsequent use in transcription, enzyme and ammonium analyses. For the long-term studies, plants were grown in rockwool cubes (Grodan, Ontario, Canada) in a greenhouse and irrigated every other day with the nutrient solutions in the same manner as described above. At maturity, shoot tissue and seeds were harvested for ammonium, amino acid and nitrogen analyses as well as for yield determinations.
Transcript analysis
Leaf and root tissues were collected from transgenic plants and null segregant controls and flash frozen in liquid nitrogen. Total RNA was extracted from tissues using the Spectrum Plant Total RNA Kit (Cat. # STRN50-1KT, Sigma, St. Louis, MO, USA). Transcript levels of the Gln-1 gene were analyzed by Northern blot hybridization (Sambrook et al. 2001). The membranes were hybridized with a 32P-labeled Gln1 gene fragment for 16–20 h at 60 °C. After hybridization, membranes were exposed to X-ray film at −80 °C.
Enzymatic assay
GS enzyme activity was determined in freshly-harvested leaf and root tissues by the synthetase reaction as described previously by O’Neal and Joy (1973). Measurement of soluble protein was performed spectrophotometrically using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard.
Evaluation of growth and biomass production
For shoot and root DW determinations, plant material was dried at 100 °C for 12 h. DW were determined for transgenic and null segregant plants at the five-leaf developmental stage. In addition, flowering time, height of the primary tiller, grain number, grain weight and vegetative shoot DW were determined in mature transgenic plants and null segregant controls at 35 dpa (primary tiller). The first long-term experiment was started on October 3, 2012 and the second long-term experiment was initiated on February 11, 2013. Both the long-term experiments were conducted in exactly the same manner, however, because of the timing of experiments, the plants experienced different temperatures, light quantity, and photoperiods as these conditions could not be fully controlled in our greenhouses.
Ammonium, amino acid and nitrogen analyses
For ammonium, amino acid and nitrogen analyses, plant material was also dried at 100 °C for 12 h. Ammonium was extracted with 60 % methanol and estimated by the salicylate-type colorimetric method described previously by Husted et al. (2000). For amino acid content determination, samples were subjected to liquid-phase hydrolysis with 6 N HCl and exposed to 100 °C for 22 h. Samples were then filtered, reconstituted in 0.4 M borate buffer and analyzed using high-performance liquid chromatography (HPLC). This analysis was performed by the Protein Chemistry Laboratory, TAMU, College Station, TX, USA. Nitrogen content was measured by the Dumas combustion method (Office of the Texas State Chemist, TAMU, College Station, TX, USA) as described by Palle et al. (2013).
Results
Generation of transgenic sorghum lines
In order to investigate the effect of GS overexpression in the improvement of sorghum NUE, we assembled a binary vector (laboratory designation: LCT93; Online resource 6) harboring the 1,071 bp long CDS of a sorghum Gln1 gene under the control of the maize Ubq promoter. Additionally, we used a second vector carrying the same gene fragment driven by the CaMV 35S promoter (laboratory designation: LCT94, Online resource 7). Both the constructs contained the hygromycin phosphotransferase (hptII) gene under the control of the CaMV 35S promoter for the selection of transgenic events. Three independent transgenic lines were generated by using the UbqP::Gln1 vector and two were obtained using the CaMV 35SP::Gln1. Two lines per vector were used for the study.
Transcription and enzyme activity
The levels of GS1 transcripts in the transgenic lines were assessed by Northern blot hybridization. Transcript accumulation was evaluated in both leaf and root tissues from T2 homozygous plants and the corresponding null segregants at the five-leaf developmental stage. Transgenic lines carrying the Gln1 gene driven by the maize Ubq promoter and grown under either low or optimal N conditions showed higher abundance of Gln1 transcripts in both leaves and roots, in comparison to the null segregants (Fig. 1). However, the transgenic plants transformed with the Gln1 gene under the control of the CaMV 35S promoter showed no major differences for the Gln1 transcripts compared to their null segregant controls in either tissue under different N conditions (Online resource 8).
Northern blot analysis of RNA from LCT93 transgenic lines and null segregant (NS) plant grown under low or optimal nitrogen conditions. a Leaves, b roots. Transcripts were detected by hybridization with 32P-labeled probes corresponding to the CDS specific to each gene (Gln1 and sorghum actin gene). Three µg of RNA were loaded in each lane
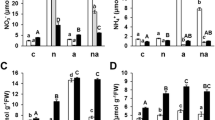
GS enzyme activity was evaluated in the leaves and roots of T2 homozygous Gln1 transformants and null segregants grown under low or optimal N conditions. GS activity was significantly higher in both leaves and roots of LCT93 plants than that in the control plants grown under either N conditions (Fig. 2). A 1.3- to 1.6-fold increase in GS activity was observed in the leaves of LCT93 transgenic plants, while a 1.3- to 2.2-fold higher activity was found in the roots. As opposed to LCT93 lines, LCT94 plants showed no significant difference in GS activity compared to the null segregants in any of the tissues analyzed and under either N treatment (Online resource 9).
Evaluation of growth, biomass production and grain yield in Gln1 overexpressing plants
To determine the effect of Gln1 overexpression on the growth and development of sorghum plants grown under low and optimal N conditions, biomass and grain production were evaluated in T2 93-2A plants at the vegetative stage and after maturity in separate experiments. No significant difference in shoot or root DW was found in the 93-2A plants harvested at the five-leaf developmental stage compared to their wild-type counterparts (Table 1). However, at 35 dpa, 93-2A plants from the experiment started on October 3, 2012 (hereafter referred to as winter experiment), grown under optimal N conditions and harvested in December 2012 showed significantly higher accumulation of biomass in the vegetative parts of the shoot than the controls (Table 2). Interestingly, the 2.1-fold increase in shoot biomass was due to the formation of more secondary tillers compared to the null segregant plants (Fig. 3). In contrast, no significant difference was observed in the DW of vegetative shoot parts between 93-2A plants and controls, grown under low N (Table 2). Similarly, with regards to the grain yield, there were no significant differences between 93-2A and control plants grown under low N conditions. The situation under optimal N conditions was more complex. The primary tillers of 93-2A transgenic plants grown under optimal N conditions exhibited a significant reduction in seed number (Table 2). However, as mentioned earlier, the transformants had a tendency to produce a secondary tiller. The number of seeds in these secondary tillers was higher than that in the primary tillers of the transformants (Online resource 10). Thus, at the termination of the experiment (35 dpa in the primary tiller), a significantly higher number of seeds was produced by the transformants compared to the null segregants. In contrast, no differences in grain yield were observed between the transformants and null segregants grown under low nitrogen conditions. Additionally, no significant difference in height was observed between transgenic plants and null segregant controls, but a slight delay in flowering was detected in transgenic individuals grown under low N conditions. A second long-term experiment was initiated on February 11, 2013 (spring experiment) and growth and development-related measurements were obtained on plants harvested on May 22, 2013. In contrast to plants harvested in the winter experiment, both, null segregant and transgenic plants from this second experiment had the tendency to produce secondary tillers that, in many cases, emerged from the middle part of the stems of primary tillers (Online resource 11A, B, C and D). At 35 dpa, 93-2A plants grown under optimal N conditions showed a statistically significant, 1.2-fold higher accumulation of the vegetative shoot biomass than the controls (Table 3). This increase in shoot biomass was due to the formation of more tillers compared to the control plants. In contrast, no significant difference in vegetative shoot biomass was observed between 93-2A plants and the controls grown under low N conditions. With regards to the grain yield, the primary tillers of 93-2A transgenic plants grown at both low and optimal N concentrations exhibited a significant reduction in seed number (Table 3). It should be noted that at the termination of this spring experiment (35 dpa in the primary tiller), only a small number of the secondary tillers had flowered and none had set seeds. No significant difference was found between transgenics and null segregants in terms of height, and a slight but significant decrease in flowering time was detected in the transgenic individuals grown under both low and optimal N conditions.
Ammonium, amino acid and nitrogen levels in Gln1 overexpressing plants
To estimate the impact of Gln1 overexpression on N metabolism in sorghum plants grown under low and optimal N conditions, the ammonium and total N contents were determined in T2 93-2A plants at the vegetative stage and at maturity (Tables 1, 2). There was no significant difference in the nitrogen levels in the leaves from young transgenic plants grown in either low or optimal N concentrations in comparison to the controls. However, there was a significant decrease in ammonium content in young leaves from 93-2A plants grown under optimal N conditions, compared to the null segregants. At maturity, there was no significant difference in the ammonium or nitrogen content in the transgenic plants compared to the controls. Although we did not expect to see major differences in the levels of amino acids between the seeds from transgenic plants and null segregants, we performed this analysis based on a report that has shown a small reduction in the levels of total amino acids in the rice seeds of GS-overexpressing plants (Cai et al. 2009). No significant difference in the concentration of amino acids was observed between the seeds of transgenic and control sorghum plants grown under optimal N conditions (Online resource 3). Interestingly, the concentration of each of the amino acids and total protein (Online resource 3 and 4) was higher in the seeds of null segregants in the winter experiment compared to the spring experiment.
Discussion
We have investigated the effects of overexpression of a Gln1 gene on growth and development in sorghum. To that end, transgenic lines carrying a sorghum Gln1 gene with 99 % of sequence homology to that of SbGln1.2 (El Omari et al. 2010) under the control of the maize Ubq promoter were generated. It has been reported that SbGln1.2 is highly expressed in sorghum roots and induced by pathogens and N deficiency (El Omari et al. 2010). Northern blot analyses and glutamine synthetase enzyme assays revealed that the UbqP::Gln1 transformants showed strong expression of the transgene in both tissues (leaves and roots) examined and under both N treatments tested (Figs. 1, 2). UbqP::Gln1 transformants grown under optimal N conditions and harvested at the vegetative stage showed lower levels of ammonium than controls, which could be due to the higher GS1 activity in these plants (Oliveira et al. 2002; Martin et al. 2006). Intriguingly, no difference in ammonium levels was observed between young transgenic and control plants growing under low N availability or between transgenics and controls at maturity (Tables 1, 2).
In contrast, Northern blot analysis on CaMV 35SP::Gln1 transformants did not show higher levels of Gln1 transcripts in either the leaves or roots of the sorghum plants grown under low or optimal N conditions (Online resource 8). Although the CaMV 35S promoter is a very strong constitutive promoter, it is also known to have lower activity in monocotyledonous plants. For instance, Kumar et al. (2011) demonstrated that the CaMV 35S promoter is not very active in sorghum regenerants upon emergence from tissue culture as well as in the older leaves of two-month old sorghum plants from the T0 generation. Nevertheless, this promoter has been used successfully to drive transgenes in monocots and the selectable marker gene, hptII, used to select sorghum transformants in this study was under the control of CaMV 35S promoter. In this regard, it should be noted that several attempts to isolate full-length cDNA of the Gln1 gene were unsuccessful and therefore only the CDS was used to assemble the overexpression constructs in the current investigation. It is known that 5′- and 3′-UTRs can have major influence on the transcript stability and translation of GS1 genes in other species (Ortega et al. 2006, 2012). Although all the mechanisms involved in the control of sorghum GS1 activity are not yet understood, it is possible that the strength of the CaMV 35S promoter was not sufficient to enhance the activity of this enzyme in the CaMV 35SP::Gln1 transformants. It will require extensive investigation with additional transgenic lines to ascertain why this set of transformants did not express the Gln1 gene.
As mentioned earlier, the maize Ubq promoter was able to drive the transcription of the Gln1 CDS to a degree that resulted in significantly higher levels of transcripts and enzyme activity in the UbqP::Gln1 transformants. Such overexpression of the Gln1 gene did not affect the growth of transgenic plants at the early stages of development (Table 1), but had a significant effect at maturity (Table 2). In the winter experiment, initiated on October 3, 2012, Gln1 overexpression lines showed a significant increase in the vegetative shoot biomass as a result of greater tillering, in comparison to the null segregant controls grown under optimal N conditions. These findings suggest that the overexpression of the Gln1 gene, tested in this study, affects the tiller formation in sorghum. Recently, Funayama et al. (2013) demonstrated that the rice Gln1;2 gene affects tillering in that species. They found that Gln1;2 knock-out mutants showed a 50 % decrease in tiller number, compared to their wild-type counterparts. The wild-type phenotype was restored by the re-introduction of Gln1;2 cDNA into the mutants. They hypothesized that the loss of Gln1;2 function may decrease the concentration of strigolactone, a phytohormone that is believed to play a role in branching and tillering. In contrast to the results obtained under optimal N levels, under low N conditions, the UbqP::Gln1 transformants did not show greater tillering response compared to their null segregant counterparts (Table 2). This suggests that the overexpression of GS1 had an effect on the physiological response of the sorghum plant to N availability. In the spring experiment, initiated on February 11, 2013, transgenic lines also exhibited a higher shoot vegetative biomass accumulation in comparison to the null segregants when they were grown under optimal N conditions but not under low N conditions (Table 3). Although the increased biomass of transformants, observed in the spring experiment was significantly higher compared to the controls, it was not as high as that in the winter experiment. This discrepancy might be a result of seasonal effects caused by the difference in day length, and, thus, the amount of daylight the plants received during each season. The plants may have also experienced different temperatures in the two experiments.
In the winter experiment, it was evident that the increase in biomass in the transgenic plants grown under optimal N availability was a result of the formation of secondary tillers (Fig. 3). Interestingly, in the spring experiment, both types of plants grown under any of the two N treatments produced multiple secondary tillers (Online resource 11). However, statistical analyses showed that in this experiment, the transgenic plants had more tillers than the controls grown under optimal N conditions (Table 3). The difference, in terms of the degree of tillering response, between the two experiments could have been due to the differences in day length, light quantity and intensity between fall and spring seasons.
There are some reports on light-dependent growth phenotype in transgenic plants overexpressing Gln1 genes. In a study conducted by Oliveira et al. (2002), tobacco plants transformed with a pea Gln1 gene and grown under different N concentrations exhibited slightly higher growth rate compared to the controls under low light (50 µmol cm−2 s−1) than under high light (200 µmol cm−2 s−1) conditions. Brauer et al. (2011) observed that Gln1-expressing transgenic rice plants exhibited increased spikelet yield when they were grown in growth chambers but not under greenhouse conditions. They suggested that this variability might be attributed to different light intensities in each of the environments, being higher in the greenhouse during the months when their experiment was conducted. Moreover, Finnemann and Schjoerring (2000) proposed a model for the mechanism of post-translational regulation of GS1 in canola (Brassica napus L.), in which the activity of the GS1 enzyme is controlled by phosphorylation that is affected by light (Miflin and Habash 2002). According to their model, in the dark, the plant ATP/AMP ratio is higher than in the light, and this favors the phosphorylation of GS1 and binding to 14-3-3 proteins, protecting the enzyme from degradation. In contrast, under light, GS1 does not undergo phosphorylation and, thereby, may be more unstable and prone to degradation. If a similar mechanism is responsible for the post-translational regulation of GS1 in sorghum, it could explain, to some degree, the differences observed in tillering and vegetative biomass increase in the transgenic plants between the winter and spring experiments. Our results, taken together with the findings from these studies, indicate that light plays an important role in the activity of GS1 enzyme and thus its impact on plant growth and development.
It is interesting that in the winter experiment, the vegetative biomass of the null segregant plants was twice as high under optimal N conditions compared to the levels under low N conditions (Table 2). On the other hand, there was no significant difference in the vegetative biomass of control plants grown under the different N levels in the spring experiment (Table 3). This result suggests that N availability affects the vegetative growth in a significant manner depending on the environmental conditions experienced by the plants in the greenhouse.
Furthermore, in the winter experiment, transformants showed a tendency to produce a secondary tiller soon after the first one under optimal N levels, therefore resulting in higher number of seeds per plant, overall, at the termination of the experiment (Table 2). However, this increase in tillering was accompanied by a reduction in the number of seeds in the primary tiller. It is possible that N remobilization within the transgenic plants occurred in a way that impaired grain set, but favored bud outgrowth and tillering, since endogenous factors involved in branching are regulated by the availability of nutrients, including nitrogen (Kebrom et al. 2013). Nonetheless, tillering in monocots is a complex phenomenon controlled by multiple hormonal, developmental and environmental factors; therefore, more extensive research needs to be conducted in order to understand in detail the role that GS1 plays in branching in these transgenic lines. In contrast to the winter experiment, where we observed higher overall seed yields in the transformants under optimal N conditions, in the spring experiment, the transgenic plants had significantly lower number of seeds under both N levels (Table 3). This is a surprising and unexpected result and points to the complexity of the interaction between climatic conditions and plant growth and development, especially in relation to N metabolism. However, despite these differences in grain number observed between transgenic and controls plants in both winter and spring experiments, there was no significant difference in the amino acid concentration between the seeds of transgenics and control plants grown under optimal N conditions (Online resource 3). The impact of the seasonal component was again evident as seen in terms of a significant difference between the protein and individual amino acid levels in the seeds obtained from the null segregant plants grown in the winter or spring in the greenhouse (Online resource 3 and 4).
Overall, this study suggests that it is possible to enhance biomass production in sorghum through the overexpression of Gln1 gene under certain conditions. A significant increase in the biomass and yield was obtained in the transformants during the winter months in the greenhouse and that too under optimal N levels. However, in the spring experiment, while there was a small but significant increase in the biomass and tillering in the transformants, their seed yields were lower than the null segregant controls. This indicates that growth and development depend on complex interactions among GS activity, N availability and environmental conditions. Thus, more extensive research is necessary to obtain higher biomass or yield in plants and to improve their NUE by the overexpression of Gln1 genes. Further work is needed to investigate the use of gene stacking strategies that involve the insertion of Gln1 genes in conjunction with one or more genes responsible for N uptake and utilization and the use of tissue-specific promoters. For instance, Katayama et al. (2009) showed that overexpression of a high-affinity nitrate transporter gene in rice resulted in enhanced vegetative growth of the plants. Thus, it will be interesting to examine the effects of expressing such transporter genes and nitrate/nitrite reductase genes along with the Gln1 gene. In conclusion, our results highlight the complexity of relationships among N availability, GS activity and environmental factors with regards to their role in growth and development and suggest the need for better understanding of additional control points before transgenic technology can be used to improve NUE of crop plants.
References
Bernard SM, Habash DZ (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol 182:608–620
Brauer EK, Rochon A, Bi YM, Bozzo GG, Rothstein SJ, Shelp BJ (2011) Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol Plant 141:361–372
Cai H, Zhou Y, Xiao J, Li X, Zhang Q, Lian X (2009) Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep 28:527–537
El Omari R, Rueda-Lopez M, Avila C, Crespillo R, Nhiri M, Canovas FM (2010) Ammonium tolerance and the regulation of two cytosolic glutamine synthetases in the roots of sorghum. Funct Plant Biol 37:55–63
Finnemann J, Schjoerring JK (2000) Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J 24:171–181
Foyer CH, Zhang H (2011) Nitrogen metabolism in plants in the post-genomic era. Annu Plant Rev 42:1–366
Fu J, Sampalo R, Gallardo F, Canovas FM, Kirby EG (2003) Assembly of a cytosolic pine glutamine synthetase holoenzyme in leaves of transgenic poplar leads to enhanced vegetative growth in young plants. Plant Cell Environ 26:411–418
Fuentes SI, Allen AJ, Ortiz A, Hernandez G (2001) Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J Exp Bot 52:1071–1081
Funayama K, Kojima S, Tabuchi-Kobayashi M, Sawa Y, Nakayama Y, Hayakawa T, Yamaya T (2013) Cytosolic glutamine synthetase 1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol 54:934–943
Gallardo F, Fu J, Cantón FR, García-Gutiérrez A, Cánovas FM, Kirby EG (1999) Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta 210:19–26
Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9:597–605
Habash DZ, Massiah AJ, Rong HL, Wallsgrove RM, Leigh RA (2001) The role of cytosolic glutamine synthetase in wheat. Ann Appl Biol 138:83–89
Hirel B, Gadal P (1982) Glutamine synthetase isoforms in leaves of a C4 plant: Sorghum vulgare. Physiol Plant 54:69–74
Husted S, Hebbern CA, Mattsson M, Schjoerring JK (2000) A critical experimental evaluation of methods for determination of NH4+ in plant tissue, xylem sap and apoplastic fluid. Physiol Plant 109:167–179
Katayama H, Mori M, Kawamura Y, Tanaka T, Mori M, Hasegawa H (2009) Production and characterization of transgenic rice plants carrying a high-affinity nitrate transporter gene (OsNRT2.1). Breed Sci 59:237–243
Kebrom TH, Spielmeyer W, Finnegan EJ (2013) Grasses provide new insights into regulation of shoot branching. Trends Plant Sci 18:41–48
Kumar V, Campbell L, Rathore K (2011) Rapid recovery- and characterization of transformants following Agrobacterium-mediated T-DNA transfer to sorghum. Plant Cell Tissue Organ Cult 104:137–146
Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, Terce-Laforgue T, Quillere I, Coque M, Gallais A, Gonzalez-Moro MB, Bethencourt L, Habash DZ, Lea PJ, Charcosset A, Perez P, Murigneux A, Sakakibara H, Edwards KJ, Hirel B (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18:3252–3274
McAllister CH, Beatty PH, Good AG (2012) Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol J 10:1011–1025
Miflin BJ, Habash DZ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53:979–987
Migge A, Carrayol E, Hirel B, Becker TW (2000) Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210:252–260
Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM (2002) Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol 129:1170–1180
O’Neal D, Joy KW (1973) Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Arch Biochem Biophys 159:113–122
Ortega JL, Moguel-Esponda S, Potenza C, Conklin CF, Quintana A, Sengupta-Gopalan C (2006) The 3′ untranslated region of a soybean cytosolic glutamine synthetase (GS1) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J 45:832–846
Ortega J, Wilson O, Sengupta-Gopalan C (2012) The 5′ untranslated region of the soybean cytosolic glutamine synthetase β1 gene contains prokaryotic translation initiation signals and acts as a translational enhancer in plants. Mol Genet Genomics 287:881–893
Palanichelvam K, Oger P, Clough SJ, Cha C, Bent AF, Farrand SK (2000) A second T-region of the soybean-supervirulent chrysopine-type Ti plasmid pTiChry5, and construction of a fully disarmed vir helper plasmid. Mol Plant Microbe Interact 13:1081–1091
Palle SR, Campbell LM, Pandeya D, Puckhaber L, Tollack LK, Marcel S, Sundaram S, Stipanovic RD, Wedegaertner TC, Hinze L, Rathore KS (2013) RNAi-mediated ultra-low gossypol cottonseed trait: performance of transgenic lines under field conditions. Plant Biotechnol J 11:296–304
Sambrook J, Russell DW, Irwin N, Janssen KA (2001) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182
Acknowledgments
The authors are thankful to Dr. James Balthrop (Office of the Texas State Chemist, TAMU) for his help with seed protein analyses and to Ms. Virginia Johnson (Protein Chemistry Laboratory, TAMU) for her help with amino acid analyses. J. U. would like to thank the Ministerio de Economía y Finanzas de Panamá, Secretaría Nacional de Ciencia, Tecnología e Innovación, and Instituto para la Formación y Aprovechamiento de Recursos Humanos for the scholarship provided during the course of her Ph.D.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Urriola, J., Rathore, K.S. Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res 24, 397–407 (2015). https://doi.org/10.1007/s11248-014-9852-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-014-9852-6