Abstract
A trypsin inhibitor from Cocculus hirsutus, commonly known as “Farid Buti” has been demonstrated to exhibit insecticidal, fungicidal, as well as nematocidal activity. The ChTI (Cocculus hirsutus Trypsin Inhibitor) gene was designed in silico and synthesized by PCR-based gene synthesis and cloned in the plant expression vector pBI121, with kanamycin, as the selectable marker. Agrobacterium strain LBA4404 was transformed with pBI121:ChTI vector for plant transformation. For developing insect-tolerant chickpea, Agrobacterium-mediated transformation of ChTI gene was performed in cultivar P-362. Twenty-day-old cotyledonary node (CN) explants were used for sonication-assisted Agrobacterium-mediated transformation. Three cycles of increasing concentrations of kanamycin were used for the selection of transformed shoots. In vitro grown transgenic chickpea shoots were grafted on decapitated stock of chickpea seedlings. After 45–50 days of acclimatization and hardening, pod development and its maturation occurred. After screening by PCR, seven transgenic events were confirmed to be positive by Southern blot hybridization analysis, showing 1–4 copies of the transgene. The quantitative expression of the ChTI gene by qRT-PCR analysis showed up to 12–17-fold change in the T1 progeny. Immunoblot analysis revealed the expression of 31 kDa and 15 kDa ChTI protein in E.coli and transgenic plants respectively. Trypsin activity assay was performed in the T1 generation and higher anti-trypsin activity was recorded. Insect tolerance against Helicoverpa armigera and Spodoptera litura were estimated by insect bioassay, wherein an overall mortality of 60–80% and weight loss (30–60% and 40–60% for Spodoptera litura and Helicoverpa armigera respectively) have been recorded in the plants of T1 generation.
Key Message
Expression of Cocculus hirsutus trypsin inhibitor (ChTI) gene in chickpea by Agrobacterium-mediated transformation, restricted the growth as well as the survival of two Lepidopteran insect pests Helicoverpa armigera and Spodoptera litura.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malnutrition is increasing at an appalling rate in the developing countries, due to the deficiency of proteins in the diet. Growing of pulses and their regular intake, can meet the daily requirement of proteins in the regions where animal proteins are less available and affordable. Consumption of chickpea (Cicer arietinum L.) a high protein crop, is effective in combating protein malnutrition and Kwashiorkor in children and adults. To attain self-sufficiency and surplus by 2050, India's total pulse production needs to reach 39 mt. In the current scenario, India holds most of the world’s area under chickpea cultivation (70%) and production (67%). The requirement of chickpea is about 16–17.5 mt by 2050 from an area of about 10.5 Mha with average productivity of 15–17 q/ha. (Dixit et al. 2019). The major obstacles in pulse production are several biotic and abiotic stresses, low plant productivity, global economic crisis, and a significant degree of fluctuation in the prices of pulses (Acharjee and Sarmah 2013).
Plants face numerous abiotic and biotic stresses in nature. Noticeably, insects impose severe threat to the survival of the plant due to their diversity, adaptability and abundance. Therefore millions of years of selection pressure exerted by insect herbivores, have given rise to the evolution of a sophisticated defense mechanism in plants (Erb and Reymond 2019).
Chickpea production is severely threatened by difficulties in managing many insect pests that attack the plant parts at all stages, starting from the seedling stage to harvest and beyond. The net yield of chickpea is severely affected by Lepidopteran insects (Helicoverpa armigera and Spodoptera litura) and diseases like wilt, Botrytis grey mold, dry root rot, Ascochyta blight, etc.) throughout crop production. As a result of larval infestation on the reproductive structures such as flowers and immature pods, reduces the crop yield significantly.
Helicoverpa armigera Hubner (Lepidoptera:Noctuidae), the gram pod borer, inflicts up to 90% mutilation in chickpea crop from the vegetative growth to the pod formation stage. H. armigera, is prominently distributed in Asia, Africa, Mediterranean regions, and Oceania. The larvae of the 1st, 2nd, and 3rd instar stage, first feed on the leaves of chickpea. A shift from foliar feeders to the developing seeds and fruits has been observed in the later stages of development. Eventually, larvae eat-up the emerging seeds inside the pod. Moreover, it causes entry holes at the basal region of chickpea pods in most of the genotypes. H. armigera larvae have characteristic longitudinal markings on lateral sides and are often dark brown, which may vary according to the food consumed by them (Yamasaki et al. 2009). Intending to overcome this issue, the cultivators are desirous of raising the strength of pesticides. However, the application of pesticides in an injudicious manner has resulted in the bioaccumulation, biomagnification, pesticide resistance, pest revival, and causes harm to the non-target beneficial organisms and the environment.
The following major strategies are considered effective for the management of pod borer pests: (1) development and use of resistant varieties of crops, (2) good agronomic practices, such as early sowing, optimum planting density, sufficient fertilizers, inter/trap crops (coriander, sunflower, sorghum, mustard, linseed, and marigold), (3) monitoring the pod borer through pheromone traps. Integrating these practices with biological control has shown promising outcomes for sustainable pod borer management, resulting in expected chickpea yield (Patil et al. 2017).
Tailoring crop plants for endogenous built-in resistance to insect pests through the introduction of insecticidal protein genes of Bacillus thuringiensis (Bt) and other insect-resistant proteins via transgenic technology and the successful commercialization of genetically modified maize, potato, tomato, cotton, canola, rice and soybean, has provided a strategy to enhance crop productivity, due to the curb in crop losses (Perlak et al. 2001; Sanahuja et al. 2011). The transgenic approach has imparted resistance to insects in different crops, e.g., Lepidopteran insects in cotton (Gossypium hirsutum) and Coleopteran and Lepidopteran insects in maize (Zea mays). Transgenic technology is superior to conventional breeding due to its advantages in widening the pool of beneficial genes, incorporating few candidate genes in a single event, thereby reducing the time required for producing a crop variety with improved agronomic traits.
Several regeneration protocols involving somatic embryogenesis and shoot organogenesis from diverse explants have been described in different cultivars of chickpea. A number of parameters like co-cultivation duration, bacterial cell density, phenolic compounds, and strain of Agrobacterium, play a critical role in determining the net transformation efficiency. To develop a non-chimeric transgenic plant, optimization of selection and screening parameters are the decisive steps for enhancing the transformation efficiency. However, there is a crucial requirement to increase the transformation efficiency and stability of transgenes in chickpea, for commercial release. Bt transgenic chickpea lines with resistance to Helicoverpa armigera for commercial release are still under development (Sanyal et al. 2005; Acharjee et al. 2010; Mehrotra et al. 2011; Asharani et al. 2011; Khatodia et al. 2014; Ganguly et al. 2014).
The higher expression of Bt toxin is responsible for reducing the growth of chickpea, which needs to be studied from a physiological point of view. However, the field-acquired resistance in H. armigera has reduced the efficacy of Bt crops for pest resistance (Tabashnik et al. 2013). In this regard, transgenic crops expressing pyramided Bt toxins against similar insect pests have been extensively applied now to delay the evolution of pest resistance (Carrière et al. 2015). However, field-evolved resistance and cross-resistance in transgenic plants expressing two different types of Bt toxins have been discovered (Gassmann et al. 2014).
Another alternate choice is by the use plant protease inhibitors (PPIs) for producing transgenic pest-resistant plants. PPIs do not have a wipe-out effect similar to synthetic pesticides and do not possess strong selection pressure, which is anticipated to impressively delay the development of resistance in the pest. The cowpea trypsin inhibitor (CpTI) gene was the first PPI gene used to produce transgenic tobacco (Hilder et al. 1987), after which many transgenic plants have been developed using the PI genes (Gatehouse et al. 1993).
Plant protease inhibitors (PPIs) are natural plant defense proteins, that have a potential function in protecting plants against herbivorous insects by cleaving their digestive proteases (Zhu-Salzman and Zeng 2015). In the same context, trypsin inhibitor activity was observed in the seed flour extract of ten selected chickpea varieties. All the varieties showed inhibitory activity in vivo and in vitro against Helicoverpa armigera (Kansal et al. 2008). In a recent study, a trypsin inhibitor from chickpea seeds was partially characterized and tested for its insecticidal activity against Helicoverpa armigera. Seventeen chickpea cultivars were categorized into three groups: with high (more than 70%), intermediate (16–70%), and very low (0–15%) trypsin inhibition activity with the 20 kDa trypsin inhibitor. Results suggest that the dynamics of the stress response is triggered by trypsin inhibitor and in turn, induces field pest resistance in the plant. In the same dimension, the dose-dependent reduction of both the larval weight and the survival was validated through feeding experiments, conducted with the 5th instar larvae (Nair et al. 2013).
Cocculus hirsutus trypsin inhibitor (ChTI), a serine proteinase inhibitor, was isolated from the shrub Cocculus hirsutus (L.) belonging to the family Menispermaceae (Rakesh and Prashant 2012). It is an 18 kDa thermo-tolerant, monomeric protein present in the vegetative parts and seeds of Cocculus hirsutus. The leaf extracts of Cocculus hirsutus has been demonstrated for insecticidal activity against Helicoverpa armigera (fruit borer) and Spodoptera litura, that infest the chickpea plant during the vegtative and fruit setting stages. The 2nd and 3rd instar larvae of H. armigera of fed with ChTI protein (5000 TIU/ml) resulted in 84.59% and 58.71% drop in the mean larval weight respectively. Subsequently, it caused 59.53% and 74% inhibition of Helicoverpa gut proteases and bovine trypsin, respectively (Bhattacharjee et al. 2009). During further evaluation, Cocculus hirsutus PI has shown to exhibit antimicrobial, antitumor, and fungicidal activity as well (Gupta et al. 2018; Thavamani et al. 2014; Bhattacharjee et al. 2009). Thus, this trypsin inhibitor of plant origin reflects a promising alternative for a combinatorial and synergistic attack against the field pests and fungal pathogens of chickpea.
Efficient plant regeneration systems amenable to gene transfer are a primary need for the development of transgenic plants. Due to recalcitrant response in vitro, and the limitation of regenerable explants for the successful delivery of T-DNA from Agrobacterium tumefaciens, still persists in chickpea. Excised pre-conditioned cotyledonary nodes (CNs) was preferably chosen as an explant for this study, as it was successfully attempted earlier for transformation in chickpea (Sanyal et al. 2005; Tripathi et al. 2013). Multiple parameters that influence Agrobacterium-mediated insertion of T-DNA were studied such as seed sterilization, age of explant, acetosyringone concentration, O.D. of Agrobacterium cell suspension, sonication duration, co-cultivation duration, cefotaxime concentration and kanamycin concentration, for the selection of putative transgenic plants. Transgenic chickpea plants expressing ChTI protein have been developed up to T1 generation and validated for resistance against field pests like H. armigera and S. litura. Development of pest resistance in chickpea, by expressing a gene from plant origin (ChTI) is the first attempt to be carried out and is investigated in this paper.
Materials and methods
Bacterial strains and plasmids
For preparation of the construct, the 363 bp CDS of ChTI (GeneBank accession no. ABN04119) was used for codon usage optimization and incorporation of the stop codon (TGA). Further, in-silico designed 14 PAGE purified oligonucleotides were assembled by PCR-based gene synthesis method. Restriction sites for KpnI and BamHI sites were incorporated at 5′ end and EcoRI and SacI site at 3′ end of the ChTI gene. The synthesized gene was cloned in pBluescript SK+ cloning vector, and clones were selected through blue-white screening. Further, the synthesized ChTI gene was cloned and expressed in the bacterial expression vector (Champion™ pET-SUMO, Thermo Fisher Scientific, USA), for expression in E. coli and in the plant expression vector pBI121, for transformation of chickpea.
Agrobacterium tumefaciens strain LBA4404 harboring the binary vector pBI121:ChTI having the kanamycin resistance (nptII) gene for plant selection, were grown in YEB medium with appropriate antibiotics at 28 °C for co-cultivation experiments, while E. coli strain DH5α was cultivated at 37 °C in LB medium with appropriate antibiotics (Sanyal et al. 2003). The binary vector pBI121:ChTI contains the nptII gene for kanamycin resistance driven by nos promoter and nos terminator. The ChTI gene is driven by a constitutive CaMV35S promoter and nos terminator (Fig. 1a, b). The plant vector was mobilized into A. tumefaciens strain LBA4404 by electroporation (Gene Pulser, Bio-Rad, USA) and further used for genetic transformation of chickpea.
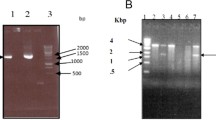
Schematic representation, characterization, and validation of the T-DNA region of the construct harboring the ChTI gene. a pBI121vector. b 2.7 kb fragment of pChTI containing the nptII gene driven by nos promoter and nos terminator.The position of BamHI and SacI on the T-DNA of the construct is used for double digestion to derive the 383 bp ChTI gene fragment. c Characterization of plasmid pBI121:ChTI in Agrobacterium tumefaciens strain LBA4404. Lane M—50 bp marker, lane one pBI121::ChTI digested with BamHI and SacI and run in 1% agarose gel. The size of the ChTI gene—383 bp. d Characterization of plasmid pBI121::ChTI in E.coli strain DH5α and Agrobacterium tumefaciens strain LBA4404 by PCR and run on an agarose gel. M-100 bp marker—PCR assay with internal primers and found desired band size of ChTI—383 bp (lane 1—DH5α, lane 2—LBA4404 lane 3—Negative control), PCR assay with 35S-P-F and NOS-T-R found desired band size of ChTI—643 bp (lane 1—DH5α, lane 2—LBA4404, lane 3—Negative control)
Sequence and structural analysis of ChTI protein
A 3-D model of ChTI protein was predicted and analyzed by the open-source package of Phyre 2.0 (Protein homology/Analogy recognition engine V 2.0). I-TASSER (Iterative Threading Assembly was used for 3-D modeling, legend binding, active site and gene ontological predictions. The SMART online tool was used for functional domain prediction. The physical and chemical parameters of protein were calculated by the ProtParam online tool (ProtParam 2017).
Plant materials and growth conditions
The mature seeds of chickpea (Cicer arietinum L.) variety Pusa 362 was procured from Crop Research Center, Gobind Ballabh Pant University of Agriculture and Technology, Pant Nagar, Uttarakhand, India. Firstly, the seeds were surface sterilized with 0.01% (w/v) HgCl2 for 5 min (Fig. S1a), and rinsed three times in sterile Milli-Q water followed by rinsing in 70% (v/v) ethyl alcohol for 1 min and rinsed three times in autoclaved Milli-Q water. Sterilized seeds were incubated in the dark for germination. The germinated seeds were incubated in appropriate medium, consisting of Murashige and Skoog salts, vitamins of B5, 3% (w/v) sucrose, 0.8% (w/v) agar and 2.0 mg/l 6-Benzylaminopurine (BAP) (Fig. S1b). The culture was incubated in the culture room maintained at 24 ±1 °C under 16 h photoperiod.
In vitro regeneration
The cotyledonary node (CN) explants were collected from 20 days old axenically grown seedlings (Fig. S1c) by removal of the multiple adventitious shoots, main root, and developed shoot buds adjoining the axillary and epicotyl region in a maximum possible way, termed as primary excision (Fig. 2a–c). These excised explants were positioned abaxially in the Petri dish containing MS medium supplemented with l-cysteine, 30 g/1 sucrose, solidified with 0.8 % (w/v) agar and incubated in dark for 24 h. Further, cutting off the emerging shoot buds from the pre-existing axillary meristem of the cotyledonary region, exposes the cells of the L2 layer by transverse secondary excision of 1.5–2.0 mm thick surface layer tissues (Fig. 2d). Following the secondary excision, the excised CNs were agroinoculated according to the optimized conditions After co-cultivation with Agrobacterium, the explants were placed abaxially on MS medium supplemented with 2.0 mg/l BAP, 100 µM acetosyringone, 30 g/1 sucrose, solidified with 0.8% (w/v) agar and incubated in dark for 48 h. Explants were transferred after 48 h into fresh MS medium supplemented with 500 mg/l cefotaxime, 30 g/1 sucrose, solidified with 0.8% (w/v) agar, and incubated for a week under 16/8 light/dark cycle. Further, explants were subcultured after 15 days interval and subsequently after 45 days of culture, the individual shoots were harvested and incubated into MS regeneration medium supplemented with zeatin (0.02 mg/l) + AgNO3 (10 mM) + chlorocholine chloride (0.2 mg/l). The individual putatively transformed shoots were micrografted onto 6–7 days old rootstock of the similar genotype, prepared by decapitating and diagonally cutting the epicotyl area of the germinated seedling. Further, the scion of the putative transgenic shoot was inserted into the vertical incision already made in the stock. Firstly, the micrograft was cultured in soilrite potting mix and irrigated with half-strength MS medium in culture room for 15 days and then were transplanted into pots filled with soilrite:loam:leaf manure (1:1:1) in the greenhouse kept at 60–65% relative humidity at 22–24 °C and 14 h photoperiod (Sanyal et al. 2003).
Explant preparation for in vitro regeneration in chickpea. a Seedlings of chickpea cultivar P-362 as plant material for the preparation of explants. b 20 day old seedlings. c Primary excised cotyledonary node (CN) as explant ready for preconditioning on MS medium supplemented with L-cysteine, 3% sucrose, solidified with 0.8% agar. d CNs excised for the second time to expose the deep-seated L2 layers. e Preparation of CNs for sonication at 42 kHz for 0–60 s. f shoot buds emerging in selection medium, supplemented with 250 mg/l cefotaxime and 100 mg/l kanamycin. g shoots subjected to the third cycle of antibiotic selection. h shoots incubated on regeneration medium. i shoots of putative transformants micro-grafted on 7 days old rootstock
Co-cultivation with Agrobacterium
The Agrobacterium cell density in the co-cultivation suspension culture with excised CNs suggested that the optical density of culture between 0.8 and 1.0, facilitates maximum shoot survival after the third antibiotic selection, higher or lower O.D., caused a considerable reduction in the number of putative transformants (Fig. S2a). About 10–12 excised CNs were co-cultivated with 10 ml A. tumefaciens suspension of OD600 0.8 supplemented with 100 µM acetosyringone (Sigma-Aldrich, USA) in liquid MS medium in 30 ml borosilicate glass tubes (Fig. 2e, Fig. S2a). The increase in the acetosyringone level beyond 100 µM significantly affected the transformation frequency in a declining manner (Fig. S2b). The expression of the ChTI gene in chickpea depends on the duration of sonication up to a larger extent. Sonication duration between 30 and 60 s for pre-conditioned CNs, create micro-wounding in the tissues, effective interaction of plant cells with Agrobacterium, and enhanced transient expression with negligible suppression of in vitro shoot regeneration. While sonication beyond 90 seconds resulted in 80–90% suppression of organogenesis and subsequent shoot development (Fig. S2c). These tubes were sonicated at 42 kHz for 30–60 s in a water bath sonicator (Branson 2510, USA) (Fig. S2c). After sonication-assisted Agrobacterium-mediated transformation (SAAT), CNs were incubated in the rotatory shaker at 75 rpm at 24 °C for 20 min. The co-cultivated CNs were blotted dry on sterile filter paper followed by their adaxial placement, on the plate of MS co-cultivation medium supplemented with 2 mg/1 BAP, 100 µM acetosyringone, and 30 g/l sucrose, solidified with 0.8% (w/v) agar. Co-cultivation was conducted for 48 h, at 24 °C under a 16 h photoperiod. Further, explants were plated on MS medium fortified with 500 mg/l cefotaxime concentration as optimized for one week (Fig. S2d, e). The selection of putative transformants was performed on a selective medium similar to MS culture medium supplementing 100 mg/1 kanamycin as optimized (Sigma-Aldrich, USA), 500 mg/l cefotaxime and 0.02 g/1 AgNO3 for the shoot proliferation (Fig. S2f). Thereafter, the 15 days old explants were transferred on to fresh selective media containing 100 mg/1 kanamycin and subjected to regeneration for 30 days with sub-culturing after 15 days (Fig. 2f). Following the selection, individual putative transgenic shoots was transferred on regeneration medium supplemented with 0.02 mg/l zeatin + 10 mM AgNO3 + 0.2 mg/l chlorocholine chloride for another 15 days (Fig. 2g, h). The emerging putative transgenic shoots were micro-grafted on the pre-grown rootstock of the control non-transgenic plant and grown in a culture room for 15–45 days, until plantlets were hardened enough for ready to survive in the greenhouse (Figs. 2i, 3a–d). Explants pre-conditioned for 24 h before co-cultivation for 48 h gives more transformation events, rather than CNs pre-conditioned beyond 24 h (Fig. S3a). Another important factor for increasing the transformation efficiency is the duration of co-cultivation with Agrobacterium. In this regard, CN explants co-cultivated for 48 h were found to be more efficient for gene transfer, than those co-cultivated for 24 and 60 h (Fig. S3b).
Hardening and acclimatization of putative transformants micro-grafted on seedling stocks for the development of the T0 plants. a Pre-acclimatization of micro-grafted shoots in a growth chamber at 22–24 °C at 60–65% relative humidity and 16 h photoperiod for 15 days. b–d T0 plants were shifted in culture room at 22–24 °C, 60–65% relative humidity and 16 h photoperiod for 30 days for the development of seeds
PCR and Southern blot hybridization analysis
Chickpea genomic DNA was isolated by the method described by (Sanyal et al. 2005). Characterization of plasmid pBI121:ChTI in Agrobacterium tumefaciens strain LBA4404 was carried out through digestion with BamHI and SacI and run in 1% agarose gel (Fig. 1c). Characterization of the plasmid pBI121:ChTI in E.coli strain DH5α and in Agrobacterium tumefaciens strain LBA4404 was done by PCR using internal primers and the desired band size for ChTI—383 bp was seen and with 35S-P-F and NOS-T-R primers the desired band size of ChTI—643 bp was observed (Fig. 1d). Seventeen regenerated transformed shoots were screened by PCR analysis for actual transgenic events through internal (ChTI forward—5′-GGTACCGGATCCATGAGTGATCTAAC-3′ and ChTI reverse—5′-GCGAACGCTATGTCCTGATA-3′) primers and with a set of flanking primers (CaMV35S forward—5′-TATTCGGCTATGACTTGGGC-3′ and ChTI reverse primer). The amplicon size of 383 bp and 483 bp were obtained with the internal and flanking primers, respectively. PCR was performed for 35 cycles, each cycle of 1 min denaturation at 95 °C, annealing at 58 °C for 30 sec, and extension at 68 °C for 30 sec followed by a final extension at 68 °C for 5 min. Amplicons were electrophoresed on 1.0% (w/v) agarose gel and documented on gel-doc (Bio-Rad, USA).
Southern blot hybridization was performed to confirm T-DNA integration in the putative transformants following the established procedures (Sambrook et al. 1989). About 10 µg of chickpea genomic DNA was digested with 50 units of HindIII (New England Biolabs, USA) for 18–24 h and electrophoresed on 0.8% (w/v) agarose at 15V and transferred on to Zeta-Probe GT nylon membrane (Bio-Rad, USA) by vacuum transfer mechanism as per manufacturer’s instructions (Bio-Rad, USA). The blots were hybridized with the 383 bp ChTI gene fragment as a probe radiolabeled with α32P-dCTP (BRIT Mumbai, India). BamHI-SacI digested fragment from pChTI vector was used as positive control. The putative transgenic events with confirmed gene integration, and validated by Southern blot hybridization, were selected to progress in the next-generation (T1) for further analysis.
Gene expression analysis of ChTI by qRT-PCR
To evaluate the expression level of ChTI gene in the transgenic events, total RNA was isolated from the leaves of the selected plants (control and transgenics) in the T1 generation by using the Spectrum plant total RNA kit (Sigma-Aldrich, USA). cDNA was synthesized using the Enhanced Avian HS RT-PCR kit (Sigma-Aldrich, USA). Further, cDNA was used as a template for qRT-PCR to assess the total transcript level by using SYBR Green PCR Master Mix (Applied Biosystems, USA) and was performed in StepOne real-time PCR system (Applied Biosystems, USA). Chickpea beta-actin gene was taken as an endogenous control. The real-time expression of the genes was estimated by using the \(2^{{ - \Delta \Delta C_{{\text{T}}} }}\) method (Livak and Schmittgen 2001). The primer sequences designed for analysing the ChTI gene and chickpea beta-actin are shown in Table S1.
Trypsin inhibition activity assay and Western blot analysis
100 mg leaf tissue from the individual T1 plant was harvested and chopped into small pieces followed by washing in cold PBS. Further, 4X-6X volumes (500–1000 µl) of trypsin activity assay buffer was added and were incubated on ice followed by homogenization in ice, and immediately centrifuged at 4 °C, for 2–5 min at maximum speed. The supernatant having total soluble protein (TSP) were collected in a fresh tube. The supernatant was further used for trypsin inhibition activity assay, as described in trypsin activity assay kit (colorimetric) (Abcam ab102531, USA) (Robles et al. 2015).
Total soluble proteins (TSPs) were incubated at 70 °C for 10 min, chilled on ice, and centrifuged at 12,000 rpm for 20 min at 4 °C (Bhattacharjee et al. 2009). Almost all the plant serine proteinase inhibitors are known to thermostable, apart from their counteracting proteinases and have negligible endogenous proteolytic activity throughout the assay. The supernatant (Heat Stable Proteins-HSPs) was used for Western blot assay. HSPs from transgenic leaves were separated on 15% SDS-PAGE. Further, the gel containing separated proteins was transferred on a PVDF membrane using a Trans-blot Semi-Dry Transfer Cell (Bio-Rad, USA), at 15V for one hour. Immunoblot was performed for the ChTI protein expressed in the T1 transgenic chickpea plants (T11 and T15), and ChTI expressed in E. coli (Bl21) by using ChTI-IgG antibodies (Chromous Biotech, Bangalore, India).
Insect bioassay
Insecticidal activity of the toxin ChTI in T1 chickpea plants was assayed by conducting a no-choice detached leaf-feeding bioassay, using the second instar neonatal larvae of H. armigera and S. litura. Larvae of H. armigera were initially reared on an artificial diet at 26 ± 2 °C, 70% relative humidity under 14 h light, and 10 h dark light regime and thereafter by feeding on castor leaves to complete their life cycle. In this experiment, about 200–250 mg of fresh leaves from transgenic and control chickpea plants, were placed in a 50 ml capped plastic bottle, and 10 neonate larvae were infested and allowed to feed on the leaves. The bottles were sealed with Parafilm and placed in the insect rearing room at 70% relative humidity, 26±2 °C, and a 16 h photoperiod, to prevent desiccation. Under the same conditions, feeding was allowed for 4 days, with the supply of fresh plant leaves on alternate days and the data was documented on larval mortality and larval weight. Feed choice assay was carried out to determine the preferential choice of leaves, when kept in the same vicinity for the larvae.
Statistical analysis
Values in the results reported are mean S.D. of three replicates, and a minimum of 50 explants was used in each experiment. The data were analyzed by using Microsoft excel (MS-Excel) and SPSS statistical package. A significant difference analysis within the means (p <0.05) was performed with the help of DMRT (Duncan's Multiple Range Test), and compared each parameter independently. Bars that contain the same letter(s) do not differ significantly.
Results
Sequence and structural analysis of ChTI protein
Retrieval of Cocculus hirsutus trypsin inhibitor sequence in FASTA format has been carried out from UniProt Primary accession number: A2TJS6 (Fig. S4). A 3-D model of protein predicted by the open-source package of Phyre 2.0 (protein homology/Analogy recognition engine V 2.0) (Fig. S5a). Secondary structure with percent disorders was evaluated and summarised (Fig. S5b). 15 Protein sequences are showing variable homology as their domain alignment indicated (Fig. S5c). Out of 15 homologues, four (c2mw7A, c6f0fB, c6f0gC, c6f0gD) have > 70% similarity as shown by their alignment (Fig. S5d) (Kelley et al. 2015).
Although I-TASSER (Iterative Threading Assembly Refinement) is a hierarchical approach to predict protein function and structure, instead of Phyre 2.0 it provides comprehensive output for a 3-D model by using meta-threading approaches in addition to ligand binding site, active site, and gene ontological predictions (Figs. S6, S7, S8) (Zhang 2009; Roy et al. 2012; Yang and Zhang 2015).
PSI search online tool has been used for functional domain analysis for homologue sequences in the database, against different profiles such as SMART, Pfam, etc. (Madeira et al. 2019) (Fig. S9). Two functional domains were annotated by the SMART online tool: the ZnF_TAZ domain and IKK beta NEMO binding domain. TAZ (Transcription Adaptor putative Zinc finger) domains are zinc-containing domains found in the homologous transcriptional co-activators CREB-binding protein (CBP) and the P300, positions predicted between 40 and 112 in the sequence, CBP and P300 are histone acetyltransferase, which catalyzes the reversible acetylation of all 4 histones in nucleosomes, eventually, regulate transcription through chromatin remodeling. However, another domain is the IKKbetaNEMO bind domain; positions are predicted between 64 to 98 in the sequence, and these proteins have a key role in inflammatory reactions. Further, this domain was validated as the binding site of IKK-beta for NEMO (Letunic and Bork 2017).
The physical and chemical parameters of the protein is calculated by the ProtParam online tool (ProtParam 2017). The estimated half-life of ChTI protein is 30 h (mammalian reticulocytes, in vitro), > 20 h (yeast, in vivo), and > 10 h (E. coli, in vivo). However, the instability index is computed to be 86.18. This classifies the protein as unstable. The grand average of hydropathicity (GRAVY) is calculated as -0.984, which indicates the hydrophilic nature of the ChTI protein.
In vitro regeneration from cotyledonary nodes explants of chickpea
To complement higher susceptibility and efficiency for Agrobacterium-mediated transformation, CNs having uncovered cells of L2 layers were used for in vitro regeneration. The CNs from 20 days old germinated seedlings of chickpea, showed the optimal response for the development and induction of adventitious shoots (Fig. S1a). After 2 weeks of culture, the development of shoot buds was initiated, and the formation of meristematic domes on the surface of excised CN was observed. These primordial structures further elongated into shoots. Micrografting of putative transgenic shoots on 6–7 days old rootstock of a similar genotype, reflects 82–85% endurance and complete plant development under the optimized glasshouse conditions (Sanyal et al. 2003).
Co-cultivation parameters for improved transformation efficiency
The transformation efficiency of Agrobacterium-mediated delivery of T-DNA into plant cells is determined by several physiological and physicochemical parameters. Alteration in parameters is taken into the account to optimize Agrobacterium-mediated gene delivery and collection of transformants using CN explants. Considering the four Agrobacterium strains (GV2260, GV3850, LBA4404, and EHA105), strain LBA4404 was found statistically significant compared to GV2260, GV3850 and EHA105 as reported in earlier studies (Sanyal et al. 2005). Based on the optimized transformation and tissue culture conditions, Agrobacterium co-cultivation was carried out with 24 h pre-conditioned CNs explants in the presence of 100 µM acetosyringone with 0.8 OD600 of Agrobacterium cell density, sonication for the 30 s followed by 48 h co-cultivation into MS medium supplemented with 2.0 mg/l BAP, 100 µM acetosyringone, 3% sucrose and 0.8% agar. Meanwhile, transformed shoots were selected with 100 mg/l kanamycin in the first and second round, followed by 150–200 mg/1 kanamycin in the third round. Following the selection cycles, 17 putative transgenic shoots were recovered and reflects a transformation frequency of 2.36% with the pChTI construct (Table S2).
Molecular analysis of T0 putative transgenic plants
Genomic DNA was isolated from all the 17 primary transformants (T0) and subsequently used for PCR screening using the internal primers and flanking primers ChTI gene, for the amplification of DNA fragments of 383 and 483 bp, respectively. The results summarized in (Fig. 4a, b) which suggested that 8 plants were PCR positive for both ChTI internal and flanking primers out of the 17 T0 putative transgenic plants evaluated, while 9 putative transformed plants did not show amplification with either set of primers. Furthermore, T0 chickpea plants were checked for gene integration as well as copy number by Southern blot hybridization analysis. Southern blot analysis with specific radiolabelled ChTI probe was performed with genomic DNA isolated from the chickpea plants (T0) showed stable integration of ChTI in 7 events (T01, T02, T03, T05, T08, T09 and T010) out of the 10 events tested (Fig. 4c). Events T01, T05 and T08 was found with single-copy insertion of the ChTI gene, T02 and T010 showed three copies and events T03 and T09 have a copy number of 4. These events, validated by Southern blot hybridization were allowed to proceed to the next-generation (T1).
Molecular analysis of transgenic eventsin chickpea. a using the internal primers of ChTI and the desired band size of 383 bp (M—100 bp ladder, NTC—non-template control, NC—negative control, PC—positive control, lane 1–8 are putative T0 transgenic events. b with flanking primers of35S-P-F and ChTI-R, and desired band size as 483 bp (M—100 bp, NTC—non-template control, NC—negative control, PC—positive control, lane 1–8 are putative T0 transgenic events. c Southern blot hybridization with the radiolabelled probe of ChTI gene (+) positive control (BamHI-SacI digested fragment from pChTI vector), (−) negative control (non-transformed chickpea plants), lines T01, T02, T03, T05, T08, T09 and T010 are the putative transgenic events hybridizing with the probe and lane T04, T06, and T07 are not, indicating the integration of ChTI gene). d qRT—PCR analysis—the relative expression of the transgene with gene-specific primers of ChTI, and the actin gene of chickpea as an endogenous control. A significant difference analysis within the means (p <0.05) was performed with the help of DMRT (Duncan's multiple range test), and compared each parameter independently. Bars that contain the same letter(s) do not differ significantly. All the given values are means of a minimum of three replicates ± SD
qRT-PCR assay
The relative expression of the ChTI was evaluated by qRT-PCR using ChTI gene-specific primers and actin gene of chickpea was utilized as an endogenous control. The fragments of 137 and 134 bp were amplified through the set of primers for ChTI and chickpea actin genes, respectively. The transcript levels in the 7 selected transgenic T1 lines expressing ChTI gene was analyzed along with the endogenous control, in triplicates. The result indicated a higher transcript level of the ChTI gene ranging from 12 to 17-fold change (Fig. 4d).
Trypsin inhibition activity assay and Western blot analysis
Total soluble protein (TSPs) was used for trypsin inhibition activity assay as described by trypsin activity assay kit (colorimetric, abcam ab102531). T1 transgenic lines expressing the ChTI gene have exhibited a higher trypsin inhibitory activity, ranging from 3100 to 5100 TIU/g (trypsin inhibition unit/gram) in comparison to control, which has very low antitrypsin activity, 900 TIU/g, due to the endogenous trypsin inhibitors (Fig. 5a). HSPs were used for Western blot analysis. HSPs isolated from the transgenic leaves (T11 and T15) and control were separated on 15% SDS-PAGE. 31 kDa ChTI protein was expressed, induced, and purified, and immunoblotted from bacteria (E.coli. BLl21 cells) (Fig. 5b (I), Fig. S 10). Immunoblot analysis using ChTI-IgG confirmed the presence of 15 kDa protein corresponding to ChTI in chickpea transgenic plants (T11 and T15) (Fig. 5b (II)).
Characterization of ChTI protein. a trypsin inhibition activity of transgenic plants expressing ChTI protein. Positively screened transgenic plant tissues (T1) were assayed for inhibitory activity of trypsin. Trypsin inhibitory activity from total soluble protein (TSP) from the leaves of control and T1 plants expressing ChTI protein was calculated as trypsin inhibitory unit/gram of tissue (TIU/g): control—900 TIU/g, 1–3300 TIU/g, 2–4200 TIU/g, 3–5100 TIU/g, 4–3800 TIU/g, 5–4600 TIU/g, 6–3100 TIU/g, 7–4400 TIU/g. (b) Immunoblot of ChTI (I) purified protein fraction from E.coli (Bl21) with Penta-His antibody lane 1—protein marker (M), lane 2—wash protein (W) from purification column, lane 3—purified ChTI protein lane 4—uninduced (UI) culture protein, lane 5-protein marker (II) Expression of ChTI in chickpea, lane 1—protein marker, lane 2 and 3—ChTI from plant protein fraction (T11 and T15), lane 4—negative control, lane 5—protein marker. A significant difference analysis within the means (p <0.05) was performed with the help of DMRT (Duncan's multiple range test), and compared each parameter independently. Bars that contain the same letter(s) do not differ significantly. All the given values are means of a minimum of three replicates ± SD
Assessment of insecticidal activity
The insecticidal activity of T1 plants expressing ChTI protein was evaluated using the leaves of individual transgenic and control chickpea plants for feeding assays, using the second instar neonate larvae of S. litura and H. armigera (Fig. 6a). Data revealed that most larvae ceased feeding after 2 days on leaves of T1 plants (Fig. 6b, d). T1 chickpea plants expressing a large amount of ChTI protein, which triggers a significant reduction in the weight of larvae from 30 to 60% and 40 to 60% for S. litura to H. armigera, respectively (Fig. 6e). Whereas, larvae challenged on the leaves of individual plants of T1 generation, reflect moderate to high (60% to 80%) mortality due to stunted growth and impaired life cycle, with early pupation of S. litura and H. armigera (Fig. 6f). Feed choice assay for determination of the larvae's preferential choice indicated that second instar larvae of S. litura to H. armigera, preferred control plants as feed choice compared to transgenic lines expressing the ChTI entomocidal protein (Fig. 6c).
Leaf feed assay for toxicity assessment on 2nd instar H. armigera and S. litura larvae. a Insect leaf feed assay was set up in 50 ml tube, with control and transgenic leaves (T1) in triplicates. b In leaf feed assay, the maximum damage is observed in control leaves, and larvae fed on transgenic leaves showed minimum damage against H. armigera and S. litura. c Feed choice assay was demonstrated for validation of preferential feeding of leaves by larvae of H. armigera and S. litura. d Insect mortality and weight loss assessment of 2nd instar S. litura and H. armigera larvae fed on leaves. 60–80% mortality and 30–60% weight loss were observed in 2nd instar larvae of S. litura and H. armigera fed on transgenic leaves, in comparison to negligible loss in larvae fed on control leaves. e, f insect weight loss and survival assessment of S. litura. g, h insect weight loss and survival assessment of H. armigera. A significant difference analysis within the means (p < 0.05) was performed with the help of DMRT (Duncan's multiple range test), and compared each parameter independently. Bars that contain the same letter(s) do not differ significantly. All the given values are means of a minimum of three replicates ± SD
Assessment of plants in T1 generation
Total of seven numbers of T0 transgenic events and four untransformed tissue culture grown control plants of chickpea were moved to the glasshouse for seed setting and for the analysis of inheritance of the transgene (ChTI) in the next generation. All the seven T0 plants were fertile and produced seeds, although the number of pods in transgenic plants was surprisingly low (4–6 per plant) in comparison to (8–10) pods in the control plants. However, the viability of T1 seeds from plants grown through tissue culture remained low, compared to seeds developed from the field-grown plants. PCR analysis of T1 transgenic plants has been carried out with internal primers and flanking primers (Fig. 7). The pattern of kanamycin resistance in T1 seeds on antibiotic-supplemented growth medium, exhibited the typical 3:1 Mendelian ratio as anticipated for single dominant gene inheritance, apart from one transgenic line, where more susceptible seeds were found, than the anticipated Mendelian ratio. The segregation pattern of expression of ChTI gene in the progenies of the promising T0 lines is shown in Table S3.
Assessment of the transgenic events of T1 generation by PCR. a Using internal primers of ChTI gene and found the desired band size of 383 bp (M 100 bp marker, NC negative control, PC positive control, lane 1–7 are putative transgenic events, lane 8: NTC non-template control). b using flanking primers with 35S-P-F and ChTI-R and found desired band size of 483 bp (M 100 bp marker, NC negative control, PC positive control, lane 1–7 are putative transgenic events, lane 8: NTC non-template control)
Discussion
Previous studies revealed that the ChTI protein has an important role in plant defense against biotic stresses, such as antimicrobial and insect resistance (Bhattacharjee et al. 2009). However, the utilization of ChTI to develop insect-resistance in an economically valuable crop is entirely unexplored. In a recent study, the expression of Cocculus hirsutus trypsin inhibitor (ChTI) in transgenic tomato and the subsequent upregulation of the endogenous plant defense response, as well as the level of antioxidants against H. armigera has been reported. The tomato plant expressing ChTI gene showed early flowering in the transgenic plants by 10 days, approximately 45% increase in the plant height, increased diameter of fruits, and delay in fruit ripening. Besides increased nutrient and antioxidant levels, 100% mortality of 2nd and 4th instar larvae of H. armigera, fed on the transgenic tomato leaves was documented (Manushree et al. 2020). This study demonstrates the first report on the expression of ChTI protein in any plant belonging to the legume family. In comparison to tomato, the regenerative potential is limited in chickpea, besides this 60–80% mortality and 30–60% weight loss were recorded from transgenic chickpea plants against the Lepidopterian insects.
Earlier, several attempts were carried out to express plant protease inhibitors in different crops and notable mortality/weight loss or both, were reported against the field pests. Transgenic tobacco plants were developed, expressing cowpea trypsin inhibitor (CpTI) and 35–40% mortality was validated against S. litura (Sane et al. 1997). Altpeter et al. (1999) developed transgenic wheat by incorporating the barley trypsin inhibitor CMe (BTI-CMe), showed resistance with 29% motality against Sitotroga cerealella. In a different study, Solanum americanum proteinase inhibitor II (PIN2) was used to develop resistance in tobacco with 50–56% mortality against H. armigera (Luo et al. 2012).
Despite gene alone stratigies, few gene pyramiding apparoches were demonstrated such as Nicotiana alata proteinase inhibitor (NaPI) and Solanum tuberosum potato type I inhibitor (StPin1A) were co-integrated in the cotton genome and 90% weight loss was documented against Helicoverpa punctigera (Dunse et al. 2010). Furthermore in another report, 40% mortality was observed against Tuta absoluta by co-expressing the serine proteinase inhibitor (BTI-CMe) and cysteine proteinase inhibitor (Hv-CPI2) in tomato (Hamza et al. 2018).
Although several attempts have been carried out to develop insect resistance in chickpea (Sanyal et al. 2005; Ignacimuthu and Prakash 2006; Gowri Neelima et al. 2008; Chakraborti et al. 2009; Acharjee et al. 2010; Asharani et al. 2011; Ganguly et al. 2014; Khatodia et al. 2014; Khatodia 2017), minimal success was observed particularly in case of S. litura and H. armigera. In this study, we have focused on using a transgene from plant origin itself, i.e., ChTI, and the development of transgenic chickpea expressing ChTI for insect resistance.
This study is focused on developing transgenic chickpea plants harboring the ChTI gene and its assessment against S. litura and the chickpea pod borer H. armigera. Although widely regarded as a highly recalcitrant crop, chickpea transformation and regeneration has shown minimal success to date (Sanyal and Amla 2008). Agrobacterium-mediated transformation and the optimized tissue culture procedures have directed this particular study to achieve a transformation efficiency up to 2.36% by using cotyledonary node as an explant. The selection of plants was made based on the kanamycin as a selection marker, utilizing an iterative procedure. After the final round of selection, 17 putative transgenic shoots were collected and subsequently micrografted for hardening and seed setting. Furthermore, the putative transgenic lines were screened through PCR, by using internal as well as flanking markers. Finally, seven PCR positive events were successfully validated through Southern blot hybridization, except one escape, possibly due to the chimeric nature of the transformant.
Real-time qPCR analysis in T1 plants revealed 12–17-fold expression upregulation of ChTI gene, which is quite impressive to confirm the potential of ChTI for the development of insect resistance in chickpea. Moreover, leaf feed assay was carried out to assess the insecticidal potential of ChTI, against two very challenging pests of this economically important legume crop, S. litura and H. armigera. T1 chickpea plants showed a significant weight loss of 30–60% and 40–60% for S. litura to H. armigera, respectively. Whereas, the 2nd instar larvae challenged on leaves of individual plants from T1 generation, have shown moderate to high (60% to 80%) mortality of S. litura and H. armigera. The immunoblot analysis and trypsin activity assay (3100–5100 TIU/g) were performed, and protein expression was validated in E. coli and in T1 generation of chickpea plants.
Albeit, transgenic chickpea plants expressing ChTI alone wouldn’t be idealy forwarded to offer 100% mortality against pod borer and leaf catterpiller. To attain 100 percent mortality and optimal weight loss, gene pyramiding apporoach alongwith protein engineering of ChTI may prove beneficial to achive further improvement in context to field pest resistance. Protein engineering through the strategies such as molecular simulation and specific localization, to enhance the functional properties of ChTI such as specific activity, stability (thermal stability, tolerance to salts and organic solvents, pH tolerance and solubility), inhibition by reaction products, and specificity against non-natural substrates.
In the future, we need to do a controlled assessment of responses by these plants for field pest tolerance. Identifying and screening of new endogenous proteases in chickpea and whether these endogenous chickpea protease inhibitors can specifically modify the resistance potential of plants against field pests, will be futuristic. It will be very interesting and crucial to identify, characterize, and validate the antimicrobial and pharmaceutical (anti-neoplastic, anti-inflammatory, anti-oxidative, anti-diabetic) potential of the ChTI protein.
Conclusions
This particular study demonstrated the role of the ChTI gene against biotic stress in an economically important leguminous crop (Cicer arietinum L.), which has not been investigated earlier in detail. This study reveals that transgenic chickpea expressing the ChTI gene can overcome the biotic stress imposed due to insects, by augmenting the plants' defense mechanism, such as anti-trypsin activity. The reduction in larval weight, mortality, and preferential feeding choice boosts the survivability in transgenic plants of chickpea against herbivory. The obtained results from the insect resistance evaluation demonstrates that the expression of ChTI offers better resistance in transgenic plants, in contrast to control plants against stress generated by insects. It was validated that the ChTI gene expression can be used to develop insect-resistant cultivars to improve crop productivity.
Change history
22 March 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11240-021-02054-x
References
Acharjee S, Sarmah BK (2013) Biotechnologically generating ‘super chickpea’for food and nutritional security. Plant Sci 207:108–116
Acharjee S, Sarmah BK, Kumar PA, Olsen K, Mahon R, Moar WJ, Moore A, Higgins T (2010) Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene. Plant Sci 178:333–339
Altpeter F, Diaz I, McAuslane H, Gaddour K, Carbonero P, Vasil IK (1999) Increased insect resistance in transgenic wheat stably expressing trypsin inhibitor CMe. Mol Breed 5(1):53–63
Asharani B, Ganeshaiah K, Kumar A, Udayakumar M (2011) Transformation of chickpea lines with Cry1X using in planta transformation and characterization of putative transformants T1 lines for molecular and biochemical characters. J Plant Breed Crop Sci 3:413–423
Bhattacharjee C, Manjunath NH, Prasad DT (2009) Purification of a trypsin inhibitor from Cocculus hirsutus and identification of its biological activity. J Crop Sci Biotechnol 12:253–260
Carrière Y, Crickmore N, Tabashnik BE (2015) Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol 33:161
Chakraborti D, Sarkar A, Mondal HA, Das S (2009) Tissue specific expression of potent insecticidal, Allium sativum leaf agglutinin (ASAL) in important pulse crop, chickpea (Cicer arietinum L.) to resist the phloem feeding Aphis craccivora. Transgenic Res 18:529–544
Dixit G, Srivastava A, Singh N (2019) Marching towards self-sufficiency in chickpea. Curr Sci 116:239–242
Dunse KM, Stevens JA, Lay FT, Gaspar YM, Heath RL, Anderson MA (2010) Coexpression of potato type I and II proteinase inhibitors gives cotton plants protection against insect damage in the field. Proc Natl Acad Sci USA 107(34):15011–15015
Erb M, Reymond P (2019) Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol 70:527–557
Ganguly M, Molla KA, Karmakar S, Datta K, Datta SK (2014) Development of pod borer-resistant transgenic chickpea using a pod-specific and a constitutive promoter-driven fused cry1Ab/Ac gene. Theor Appl Genet 127:2555–2565
Gassmann AJ, Petzold-Maxwell JL, Clifton EH, Dunbar MW, Hoffmann AM, Ingber DA, Keweshan RS (2014) Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc Natl Acad Sci USA 111:5141–5146
Gatehouse AM, Shi Y, Powell KS, Brough C, Hilder VA, Hamilton WD, Newell CA, Merryweather A, Boulter D, Gatehouse JA (1993) Approaches to insect resistance using transgenic plants. Philos Trans R Soc Lond Ser B Biol Sci 342:279–286
Gowri Neelima M, Ramu S, Sreevathsa R, Rani A, Kumar A, Gayatri M (2008) In planta transformation strategy to generate transgenic plants in chickpea: proof of concept with a cry gene. J Plant Biol (New Delhi, India) 35:201–206
Gupta VK, Kaushik A, Chauhan DS, Ahirwar RK, Sharma S, Bisht D (2018) Anti-mycobacterial activity of some medicinal plants used traditionally by tribes from Madhya Pradesh, India for treating tuberculosis related symptoms. J Ethnopharmacol 227:113–120
Hamza R, Pérez-Hedo M, Urbaneja A, Rambla JL, Granell A, Gaddour K, Beltrán JP, Cañas LA (2018) Expression of two barley proteinase inhibitors in tomato promotes endogenous defensive response and enhances resistance to Tuta absoluta. BMC Plant Biol 18(1):1–14
Hilder VA, Gatehouse AM, Sheerman SE, Barker RF, Boulter D (1987) A novel mechanism of insect resistance engineered into tobacco. Nature 330:160
Ignacimuthu S, Prakash S (2006) Agrobacterium-mediated transformation of chickpea with α-amylase inhibitor gene for insect resistance. J Biosci 31:339–345
Kansal R, Kumar M, Kuhar K, Gupta RN, Subrahmanyam B, Koundal KR, Gupta VK (2008) Purification and characterization of trypsin inhibitor from Cicer arietinum L. and its efficacy against Helicoverpa armigera. Braz J Plant Physiol 20:313–322
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845
Khatodia S (2017) Helicoverpa resistant chickpea plants: from Bt toxins to plant-mediated RNAi. Ekin Journal of Crop Breeding and Genetics 3:52–60
Khatodia S, Kharb P, Batra P, Chowdhury VK (2014) Development and characterization of transgenic chickpea (Cicer arietinum L.) plants with cry1Ac gene using tissue culture independent protocol. Int J Adv Res 2:323–331
Letunic I, Bork P (2017) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496
Luo M, Ding LW, Ge ZJ, Wang ZY, Hu BL, Yang XB, Sun QY, Xu ZF (2012) The characterization of SaPIN2b, a plant trichome-localized proteinase inhibitor from Solanum americanum. Int J Mol Sci 13(11):15162–15176
Madeira F, Ym Park, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641
Manushree V, Devaraj V, Prasad D (2020) Expression of Cocculus hirsutus trypsin inhibitor promotes endogenous defensive response against Helicoverpa armigera and enhanced levels of antioxidants. Afr J Plant Sci 14:65–82
Mehrotra M, Singh AK, Sanyal I, Altosaar I, Amla D (2011) Pyramiding of modified cry1Ab and cry1Ac genes of Bacillus thuringiensis in transgenic chickpea (Cicer arietinum L.) for improved resistance to pod borer insect Helicoverpa armigera. Euphytica 182:87
Nair M, Sandhu SS, Babbar A (2013) Purification of trypsin inhibitor from seeds of Cicer arietinum (L.) and its insecticidal potential against Helicoverpa armigera (Hübner). Theor Exp Plant Physiol 25:137–148
Patil SB, Goyal A, Chitgupekar SS, Kumar S, El-Bouhssini M (2017) Sustainable management of chickpea pod borer: a review. Agron Sustain Dev 37:20
Perlak FJ, Oppenhuizen M, Gustafson K, Voth R, Sivasupramaniam S, Heering D, Carey B, Ihrig RA, Roberts JK (2001) Development and commercial use of Bollgard® cotton in the USA—early promises versus today’s reality. Plant J 27:489–501
ProtParam E (2017) ExPASy-ProtParam tool
Rakesh T, Prashant T (2012) Effects of Cocculus hirsutus leaves extract on Freund’s complete adjuvant and formaldehyde-induced arthritis. Int Res J Pharm 3:267–270
Robles L, Vaziri ND, Li S, Takasu C, Masuda Y, Vo K, Farzaneh SH, Stamos MJ, Ichii H (2015) Dimethyl fumarate ameliorates acute pancreatitis in rodent. Pancreas 44:441–447
Roy A, Yang J, Zhang Y (2012) COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res 40:W471–W477
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P (2011) Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotech J 9:283–300
Sane VA, Nath P, Sane A, Sane PV (1997) Development of insect-resistant transgenic plants using plant genes: expression of cowpea trypsin inhibitor in transgenic tobacco plants. Curr Sci 72(10):741–747
Sanyal I, Amla D (2008) Genetic transformation of chickpea (Cicer arietinum L) using cotyledonary node explants. Handbook of new technologies for genetic improvement of legumes. The Haworth Press, Taylor & Francis Group, Boca Raton, pp 147–158
Sanyal I, Singh A, Amla D (2003) Agrobacterium tumefaciens-mediated transformation of chickpea (Cicer arietinum L.) using mature embryonic axes and cotyledonary nodes. Indian J Biotechnol 2:524–532
Sanyal I, Singh AK, Kaushik M, Amla DV (2005) Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) with Bacillus thuringiensis cry1Ac gene for resistance against pod borer insect Helicoverpa armigera. Plant Sci 168:1135–1146
Tabashnik BE, Brévault T, Carrière Y (2013) Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol 31:510
Thavamani BS, Mathew M, Palaniswamy DS (2014) Anticancer activity of Cocculus hirsutus against Dalton’s lymphoma ascites (DLA) cells in mice. Pharm Biol 52:867–872
Tripathi L, Singh AK, Singh S, Singh R, Chaudhary S, Sanyal I, Amla DV (2013) Optimization of regeneration and Agrobacterium-mediated transformation of immature cotyledons of chickpea (Cicer arietinum L.). Plant Cell Tiss Org Cult (PCTOC) 113:513–527
Yamasaki A, Shimizu K, Fujisaki K (2009) Effect of host plant part on larval body-color polymorphism in Helicoverpa armigera (Lepidoptera: Noctuidae). Ann Entomol Soc Am 102:76–84
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181
Zhang Y (2009) I-TASSER: fully automated protein structure prediction in CASP8. Proteins Struct Funct Bioinform 77:100–113
Zhu-Salzman K, Zeng R (2015) Insect response to plant defensive protease inhibitors. Ann Rev Entomol 60:233–252
Acknowledgements
The authors are grateful to the Director, CSIR-NBRI, Lucknow, for infrastructural support. The authors are grateful for fellowships to UGC, New Delhi (AP, SK), CSIR, New Delhi (RY, AK, PS), and DST (AY), New Delhi. The research work was carried out under the CSIR-NBRI In-house project OLP104. NBRI manuscript no. CSIR-NBRI_MS/2021/02/06.
Author information
Authors and Affiliations
Contributions
Conceptualization: AP, RY & IS, Formal analysis: AP, AK, RY, Funding acquisition: IS, Investigation: AP, RY, AK, SK, PS, AY, Methodology: AP & RY, Project administration: IS, Resources: IS, Supervision: IS, Validation: AP, Writing - original draft: AP, Writing - review & editing: IS and AP.
Corresponding author
Ethics declarations
Conflict of interest
This is to state that all authors declare no conflict of interest.
Additional information
Communicated by Sergio J. Ochatt.
The original version of this article has been revised: The following missing sections have been included: Acknowledgements, Author contributions, and Declaration.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pandey, A., Yadav, R., Kumar, S. et al. Expression of the entomotoxic Cocculus hirsutus trypsin inhibitor (ChTI) gene in transgenic chickpea enhances its underlying resistance against the infestation of Helicoverpa armigera and Spodoptera litura. Plant Cell Tiss Organ Cult 146, 41–56 (2021). https://doi.org/10.1007/s11240-021-02041-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-021-02041-2