Abstract
The advent of high-throughput RNA Sequencing (RNA-Seq) datasets revealed that a large fraction of transcribed RNAs does not code for proteins. However, studies suggest that these RNAs have various regulatory roles. The long non-coding RNAs (lncRNAs) (length > 200 nucleotides) is the largest family of the non-coding RNA (ncRNAs) and the largest portion among various types of ncRNAs. LncRNAs regulate the gene expression at transcriptional, post-transcriptional and epigenetic levels through different mechanistic themes and interaction with all kind of biomolecules like DNA, RNA and proteins. The lncRNAs have important roles in the regulation of reproduction and sex determination, modulation of transcriptional patterns and protein activities as well as alteration of RNA processing events in animals and plants. Debates are still going on the role of lncRNAs especially in plants as the biological data are increasing with time. In this review, mechanisms of action, biological roles, classification on the basis of genomic locations of plant lncRNAs have been discussed in detail.
Key message
Despite being the largest group of non-coding RNA (ncRNA) family, the long non-coding RNAs (lncRNAs) are least characterized in terms of regulatory functions. In this review, updated information has been provided regarding biological roles and mechanism of action of plant lncRNAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although approximately 90% of the genomes are transcribed into RNA (Costa 2010; Pertea 2012), only ~ 1–2% of the transcribed genome encodes for various proteins and the major portion (98–99%) remains non-coding (Fang et al. 2018). These non-coding RNAs (ncRNAs) are of two types—housekeeping and regulatory. The housekeeping or structural ncRNAs consist of tRNA, rRNA, snRNA and snoRNA and show constitutive expression in the cell (Ponting et al. 2009). The regulatory ncRNAs again are of two types based on their length. Short regulatory ncRNAs including miRNA (22–23 bp), siRNA (19–25 bp) and piRNA (26–31 bp) and long regulatory ncRNAs are also known as long noncoding RNAs (lncRNAs) (> 200 bp) (Quan et al. 2015). The lncRNAs are the most recently identified member among different ncRNAs. The lncRNAs are reported to constitute 80% of the total ncRNAs (Zhang et al. 2013a). Previously, it was thought that lncRNAs contribute only in the transcriptional noise in the cell (Ponjavic et al. 2007). However, in the present scenario, these are known to play a significant role at different molecular levels in the cells. On the basis of their relationship with neighbouring protein-coding genes, the lncRNAs are further divided into different classes (Rinn and Chang 2012). The predictive measures of lncRNAs are of > 200 bp and having the capacity of coding open reading frame (ORF) of < 100 amino acid residues. These lncRNAs lack a significant ORF, initiation codon, 3′ UTRs and a termination codon. The process of biogenesis of lncRNAs is same as protein-coding mRNAs. Most of the lncRNAs are transcribed by RNA polymerase II, like mRNAs but some lncRNAs have also been reported to be transcribed by RNA Polymerase III (Wu et al. 2012b). The lncRNAs have all the properties like polyadenylation at 3′ end, 5′ capping and splicing similar to mRNAs except the potential of coding a protein (Quan et al. 2015). The lncRNAs participate in specifically three modes of regulation of gene expression such as transcriptional, post-transcriptional and epigenetic control in the cis and trans manner (Quan et al. 2015; Zhang et al. 2014a). A large proportion of lncRNAs (42%) are reported to be involved in transcriptional regulation (Quek et al. 2015). In transcriptional regulation, the lncRNAs affect the transcription of genes through transcriptional interference. In the cell, the epigenetic and transcriptional regulations occur in the nucleus whereas the post-transcriptional regulations/modifications take place in the cytoplasm (Zhang et al. 2014a, b, c). The higher percentage of ncRNAs in different eukaryotes such as human, mouse, yeast and Drosophila suggest its abundance and hotspot for future research. The genes of the organisms and their regulation of the expression are the most important factors governing changes in the morphology, anatomy or physiology (Bard 2014) and show a quick response against any kind of variation in the cell. The expression of genes is governed by different regulatory elements, mainly transcription factors and ncRNAs. The family of ncRNAs is responsible for rapid changes in the gene expression at every level of genomic processing. The research on plant lncRNAs is also an emerging area of performing various regulatory mechanisms. In this review, we describe the updated information related to the potential regulatory roles of one of the important ncRNAs i.e. lncRNAs in plants.
lncRNA classification
Though the lncRNAs were considered as the dark matter of the genome, these are currently explored as crucial gene regulators. Based on the location and orientation to the nearest protein-coding gene in the genome, the lncRNAs can be classified into five types (Ma et al. 2013). Sense lncRNAs are present on the same strand of protein-coding genes whereas Antisense lncRNAs are located on the opposite strand of the protein-coding genes. Bidirectional lncRNAs are located on the opposite strand of the protein-coding genes whose transcription is initiated less than 1000 bp away. In addition, Intronic lncRNAs and Intergenic lncRNAs (lincRNA) classes of lncRNAs are located in the introns of the protein-coding gene and between two protein-coding genes respectively.
Mechanistic themes of lncRNAs
The ncRNAs are involved in diverse mechanisms of gene expression regulation. The mechanistic themes of the regulation of small RNAs such as miRNAs (Pillai 2005; Sanchita et al. 2018; Tiwari et al. 2014) and siRNAs (Carthew and Sontheimer 2009) are very well known. These are involved in gene silencing at transcriptional and post-transcriptional levels through specific base pairing with their corresponding targets (mRNA). The lncRNAs regulate the expression of genes by various mechanisms. They interact with different biomolecules such as DNA, RNA and proteins through base pairing and interaction mechanisms. The understanding of the mechanism of action of lncRNAs is necessary for the rapid discovery and their biological utilization. Various processes are involved in the processing of lncRNAs in the cells to perform different functions (Rinn and Chang 2012). The known mechanism of actions has already been reviewed extensively (Wang and Chang 2011). Most of the lncRNAs have been shown to work as Enhances (cell-type specific expression and respond to diverse stimuli), Decoys (hinders the access of regulatory proteins to DNA or miRNA to mRNAs, Guides (localizes specific protein complexes) and Scaffolds (brings two or more proteins into discrete complexes).
Through these mechanisms, the lncRNAs are reported to perform various roles at different levels of biological functions in the plants like vernalization, protein re-localization, fertility, photomorphogenesis and phosphate homeostasis. Based on the consistent research and development in the area of regulatory lncRNAs in different organisms, it is believed that it will add more mechanistic themes in the future.
Functionally known biological roles of lncRNAs
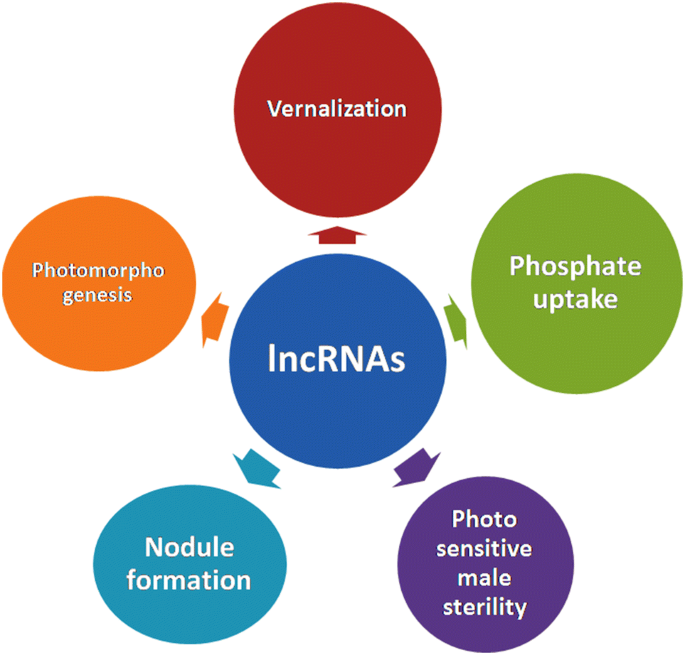
The functional roles of lncRNAs in human and animals have been studied extensively. In plants, they also play an important functional role in terms of plant’s growth and development. In plants, a limited number of experimentally identified lncRNAs are reported and these include Enod40, COOLAIR, COLDAIR, LDMAR, IPS1 and HID1 etc. These lncRNAs are reported to be involved in various processes including nodule formation, vernalization, photo-sensitive male sterility, phosphate uptake and photomorphogenesis (Fig. 1).
Vernalization
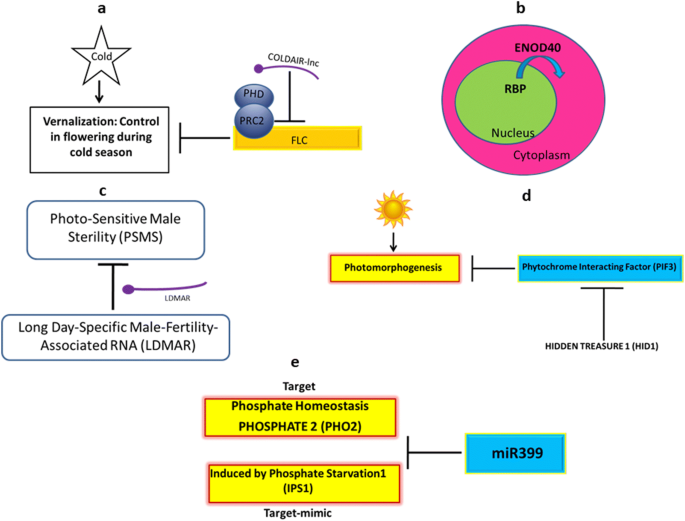
The vernalization or cold dormancy is the process of control in flowering time during winter/cold season. The Flowering Locus C (FLC) is the key repressor of flowering. During cold temperature, the expression of FLC is suppressed leading to the enhancement in flowering. Therefore, silencing of FLC gene is required to maintain the process of vernalization. Studies suggest that COOLAIR (Cold-induced antisense intragenic RNA) and COLDAIR (Cold-assisted intronic noncoding RNA) lncRNAs present at the same locus of FLC gene silence FLC via promoter interference and histone modification, respectively (Heo and Sung 2011; Zhang and Chen 2013). The process of regulation of vernalization by these lncRNAs has been described in Fig. 2a.
Nodule formation
Enod40 (early nodulin 40) was the first plant lncRNA identified in Glycine max and Medicago sativa (Crespi et al. 1994; Yang et al. 1993). It regulates the symbiosis between microbes and leguminous plants for nodule organogenesis. It also facilitates the protein relocalization or transportation of RNA binding protein (RBP) from nuclear speckles to cytoplasmic granules during nodulation (Fig. 2b). Although Enod40 does not contain long ORF, it encodes short peptides. However, the general mechanism of action is reported to be achieved through its RNA molecule rather than the peptide.
Photo-sensitive male sterility
Two varieties of japonica rice are reported to show photo-sensitive male sterility (PSMS). As a result the pollens become completely sterile under long day growth condition while viable in short day condition (Ding et al. 2012). The LDMAR (long day male fertility associated RNA) is a 1236 bp lncRNA that controls the PSMS and the plant remain fertile in long day condition. A high amount of LDMAR is reported to be required for maintaining male fertility under long day condition in Rice (Fig. 2c).
Photomorphogenesis
Photomorphogenesis is the effect of light on the growth and development of plants. The lncRNA named Hidden Treasure (HID1) is responsible for the negative regulation of Phytochrome Interacting factor (PIF3) protein and positive regulation of photomorphogenesis. PIF3 is defined as the central hub in the phytochrome-mediated light signaling pathways controlling seedling photomorphogenesis (Zhang et al. 2013b). PIF3 encoding basic helix-loop-helix transcription factor is a well-known key repressor of photomorphogenesis that modulates light response. HID1 negatively regulates the expression of PIF3 gene through binding at the promoter region of PIF3 gene (Wang et al. 2014). HID1 forms a large nuclear protein-RNA complex and associates with the chromatin of the first intron of PIF3 to repress transcription of PIF3 suggesting transcriptional control by lncRNAs. The regulation of photomorphogenesis by lncRNAs may give an interesting area of research (Fig. 2d).
Phosphate uptake
Phosphate is required for the proper growth and development of the plants. In plants, PHOSPHATE2 (PHO2) coding for ubiquitin-conjugating E2 enzyme is responsible for the phosphate uptake or homeostasis mechanism. One of the known miRNAs, mir399 is responsible for the suppression of PHO2 gene leading to a reduction in phosphate uptake. The lncRNA IPS1 (Induced by Phosphate Starvation1) is induced to block the suppression of miR399 and facilitate the mechanism of phosphate uptake. IPS1 acts as a target mimic for miR399 leading to negative regulation (Fig. 2e). In terms of the post-transcriptional control of lncRNAs, the regulation of phosphate uptake is one of the examples. Thus, leading to stop the cleavage effect of miRNA and leads to ultimate phosphate uptake. The regulatory role of IPS1 is reported in Arabidopsis thaliana (Franco-Zorrilla et al. 2007).
Plant lncRNAs identification: transcriptome analysis
The prediction and identification of lncRNAs are growing in an increasing manner. The species- as well as tissue-specific lncRNAs are also being identified. They have also been explored for their response in various biotic as well as abiotic stresses. In plants, most of the lncRNAs are identified through computational methods. The first computational prediction of lncRNA was reported in Medicago truncatula in 2007 using EST and genomic sequence data (Wen et al. 2007). The identified lncRNAs in different plants along with their numbers and type of sequence data has been listed in Table 1. In each plant, the steps of identification are almost the same as mentioned above. Till date, no software or tool is available which is specialized for the computational identification at a single platform. Therefore, a bioinformatics pipeline is required to be integrated with all the steps of analysis. The number of identified lncRNAs varied considerably ranging from 76 to 23,324 in Arabidopsis thaliana and Medicago truncatula, respectively. The lncRNAs were also found to have lower expression level and shorter lengths as compared to protein-coding genes (Li et al. 2015b; Song et al. 2016). The spatial expression of lncRNAs has been reported in Triticum aestivum, Digitalis purpurea and Salvia miltiorrhiza. No reports are available on the temporal expression of lncRNAs in plants. Since very less information of the experimental identification of lncRNAs in various organisms is available, the bioinformatics prediction plays an important role. Through a series of steps of bioinformatics pipeline, one can find the predicted lncRNAs from whole transcriptomic as well as genomic data. Although research is going on in the area of identification of lncRNAs, their functional characterization is lagging behind or comparatively slow. As previously pointed out, it is a challenge to understand how the molecular functions of lncRNAs affect the organisms (Wilusz et al. 2009). Numerous molecular functions of lncRNAs are reported but there is a lack of understanding of its mechanism of action for a specific molecular function.
Putative functions of lncRNAs
Biotic and biotic stresses
Abiotic stress is the adverse effect of environmental conditions on the organisms (Cramer et al. 2011). Various abiotic stresses including drought, salt, cold and heat stress effect the yield and crop productivity (Latha et al. 2019). In addition, biotic stress is an additional challenge to the plants against damages through pathogen or herbivore attack (Rejeb et al. 2014). The plants being sessile organism have developed mechanisms to withstand the extremes of these stresses. The mechanism of tolerance of plants is due to various regulatory elements responsible for the regulation of corresponding genes. LncRNAs being one of the important regulatory elements are also responsible for playing regulatory role against various environmental responses. The mechanism of action of lncRNAs under the influence of various stress conditions has been studied in plants. The stress responding lncRNAs reported from various plants are listed in Table 2. In Arabidopsis thaliana, the identified lncRNAs were found to be antisense to protein-coding mRNAs suggesting their cis-regulatory role and precursor for miRNA, siRNA and tasiRNAs. The accumulation of 28% of total identified lncRNAs in the root tissues was altered by phosphate starvation, salt and water stress. These lncRNAs have shown over and under expression due to the effect of stress conditions (Ben Amor et al. 2009). In another report, Arabidopsis thaliana was utilized for the identification of lncRNAs in response to heat, cold, salt, drought and high light stresses (Di et al. 2014). The differential expression studies of lncRNAs have resulted in the analysis of motifs responsible for the stress responses such as UUC motif responding to salt and AU-rich stem-loop responding to cold. The developmental regulation and stress response of lncRNAs have also been studied in Triticum aestivum, 125 putative and non-conserved lncRNAs have been identified. Some of them were small RNA (miRNA and siRNA) precursors. The tissue-dependent expression pattern study has resulted in their response to powdery mildew and/or heat stress indicating their role in abiotic and biotic stresses (Xin et al. 2011).
The regulatory roles of lncRNAs in drought stress condition has been studied in Setaria italica. It was shown that around 3% of the total identified lncRNAs responded to PEG-induced drought stress. These lncRNAs were also reported to be affecting neighbouring protein-coding gene expression in response to drought stress (Qi et al. 2013). The lncRNAs responding to osmotic and salt stresses have been studied in Medicago truncatula. The co-expression analysis of stress responding lncRNAs and protein-coding genes results in their involvement in regulating responses and adaptation to osmotic and salt stresses in leaves and root tissues of plants (Wang et al. 2015). In Zea mays, a total of 664 transcripts were identified as drought responsive lncRNAs in leaf tissues (Zhang et al. 2014b). Further validation of the response of these lncRNAs was performed in leaf and root tissues at different time intervals of drought stress through RT-qPCR. In Brassica rapa, 67 and 192 genes were found to be regulated by lncRNA in cold and heat stresses, respectively through co-expression network analysis among protein-coding genes and lncRNAs. Several common genes were also identified to be targeted by cold as well as heat responding lncRNAs indicating their crosstalk with these stresses (Song et al. 2016).
A total of 504 lncRNAs were found to be drought responsive, eight of which were confirmed by RT-qPCR in Populus trichocarpa (Shuai et al. 2014). The role of lncRNAs has also been studied in some medicinally important plants such as Digitalis purpurea and Salvia miltiorrhiza. D. pupurea is a rich source of cardiac glycosides helpful in heart failure treatment (Warren 2005). The biosynthetic pathways of cardiac glycosides comprise of terpenoid backbone biosynthesis, steroid biosynthesis and cardenolide biosynthesis. The expression analysis of identified lncRNAs was performed in cold and dehydration stresses to study their responses in different time periods of these stresses. Among 27 lncRNAs considered for the study, 24 and 27 have shown more than twofold changes between at least two-time points of cold and dehydration treatments, respectively (Wu et al. 2012a). Some lncRNAs have shown a similar response to both the stresses results in the fact that cold and dehydration signaling networks are probably overlapped.
Expression of lncRNAs has been shown to be modulated by different elicitors. To study the response of methyl jasmonate, 17 lncRNAs were analysed in leaf tissues of S. miltiorrhiza through qRT-PCR. The expression of 15, out of 17 lncRNAs, was significantly altered in at least one-time point of methyl jasmonate treatment. Majority of lncRNAs resulted in the down-regulation in different time points of treatment (Li et al. 2015a). The drought stress-responsive lncRNAs have also been reported in Musa acuminata leaf tissues (Muthusamy et al. 2015). All the stress-responsive lncRNAs exhibit distinct expression patterns in drought tolerant and susceptible cultivars. In another report on Musa acuminata, lncRNAs were predicted from the root tissues after infection of Fusarium oxysporum.
Overall, 5294 novel lncRNAs were identified and reported to be involved in plant-pathogen interactions, biosynthesis and transduction of auxin, ethylene, salicylic acid (SA) and jasmonic acid (JA), and the regulation of pathogenesis-related (PR) genes (Li et al. 2017). Another example of biotic stress-responsive lncRNAs is reported in Brassica napus infected with Sclerotinia sclerotiorum causing stem rot (Joshi et al. 2016). The salt stress responding lncRNAs were identified in Gossypium hirsutum. Out of 1117 predicted lncRNAs, 44 have shown differential expression in salt stress condition (Deng et al. 2018). These reports extend the understanding of current view on the role of lncRNAs as universal ribo-regulators in response to various stresses. The lncRNAs are reported to be expressed in roots, stems, leaves and flowers, independently. The tissue-specific expression of lncRNAs suggests that the growth and development of every tissue is being regulated by specific lncRNA.
Secondary plant product biosynthesis
In addition to the regulatory effect on stress responses, the lncRNAs have also been reported to play a significant role in secondary metabolism of medicinally important plants. It is very well known that secondary metabolites produced from medicinal plants are pharmacologically active constituents. These compounds are of different groups such as alkaloids, terpenoids and phenylpropanoids with corresponding specific functional properties. In Digitalis purpurea, some of the identified lncRNAs have shown similarity with protein-coding genes, 4-Hydroxy-3-methyl-but-2-en-1-yl diphosphate synthase, Solanesyl diphosphate synthase, Dihydroflavonal-4-reductase, Phytoene dehydrogenase and Aromatic amino acid decarboxylase involved in the biosynthesis of terpenoids or cardiac glycosides (Wu et al. 2012a). The identified similarity may signify the regulatory effect of these lncRNAs on cardiac glycosides leading to enhanced production as well as growth and development of plants. The interaction mechanism between lncRNAs and corresponding protein coding genes involved in secondary metabolism might give a clear vision on the regulation pattern of lncRNAs. Although Panax ginseng and Salvia miltiorrhiza are also important medicinal plants and lncRNAs have been identified, no information is available on the role of identified lncRNAs in secondary metabolic pathways. There is no report available regarding the mechanism of action of any lncRNAs involved in the regulation of expression of genes involved in secondary metabolite production. Besides, participating in secondary metabolism, the lncRNAs have also been reported for their significant role in fruit ripening processes.
Fruit ripening processes
The role of lncRNAs in fruit ripening is analyzed in two plants such as Actinidia chinensis (Kiwi fruit) and Solanum lycopersicum (Tang et al. 2016; Zhu et al. 2015). The combination of sugars, organic acids and free amino acids contribute to kiwi fruit flavor. The dietary antioxidants of kiwifruit are represented by carotenoids, chlorophyll and anthocyanins. The lncRNAs have been studied for their involvement in fruit development and ripening in A. chinensis (Tang et al. 2016). Study utilized genomic data from the nine different RNA-Seq libraries and identified 7051 lncRNAs that were not annotated in the draft genome This study led to the understanding of involvement of lncRNAs in fruit development and ripening process in kiwifruits. One more example explaining the role of lncRNAs in fruit ripening have been reported in Solanum lycopersicum. The identified lncRNAs have shown significant up- and down-regulation indicating their involvement in the regulation of fruit ripening. Further, silencing of two lncRNAs has resulted in delayed ripening of tomato (Zhu et al. 2015).
Target mimicry for miRNAs
The miRNAs are endogenous regulatory small RNAs playing a significant role in plant growth, development and stress responses (Sharma et al. 2015, 2016; Chen et al. 2018). The mature miRNAs function through the degradation of their target genes through cleavage or translational inhibition. The miRNAs function through the negative regulation of corresponding genes. The lncRNAs act as target mimic to hinder the complementarity of miRNAs with its target gene and inhibits the negative regulatory effect of miRNAs. In the process of target mimicry, the lncRNAs serve as decoy/sponges for corresponding miRNAs. The target mimicry phenomenon was first discovered in 2007 with the identification of IPS1 as an endogenous target mimic for miR399 in Arabidopsis thaliana (Franco-Zorrilla et al. 2007). The PHO2 gene encodes ubiquitin-conjugating E2 enzyme and plays a significant role in phosphate starvation in plants. MiR399 binds through sequence complementarity with PHO2 genes and suppresses its expression. During phosphate deprivation, the lncRNA IPS1 are induced to mimic the PHO2 for miR399 binding. The specific regions binding with miR399 are reported to be conserved among 13 plant families (Rymarquis et al. 2008). miRNAs are reported to perfectly pair at their 9th to 11th positions with corresponding targets resulting in the effective cleavage of targets (Zheng et al. 2012). The 24 nucleotide region of IPS1 binds with miR399 with a bulge between 10 and 11 base position of miRNA resulting into cleavage of IPS1. The expression of PHO2 increases, in turn. By keeping the view of target mimic, many studies are being done in the area of artificial target mimicry. The predicted endogenous target mimics have also been identified for miR160 and miR166 in Arabidopsis thaliana and Oryza sativa (Wu et al. 2013). The overexpression of these eTMs causes the increased expression of corresponding miRNA targets. It has also been found that the target mimic regions of these lncRNAs complementary to miRNAs were highly conserved whereas the sequences outside this region are highly diverse. The artificial target mimics have also been designed against miR156 and miR319 through mutation of IPS1. These mutated IPS1 have resulted in an increased level of corresponding target genes (Rymarquis et al. 2008). A database on plant endogenous target mimics (PeTMbase) has also been reported (Karakulah et al. 2016). For the formation of this database, the plant miRNAs from miRBase (Kozomara and Griffiths-Jones 2014), a well-known database of miRNAs was considered. The lncRNAs were utilized through either computational identification from transcriptome data or lncRNA databases such as GREENC (Paytuvi Gallart et al. 2016) and PNRD (Yi et al. 2015). In present scenario, the target mimic technology is being utilized for the genetic engineering of specific gene or a group of genes that leads to the crop improvement.
Computational prediction of lncRNAs in plants
The computational or in silico prediction methods reduce the time complexity of the experimentation processes. These methods, have been utilized for the predictive analysis of lncRNAs in several plants such as Zea mays, Arabidopsis thaliana, Triticum aestivum etc. In the computational prediction, involvement of several steps leads to identification of the lncRNAs are involved. In Step 1, the prediction starts with filtering of sequences based on its length and sequences > 200 bp are retained for the further analysis. Step 2 analysis is carried out to ascertain that ncRNA does not code for protein or does not contain a long ORF. If ORF is identified, the length of the coded sequence should be < 100 aa. In Step 3, the transcripts identified by Step 2 are used for the homology search with the SWISS-PROT database (e value < 0.001).
Databases and web servers of plant lncRNAs
The lncRNA-related study has been expanded to plant species and the number of identified plant lncRNAs has dramatically increased in the past years. So there is a need to collect the identified lncRNAs in a user-friendly environment. There are many databases which have been developed with the collection of plant lncRNAs (Table 3). The first database consisting of plant lncRNAs was developed in 2011 (Amaral et al. 2011) and updated in 2015 (Quek et al. 2015). The lncRNAdb is a database of lncRNAs of different plants such as Arabidopsis thaliana, Brassica rapa, Vitis vinifera, Solanum lycopersicum, Glycine max, Oryza sativa, Populus tremula, Medicago truncatula, M. sativa. LncRNAdb includes a variety of annotations for eukaryotic lncRNAs, including gene expression data, evolutionary conservation, structural information, genomic context, subcellular localization, functional evidence, links to the primary literature and the transcript sequence. PLncDB (Jin et al. 2013) contains 16227 lncRNAs identified from intergenic regions of Arabidopsis thaliana genome. This database provides a collection and integration of lncRNAs from different data resources such as TAIR, lncRNAdb, NONCODE etc., expression levels in various samples including different tissues, developmental stages, mutants and stress treatments, the epigenetic modifications (DNA methylations and histone modifications) and a collection of siRNA sequencing dataset across the whole genome. PNRD (Yi et al. 2015) is a database of all kind of plant noncoding RNAs such as miRNAs, snoRNAs, snRNA, tasiRNA, tRNA including lncRNAs. The lncRNAs have been reported from 20 plant species. PLNlncRbase (Xuan et al. 2015) is a detailed repository of experimentally identified plant lncRNAs. This database consists of 1187 lncRNAs from 43 plant species collected from nearly 200 published articles. Through this database, the potential biological roles, sequences, expression patterns and the probable target genes of lncRNAs could be identified. The NONCODE 2016 (Zhao et al. 2016) consists lncRNAs of different organisms including one plant, Arabidopsis thaliana. Around 3853 lncRNAs have been reported in Arabidopsis thaliana. The earlier versions of NONCODE v3 (Bu et al. 2012) and v4 (Xie et al. 2014) have been developed consisting of lncRNAs of human and mouse only. GREENC (Paytuvi Gallart et al. 2016) contains more than 120000 annotated lncRNAs from 37 plant species. This database provides information on the sequence, genome position, coding potential and folding energies of lncRNAs. This database includes computationally identified lncRNAs through an inbuilt pipeline. CANTATAdb (Szczesniak et al. 2016) is also the collection of computationally identified lncRNAs from 10 model plants.
Various bioinformatics pipelines and web servers are also available for the identification of lncRNAs (Table 4). Annocript is reported for the computational identification as well as annotation of lncRNAs from transcriptome data (Musacchia et al. 2015). One more interactive platform, lncRNA-screen is also available for the computational screening of lncRNAs in genomics data. This server is currently available on biRxiv, the preprint server for biology (Gong et al. 2017). An integrated bioinformatics pipeline, Sebnif is also available for the identification of high-quality single-exonic lincRNAs (Sun et al. 2014). UClncR is a pipeline for lncRNA detection through RNA-seq alignment file. In the processing, it performs transcript assembly, predicts and annotates both known and novel lncRNAs (Sanchita et al. 2018; Tiwari et al. 2014).
Conclusions and future perspectives
The functions of small RNAs and proteins can be identified based on their sequences and structures. However, besides having long sequence and well-identified stem and loop structures, the lncRNAs could not be inferred for their functions. Despite being the largest group of ncRNA family, the lncRNAs are least characterized in terms of regulatory function and the question remains unclear how a particular lncRNA function in the cell at the molecular level. Although, it is well known that lncRNAs play a significant role in stress responses, secondary metabolism and fruit ripening in plants, the detailed studied to establish their mode of action in regulating these processes are still required. Therefore, there is a requirement to take up these continuously increasing predicted lncRNAs for their functional categorization. This is one of the challenges for not achieving the functional annotation of identified lncRNAs. By keeping in mind the regulatory importance of lncRNAs in the secondary metabolism of medicinal plants and ripening of fruits, more research is required in the present scenario.
References
Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS (2011) lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39:D146–151
Bard J (2014) Generating anatomical variation through mutations in networks—implications for evolution. J Anat 225:123–131
Ben Amor B, Wirth S, Merchan F, Laporte P, d’Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, Vaucheret H, Thermes C, Crespi M (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19:57–69
Boerner S, McGinnis KM (2012) Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE 7:e43047
Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, Xiao H, Liao Q, Luo H, Zhao G, Zhao H, Liu Z, Liu C, Chen R, Zhao Y (2012) NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res 40:D210–215
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Chen J, Quan M, Zhang D (2015) Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 241:125–143
Chen X, Liu Z, Shi G, Bai Q, Guo C, Xiao K (2018) MIR167a transcriptionally regulates ARF6 and ARF8 and mediates drastically plant Pi-starvation response via modulation of various biological processes. Plant Cell Tiss Org 133:177–191
Costa FF (2010) Non-coding RNAs: meet the masters. BioEssays 32:599–608
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Crespi MD, Jurkevitch E, Poiret M, d’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13:5099–5112
Deng F, Zhang X, Wang W, Yuan R, Shen F (2018) Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol 18:23
Di C, Yuan J, Wu Y, Li J, Lin H, Hu L, Zhang T, Qi Y, Gerstein MB, Guo Y, Lu ZJ (2014) Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J 80:848–861
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci 109:2654–2659
Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, Zhao L, Li X, Teng X, Sun X, Sun L, Zhang MQ, Chen R, Zhao Y (2018) NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res 46:D308–D314
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Gong Y, Huang HT, Liang Y, Trimarchi T, Aifantis I, Tsirigos A (2017) lncRNA-screen: an interactive platform for computationally screening long non-coding RNAs in large genomics datasets. BMC Genomics 18:434
Hao Z, Fan C, Cheng T, Su Y, Wei Q, Li G (2015) Genome-wide identification, characterization and evolutionary analysis of long intergenic noncoding RNAs in cucumber. PLoS ONE 10:e0121800
Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331:76–79
Jin J, Liu J, Wang H, Wong L, Chua NH (2013) PLncDB: plant long non-coding RNA database. Bioinformatics 29:1068–1071
Joshi RK, Megha S, Basu U, Rahman MH, Kav NN (2016) Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS ONE 11:e0158784
Karakulah G, Yucebilgili Kurtoglu K, Unver T (2016) PeTMbase: a database of plant endogenous target mimics (eTMs). PLoS ONE 11:e0167698
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Latha GM, Raman KV, Lima JM, Pattanayak D, Singh AK, Chinnusamy V, Bansal KC, Sambasiva Rao KRS, Mohapatra T (2019) Genetic engineering of indica rice with AtDREB1A gene for enhanced abiotic stress tolerance. Plant Cell Tiss Org 136:173–188
Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, Evans MM, Scanlon MJ, Yu J, Schnable PS, Timmermans MC, Springer NM, Muehlbauer GJ (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15:R40
Li D, Shao F, Lu S (2015a) Identification and characterization of mRNA-like noncoding RNAs in Salvia miltiorrhiza. Planta 241:1131–1143
Li ZF, Zhang YC, Chen YQ (2015b) miRNAs and lncRNAs in reproductive development. Plant Sci 238:46–52
Li WB, Li CQ, Li SX, Peng M (2017) Long noncoding RNAs that respond to Fusarium oxysporum infection in ‘Cavendish’ banana (Musa acuminata). Sci Rep 7:16939
Lv Y, Liang Z, Ge M, Qi W, Zhang T, Lin F, Peng Z, Zhao H (2016) Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genomics 17:350
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10:925–933
Musacchia F, Basu S, Petrosino G, Salvemini M, Sanges R (2015) Annocript: a flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics 31:2199–2201
Muthusamy M, Uma S, Backiyarani S, Saraswathi MS (2015) Genome-wide screening for novel, drought stress-responsive long non-coding RNAs in drought-stressed leaf transcriptome of drought-tolerant and -susceptible banana (Musa spp) cultivars using Illumina high-throughput sequencing. Plant Biotechnol Rep 9:279–286
Paytuvi Gallart A, Hermoso Pulido A, Martinez Anzar, de Lagran I, Sanseverino W, Aiese Cigliano R (2016) GREENC: a Wiki-based database of plant lncRNAs. Nucleic Acids Res 44:D1161–1166
Pertea M (2012) The human transcriptome: an unfinished story. Genes 3:344–360
Pillai RS (2005) MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11:1753–1761
Ponjavic J, Ponting CP, Lunter G (2007) Functionality or transcriptional noise? evidence for selection within long noncoding RNAs. Genome Res 17:556–565
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641
Qi X, Xie S, Liu Y, Yi F, Yu J (2013) Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol Biol 83:459–473
Quan M, Chen J, Zhang D (2015) Exploring the secrets of long noncoding RNAs. Int J Mol Sci 16:5467–5496
Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME (2015) lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43:D168–173
Rejeb IB, Pastor V, Mauch-Mani B (2014) Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants 3:458–475
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166
Rymarquis LA, Kastenmayer JP, Huttenhofer AG, Green PJ (2008) Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci 13:329–334
Sanchita, Trivedi R, Asif MH, Trivedi PK (2018) Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation. RNA Biol 15:1433–1439
Sharma D, Tiwari M, Lakhwani D, Tripathi RD, Trivedi PK (2015) Differential expression of microRNAs by arsenate and arsenite stress in natural accessions of rice. Metallomics 7:174–187
Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK (2016) MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol 171:944–959
Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, Xia X, Yin W (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot 65:4975–4983
Song X, Liu G, Huang Z, Duan W, Tan H, Li Y, Hou X (2016) Temperature expression patterns of genes and their coexpression with LncRNAs revealed by RNA-Seq in non-heading chinese cabbage. BMC Genomics 17:297
Sun K, Zhao Y, Wang H, Sun H (2014) Sebnif: an integrated bioinformatics pipeline for the identification of novel large intergenic noncoding RNAs (lincRNAs)–application in human skeletal muscle cells. PLoS ONE 9:e84500
Sun Z, Nair A, Chen X, Prodduturi N, Wang J, Kocher JP (2017) UClncR: Ultrafast and comprehensive long non-coding RNA detection from RNA-seq. Sci Rep 7:14196
Szczesniak MW, Rosikiewicz W, Makalowska I (2016) CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol 57:e8
Tang W, Zheng Y, Dong J, Yu J, Yue J, Liu F, Guo X, Huang S, Wisniewski M, Sun J, Niu X, Ding J, Liu J, Fei Z, Liu Y (2016) Comprehensive transcriptome profiling reveals long noncoding RNA expression and alternative splicing regulation during fruit development and ripening in Kiwifruit (Actinidia chinensis). Front Plant Sci 7:335
Tiwari M, Sharma D, Trivedi PK (2014) Artificial microRNA mediated gene silencing in plants: progress and perspectives. Plant Mol Biol 86:1–18
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914
Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci U S A 111:10359–10364
Wang TZ, Liu M, Zhao MG, Chen R, Zhang WH (2015) Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol 15:1–13
Warren B (2005) Digitalis purpurea. Am J Cardiol 95:544
Wen J, Parker BJ, Weiller GF (2007) In silico identification and characterization of mRNA-like noncoding transcripts in Medicago truncatula. Silico Biol 7:485–505
Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23:1494–1504
Wu B, Li Y, Yan H, Ma Y, Luo H, Yuan L, Chen S, Lu S (2012a) Comprehensive transcriptome analysis reveals novel genes involved in cardiac glycoside biosynthesis and mlncRNAs associated with secondary metabolism and stress response in Digitalis purpurea. BMC Genomics 13:15
Wu J, Okada T, Fukushima T, Tsudzuki T, Sugiura M, Yukawa Y (2012b) A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biol 9:302–313
Wu HJ, Wang ZM, Wang M, Wang XJ (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 162:1875–1884
Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y (2014) NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res 42:D98–103
Xin M, Wang Y, Yao Y, Song N, Hu Z, Qin D, Xie C, Peng H, Ni Z, Sun Q (2011) Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol 11:1
Xuan H, Zhang L, Liu X, Han G, Li J, Li X, Liu A, Liao M, Zhang S (2015) PLNlncRbase: a resource for experimentally identified lncRNAs in plants. Gene 573:328–332
Yang WC, Katinakis P, Hendriks P, Smolders A, de Vries F, Spee J, van Kammen A, Bisseling T, Franssen H (1993) Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J 3:573–585
Yi X, Zhang Z, Ling Y, Xu W, Su Z (2015) PNRD: a plant non-coding RNA database. Nucleic Acids Res 43:D982–989
Zhang YC, Chen YQ (2013) Long noncoding RNAs: new regulators in plant development. Biochem Biophys Res Commun 436:111–114
Zhang J, Mujahid H, Hou Y, Nallamilli BR, Peng Z (2013a) Plant long ncRNAs: a new frontier for gene regulatory control. Am J Plant Sci 4:1038–1045
Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, QuaIL PH (2013b) A Quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9:e1003244
Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W (2014a) The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 547:1–9
Zhang W, Han Z, Guo Q, Liu Y, Zheng Y, Wu F, Jin W (2014b) Identification of maize long non-coding RNAs responsive to drought stress. PLoS ONE 9:e98958
Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, Li QF, Qu LH, Shu WS, Chen YQ (2014c) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15:512
Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, Chen R (2016) NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44:D203–208
Zheng Y, Li YF, Sunkar R, Zhang W (2012) SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Res 40:e28
Zhu B, Yang Y, Li R, Fu D, Wen L, Luo Y, Zhu H (2015) RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J Exp Bot 66:4483–4495
Acknowledgements
The financial grant given by SERB, DST, New Delhi in the form of National-Post Doctoral Fellowship to Sanchita is highly acknowledged. Authors acknowledge Council of Scientific and Industrial Research (CSIR), New Delhi, in the form of NCP project (MLP026). P.K.T. also acknowledges Department of Biotechnology, New Delhi for the financial support in form of TATA Innovation Fellowship (GAP3445).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Sergio J. Ochatt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sanchita, Trivedi, P.K. & Asif, M.H. Updates on plant long non-coding RNAs (lncRNAs): the regulatory components. Plant Cell Tiss Organ Cult 140, 259–269 (2020). https://doi.org/10.1007/s11240-019-01726-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01726-z