Abstract
Carica papaya L. plantlets, normally exhibit low rooting capacity when cultured in vitro. It has been suggested in other species that auxin concentration at root tissues, is the result of a reflux system driven by auxin influx transporters (AIT; AUX1/LAX) and auxin efflux transporters (AET; PIN), that regulates the mechanism of initiation and development of lateral roots. Therefore, in the present paper, we studied the structure, phylogeny and the expression patterns of the whole family of AIT and AET in C. papaya, and their possible relation with the limited capacity to generate adventitious roots of in vitro cultured papaya plantlets. We found 4 AUX1/LAX genes (CpAUX1, CpLAX1, CpLAX2, CpLAX3) and 6 PIN genes (CpPIN1, CpPIN2, CpPIN3, CpPIN4, CpPIN5, CpPIN6) within the genome of C. papaya. The expression patterns and levels of those genes were studied in stem-base and root tissues from C. papaya cv. Maradol plants under four different treatments: (1) in vitro plantlets without IBA (that did not generate roots), (2) in vitro plantlets treated with 2 mg L−1 IBA (that did generate roots), (3) de-rooted seedlings treated with the same concentration of IBA (that also generated adventitious roots), and (4) intact seedlings used as controls. Histological studies made on the stem base and root tissues from all treatments showed that the IBA-induced roots were histologically equivalent, to those naturally formed in intact seedlings. In vitro plantlets non-treated with IBA had low expression of all auxin transporters genes in stem-base tissues and they were unable to produce roots. On the contrary, in vitro plantlets treated with IBA experienced a marked increase in the expression of most auxin transporters genes, in particular of CpLAX3 and CpPIN2, and they were capable to produce roots. Those roots generated in the IBA-treated in vitro plantlets, showed expression levels and patterns of auxin transporter genes, equivalent to those shown in both, the IBA-treated de-rooted seedlings, and in the naturally formed roots from intact seedlings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most common form of propagating Carica papaya is by seeds (Jiménez et al. 2014), however it can also be micropropagated by in vitro tissue culture techniques, with the advantage of obtaining genetically-homogeneous in vitro plantlets (Talavera et al. 2007). Although there are reports of micropropagation of C. papaya (Drew and Miller 1993; Islam et al. 1993; Magdalita et al. 1997; Yu et al. 2000), the in vitro rooting and ex vitro acclimatization, are fundamental to the establishment of an efficient and low cost large scale micropropagation protocol. Limited rooting in papaya micropropagated plantlets might result in high (70 %) plant mortality during the ex vitro stage, before they can be planted in the field (Malabadi et al. 2011). Therefore, high rooting and efficient acclimatization represent a problem in commercial micropropagation of C. papaya cultivars, in particular in the cv. Maradol.
The phytohormone auxin, which is synthesized mainly in the shoots and translocated to the sites of action, plays critical roles in regulating plant development and growth (Quint and Gray 2006; Teale et al. 2008; Hoffmann et al. 2011; Chen et al. 2014; Shen et al. 2015). In the plant, auxins may be distributed in two interconnected transport systems: first, a fast nondirectional stream in the phloem along with photosynthetic assimilates, and a second one, that is slow and directional cell-to-cell polar auxin transport (PAT). In this way , PAT distributes auxin in a precise manner, that is critically important for the formation of local auxin maxima, mainly in developing tissues (Adamowski and Friml 2015). Coordinated action of auxin biosynthesis, perception, signaling and PAT are required to form normal lateral roots (Lavenus et al. 2013). In Arabidopsis thaliana the expression of the auxin-responsive reporter gene DR5, decreased after plants were exposed to the auxin efflux inhibitor 1-N-naphthylphthalamic acid, indicating that these auxin responses depend on PAT (Himanen et al. 2002; De Smet et al. 2007; Moreno-Risueno et al. 2010). Ivanchenko et al. (2015), showed that the tomato diageotropica mutant (dgt; deficient of PAT), is not capable to form lateral roots, demonstrating that normal PAT fluxes along the root tip are required for lateral root formation. In roots, the natural auxin indolacetic acid (IAA) moves acropetally (from apical regions to the root apex) and basipetally (from the root apex towards apical zones) by outer layers of the root, and PAT is carried out by the auxin influx (AIT) and auxin efflux (AET) transporter proteins (Muday and DeLong 2001). A number of AIT, such as AUX1/LAX (AUX1-like), and AET, such as PINFORMED (PIN) genes, have been isolated and characterized in A. thaliana (Vanneste and Friml 2009; Ugartechea-Chirino et al. 2010).
Within the A. thaliana genome, four genes encode for AIT (AUX1, LAX1, LAX2 and LAX3). The size of those genes are: AtAUX1, 4188 pb; AtLAX1, 3942 pb; AtLAX2, 2547 pb; AtLAX3, 2400 pb, while their exons number are: 9, 9, 6 and 7, respectively (NCBI, http://www.ncbi.nlm.nih.gov). The cellular entry of auxins is performed through AUX1 and its homologues LAX1, LAX2 and LAX3 which encode highly conserved transmembrane proteins with permease activity. AIT proteins (AUX1/LAX) family readily transport auxin across cell membranes (Ugartechea-Chirino et al. 2010) and they are involved in many post-embryonic developmental processes such as: gravitropism, phyllotaxis and lateral root formation (Parry et al. 2001b; Marchant et al. 2002; Reinhardt et al. 2003; Swarup et al. 2005; Dubrovsky et al. 2006; Bainbridge et al. 2008; Laskowski et al. 2008; Swarup et al. 2008). AUX1/LAX proteins, range between 487 and 553 aminoacids (AA) and they show a highly conserved central region with 10 transmembrane helices. LAX proteins are rich in acidic AA in its N-terminal and they are rich in proline at its C-terminal (Shen et al. 2010). In addition, in A. thaliana, eight genes of the PINFORMED family (PIN 1 to PIN 8; Gälweiler et al. 1998; Müller et al. 1998), are involved in the facilitation of AET, which is an essential step in PAT (Sawchuk and Scarpella 2013). These proteins were named after the creation of the first mutant pin1, that develops pin-shaped inflorescences and they are defective in PAT and showed loss of function in the PIN1 gene. The size of those genes are: AtPIN1, 3506 bp; AtPIN2, 4043 bp; AtPIN3, 3342 bp; AtPIN4, 3224 bp; AtPIN5, 1892 bp; AtPIN6, 3802 bp; AtPIN7, 2294 bp; AtPIN8, 1779 bp, while their exons numbers are: 6, 9, 6, 6, 5, 7, 6, 6, respectively (NCBI, http://www.ncbi.nlm.nih.gov).
In the present work, we performed an in silico characterization of the whole families of AUX1/LAX and PIN in the C. papaya genome, and primers were designed for those genes. Experiments were then designed to evaluate different auxin-treatments that allowed efficient adventitious root formation on in vitro plantlets of C. papaya cv. Maradol versus treatments that did not promote adventitious root formation. The expression patterns of CpAUX1/LAX and CpPIN genes were then analyzed in plantlets under those contrasting treatments, in order to determine if a correlation exists between changes in the expression patterns of CpAUX1/LAX and CpPIN genes, with the capacity of auxin-treated in vitro plantlets of C. papaya, to form adventitious roots.
Materials and methods
Plant material
Seedlings
Fifty C. papaya cv. Maradol seeds were germinated and 2 days after germination, stem base (SB) and root (R) tissues from those seedlings were collected. All seedlings were placed in the greenhouse in nursery plastic trays containing a peat-moss:agrolite (2:1) substrate. Seedlings were watered with 10 mL of sterile distilled water every 48 h, with the addition of 1 g L−1 Benlate fungicide. The greenhouse conditions were: temperature (T) of 35 ± 1 °C, relative humidity (RH) of 70 % and PPFD of 750 µmol m−2s−1.
In vitro plantlets
Four hundred and twenty in vitro shoots originally obtained from 1 year-old plants of C. papaya cv. Maradol plants, were cultured in vitro inside culture flasks (with three plants each), containing sterilized peat-moss:agrolite (2:1) substrate, 2.21 g L−1 MS salts (Murashige and Skoog 1962), 10 g L−1 sucrose, but with 14 different auxin treatments (IBA, NAA or IBA + NAA) in experiment 1 (T1–T14; Table 1), or with (2 g L−1) and without IBA in experiment 2 (D1–D4; Table 2). Shoots from all treatments were cultured in growth rooms (T = 25 °C, PPFD = 300 µmol m−2s−1 and 12 h photoperiod). At the end of the 21 days exposure to each treatment, plant height, leaves number, root number, root length and rooting percentage were recorded. Then, the plantlets from each treatment were transferred to plastic trays containing peat-moss:agrolite (2:1) as substrate and 1 g L−1 of fungicide (Benlate) and covered with a dome (to maintain a high relative humidity), under greenhouse conditions (T = 35 ± 1 °C, RH = 70 % and PPFD = 750 µmol m−2s−1). After growing in the greenhouse for further 42 days, plant survival percentage was evaluated for each treatment.
IBA treatment on de-rooted seedlings of C. papaya
Fifteen seeds C. papaya cv Maradol were germinated and the resulting intact seedlings were used as controls for anatomical studies and plant height, leaves number, root number, root length and rooting percentage were recorded at day 0 (C1) and 21 days after germination (C2). Another fifteen seedlings were germinated and 2 days after germination, the resulting seedlings were de-rooted (i.e. roots were cut off at the stem-base). The cut end stems of the de-rooted seedlings were rinsed with 10 mL of sterile distilled containing 1 g L−1 fungicide (Benlate) and 2 mg L−1 IBA was added as adventitious root inducer. Those seedlings were measured 10 days after roots were removed and new adventitious roots were formed (day 0, C3), and 21 days after (C4) (Table 2). Seedlings from the four treatments (C1–C4) were then transferred to plastic trays containing peat-moss:agrolite (2:1) as substrate and 1 g L−1 of fungicide (Benlate), under greenhouse conditions (T = 35 ± 1 °C, RH = 70 % and PPFD = 750 µmol m−2s−1), where the de-rooted seedlings took 10 days to generate new adventitious roots (considered as day 0; C3). After growing for further 21 days under those conditions, previously de-rooted seedlings, now bearing new adventitious roots (C4), were sampled for growth, anatomical studies and gene expression analysis.
In silico cloning
The A. thaliana AUX1/LAX and PIN nucleotide sequences were obtained from Phytozome database (http://phytozome.net), and their corresponding homologous sequences were identified within the genome of C. papaya cv. Sun Up. Sorting and identification of those sequences with higher percentage of identity were made with Blast Parser v1.2 program (http://geneproject.altervisa.org). The sequences were selected with the following parameters: coding sequences with E values <10−14, sequences with percentages of identity and similarity greater than 60 % within the genome of C. papaya cv. Sun Up. The selected genomic sequences were translated in all six possible reading frames; using the algorithms software of gene prediction FGENESH program (http://linux1.softberry.com/all.htm) and the genetic code of dicotyledonous plants (A. thaliana). These predicted protein sequences were edited with the TRANSLATE program with the output format that includes the protein sequence. Using ortholog genes from C. papaya cv. Sun Up, specific oligonucleotides were designed for the qRT-PCR expression analysis of CpAUX1/LAX and CpPIN genes in C. papaya cv. Maradol. The oligonucleotides were designed with Primer Premier ver. 6. Program (http://www.premierbiosoft.com/primerdesign/).
Protein alignment and phylogenetic analysis
The multiple alignment and phylogenetic trees were constructed using the sequences AtAUX1/LAX and AtPIN proteins from A. thaliana and orthologs CpAUX1/LAX and CpPIN proteins from C. papaya cv. Sun Up. The phylogeny was obtained by Neighbor-Joining distance based method with 1000 bootstrap replications, and the calculated evolutionary distances were obtained from the Poisson correction evolutionary model, gaps and missing data were treated with pairwise deletion. Edition of the alignment was performed with BioEdit program (Hall 1999). Both trees were constructed with MEGA 5 program (Tamura et al. 2011). AIT and AET families members were aligned with ClustalX 1.81 (Blosum Weight Matrix; Gap Opening Penalty: 5; Gap Extension Penalty: 0.20; Thompson et al. 1997).
RNA isolation and qRT-PCR
Total RNA was extracted from SB and R tissues from seedlings (intact or de-rooted) and from in vitro plantlets, exposed or not to 2 mg L−1 of IBA, using the CTAB protocol from Idrovo et al. (2012). The concentration and purity of RNA samples were determined with a NanoDrop™ 1000 Spectrophotometer (Thermo Scientific NanoDrop Technologies, LLC, Wilmington, DE, USA). For first-strand cDNA synthesis, total RNA (5 mg) and 200 U of Superscript III reverse transcriptase were used, following the manufacturer’s protocol (Invitrogen/Life Technologies, CA, USA). qRT-PCR was performed in a thermocycler STEP ONE SYSTEM and StepOne Software v2.3 (Applied Biosystems, Foster City, CA, USA). Expression levels were calculated relative to the expression level of C. papaya elongation factor 1-α (CpEF1α) using the ΔΔCt method (Applied Biosystems). The specificity of the reactions was confirmed by the standard melt curve method. Each assay was repeated at least three times. Statistical analysis was performed using STATGRAPHICS Plus (http://www.statgraphics.com). One-way analysis of variance (ANOVA) was used to evaluate the relative expression levels of CpAUX1/LAX and CpPIN genes found in SB and R tissues from plants from the different treatments.
Histological analysis
Fresh samples from SB and R tissues from seedlings or in vitro plantlets of C. papaya cv Maradol, were collected at day 21 of the rooting stage. Subsequently, cross-sections of SB at a distance of 0.2 cm from the roots, and from R at a distance of 0.2 cm from the stem-base were used. All samples from SB and R from seedlings, de-rooted seedlings, and in vitro plantlets were treated according to the protocol of Berlin and Miksche (1976). Tissues were infiltrated and embedded in plastic resin (JB-4 Glycol Methacrylate, Polysciences, Los Angeles, CA, USA). Transverse sections 3 µm thick were cut using a microtome (Microm HM 325; Thermos Scientific, Walldorf, Germany). Sections were stained with Schiff reagent as described by McManus (1961) and 7 % (w/v) Naphthol blue black and mounted in Poly-mount (Polysciences Inc., Warrington, PA, USA). Series of sections from SB and R tissues were then observed and photographed, using a microscope (Axio scope A1, Carl Zeiss, Germany).
Statistical analysis
All in vitro treatments were analyzed as completely randomized designs. Each experiment was repeated three times using 10 replicate culture flasks per treatment (each with three plantlets). Data were analyzed using one-way ANOVA, and the level of Least Significant Difference was determined using Tukey multiple range test at *p < 0.05 for comparing treatments means. The program used was STATGRAPHICS Plus (http://www.statgraphics.com) and graphs were performed using the Sigma Plot ver. 11.0 Program.
Results and discussion
Although some species easily form adventitious roots under conditions of in vitro culture, for many other species, such as C. papaya, the formation of roots under in vitro culture conditions is limited (Yu et al. 2000). It is known that auxins and possibly auxin transporters, among other factors, might be involved in the limited capacity to develop roots under in vitro culture conditions. In other species such as A. thaliana, the expression of auxin transporters genes in roots and leaves have been studied, however, little information on the expression levels of auxin transporter genes is available for tropical species such as C. papaya. Particularly, little information is available for gene expression studies of auxin transporter genes in the stems of tissue cultured plantlets. Therefore, it was important to gain understanding on the expression of auxin transporters genes at the stem base of C. papaya plantlets, under conditions that may promote the formation of adventitious roots.
Exogenous auxin promoted adventitious root formation on in vitro plantlets of C. papaya cv. Maradol
All in vitro plantlets at the beginning of the different treatments had an average plant height (PH) of 2.79 cm. At the end of the 21 days exposure to the different treatments, plantlets reached a PH of 3.21 cm, and they had a leaf number (LN) of 4.23, on average. In terms of root number (RN) and root length (RL), when in vitro plantlets were not treated with any plant hormone (T1 and T6, Table 1), they were not able to generate any adventitious roots, causing high mortality at 42 days after being transferred to the greenhouse. On the contrary, when plantlets were treated with 2 mg L−1 IBA (T4), they formed the highest RN (5.78), the highest RL (3.85 cm), the best rooting percentage (96.5 %) and the highest survival ex vitro (88.9 %). The addition of NAA alone to the culture medium did result in limited rooting and low survival (T7–T10, Table 1). In combination (IBA + NAA; T11–T14), plantlets did show intermediate rooting percentage and survival. In summary, these results pointed out that IBA was the best phytohormone to induce root formation and that 2 mg L−1 IBA was the ideal concentration to achieve high adventitious rooting in this papaya cultivar. Also, that rooting is critical for in vitro plantlets of papaya cv. Maradol, as those treatments with low rooting had very low ex vitro survival. On the contrary, plantlets treated with IBA (T4, Table 1), that had the best rooting number and the longest roots, also showed the highest plant ex vitro survival.
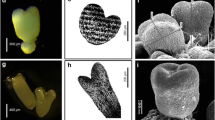
Those results were confirmed in a second experiment, now comparing in vitro plantlets (Table 2, D1–D4; Fig. 1c, d, g, h) with intact or de-rooted seedlings (Table 2, C1–C4; Fig. 1a, b, e, f). Again, those in vitro plantlets that were not treated with IBA, did not generate roots and exhibited high mortality ex vitro (Table 2, D2). In contrast, in vitro plantlets treated with 2 mg L−1 IBA induced high rhizogenesis (93.3 %) and exhibited high ex vitro survival (90.8 %; Table 2, D4). In order to evaluate if the process was also observed at natural, non in vitro, conditions, seedlings were de-rooted, and the same concentration of IBA (2 mg L−1) was applied to the cut end of the de-rooted seedlings, and their capacity to generate new adventitious roots was evaluated. As shown in Table 2, C1–C4 and Fig. 1a, b, e, f, intact seedlings (used as controls) at day 21 reached a PH of 17.5 cm, a LN of 10.24, a RN of 16.77, RL of 12.29 cm, 100 % rooting and 100 % survival (Table 2, C2; Fig. 1e). On the other hand, de-rooted seedlings treated with IBA, showed at day 21, a PH of 8–89 cm, a LN of 8.52, RN of 14.54, RL of 6.61 cm, high rooting (85.2 %) and high survival (91.1 %) (Table 2, C4; Fig. 1f). Our results are consistent with previous reports where the exogenous application of auxins (IBA and NAA) also induced high rooting in in vitro papaya shoots of a different cultivar (Drew 1987). Also Drew et al. (1993) reported that rooting in papaya of a different cultivar, can be promoted by a short exposure (3 days) to IBA. In other micropropagated species, the use of different concentrations of IBA in the rooting medium of Rumex palustris and Rumex thyrsiflorus promoted high rooting (Visser et al. 1995).
Views from intact C. papaya cv. Maradol seedlings, 2 days after germination (day 0; a) and 21 days after (day 21; e); de-rooted seedlings 2 days after germination (day 0; b), and 21 days after being treated with 2 mg L−1 IBA (day 21; f). In vitro plantlet not treated with IBA at day 0 (c) and at day 21 when no adventitious roots formation occurred (g); in vitro plantlets treated with IBA 2 mg L−1 at day 0 (d) and at day 21 (h). The 2 cm scale bar applies for all figures except for e, that it has its own scale bar of 15 cm that applies only for e
In silico cloning, alignment and phylogenetic analysis of CpAUX1/LAX and CpPIN genes and proteins
Within the C. papaya cv. Sun Up genome, we found 4 AUX1/LAX and 6 PIN nucleotide sequences with TBLASTX. All sequences had percentage identities from 66 to 95 % and E value between 7E−11 and 1E−180 (Table 3). Both families were classified and named based on the percentage identity and phylogeny with A. thaliana AUX1/LAX and PIN genes, a dicot species whose members have all been well characterized at the genomic level. The papaya protein sequences were named as: CpAUX1, CpLAX1, CpLAX2, CpLAX3 for the AUX1/LAX family and CpPIN1, CpPIN2, CpPIN3, CpPIN4, CpPIN5 and CpPIN6 for the PIN family (Tables 3, 4). In relation to the pair-wise comparison analysis of the predicted protein sequences of AtAUX1/LAX family, CpAUX1 showed the highest percentage identity with AtAUX1 (95.1 %). CpLAX1 showed a high percentage identity with AtLAX1 (94.06 %), while CpLAX2 showed a 94.3 % of identity with AtLAX2, and finally CpLAX3 presented an 88.1 % of identity in relation to AtLAX3 (Table 4). In the PIN family, the AA sequences CpPIN1 reached the highest identity with AtPIN1 (89.08 %), CpPIN2 showed greater identity with AtPIN2 (85.06 %), CpPIN3 showed an 84.46 % of identity with AtPIN3, similarly CpPIN4 showed higher identity with AtPIN4 (91.95 %), CpPIN5 showed a 69.52 % identity with AtPIN5, whereas CpPIN6 showed the highest percent identity (69.92 %) with AtPIN8 (Table 4).
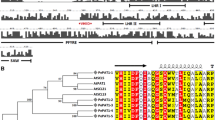
The AA sequences alignments of the four CpAUX1/LAX proteins (CpAUX1, CpLAX1, CpLAX2 and CpLAX3) are shown in Fig. 2, while those of the six CpPIN proteins (CpPIN1, CpPIN2, CpPIN3, CpPIN4, CpPIN5 and CpPIN6) are shown in Fig. 3. The CpAUX1/LAX and CpPIN proteins sequences were analyzed with different sequence multiple alignment and phylogenetic analysis tools, to develop a robust evolutionary model of transporters. Our results, regarding the alignment of AUX1/LAX predicted proteins in C. papaya showed an identical profile, length ranged between 387 and 491 AA and high percentage of identity values, equivalent to their counterparts in A. thaliana. Therefore, it is likely that CpAUX1, CpLAX1, CpLAX2 and CpLAX3 genes are homologs. In addition, it was also observed the characteristic 10 transmembrane helix regions, acidic AA on its N-terminus and proline-rich AA on its C-terminus, all of these regions were well conserved among all sequences (Fig. 2). From the alignment of CpPIN predicted protein sequences, we identified LD proteins (CpPIN 1–4) that showed three TPRXS (N/S) conserved motifs and the hydrophilic loop (crucial to polarity maintenance). The polar location apical–basal of the PIN proteins determines the direction of flow of auxin controlled by the reversible phosphorylation of PIN hydrophilic loops (PINHL; Friml et al. 2004). In addition, two proteins that lack the TPRXS (N/S) motif and the hydrophilic loop, were identified as SD proteins and named as CpPIN5 and CpPIN6 (Fig. 3).
ClustalX alignment of the deduced amino acid sequences of C. papaya and A. thaliana AUX1/LAXs orthologs. Identical amino acids are shaded in black and conservative substitutions are shaded in grey while the other amino acids are different. The regions predicted to form transmembrane helix in the primary structure of CpAUX1/LAXs proteins are marked by red helix. Alignment was made with MEGA5 (Tamura et al. 2011) and edition was made with BioEdit (Hall 1999)
ClustalX alignment of the deduced amino acid sequences of C. papaya and A. thaliana PIN orthologs. Identical amino acids are shaded in black and conservative substitutions are shaded in grey while the other amino acids are different. The double line indicates the characteristic signature the three TPRXS from group PIN long-distance, and the central phospho-serines in the motifs are indicated in red. Two hydrophobic domains in the CpPIN proteins are underlined with solid black boxes while the hydrophilic loop region is underlined with solid blue box. The predicted transmembrane helix-formed regions in the primary structure of CpPIN proteins are indicated by red helix. Alignment was made with MEGA5 (Tamura et al. 2011) and edition was made with BioEdit (Hall 1999)
Our phylogenetic trees exhibit structural similarities between the predicted proteins AUX1/LAX (Fig. 4) and PIN (Fig. 5) from C. papaya and those from A. thaliana. The phylogenetic tree from CpAUX1/LAX predicted proteins showed two subclades. CpAUX1 and CpLAX1 that were positioned in the same subclade I with their counterparts AtAUX1 and AtLAX1 from A. thaliana. Moreover, the proteins CpLAX2 and AtLAX2 are closely related to each other, and for the case of CpLAX3 it shows a high percentage identity (88.10 %) with AtLAX3, these were grouped in the same subclade II (Fig. 4). PIN phylogenetic tree showed two subclades, PIN proteins were grouped as follows: subclade I formed by AtPIN1, AtPIN2, AtPIN3, AtPIN4, AtPIN6, AtPIN7, CpPIN1, CpPIN2, CpPIN3, and CpPIN4; subclade II formed by AtPIN5 and CpPIN5, AtPIN8 and CpPIN6. In this sense, CpPIN4 was grouped with its orthologs AtPIN3, AtPIN4 and AtPIN7. In the same way, CpPIN1 was grouped with its ortholog AtPIN1, and CpPIN2 was grouped with AtPIN2, these proteins were found in the same clade, identified as LD transporters. The phylogenetic tree showed that CpPIN5 gene presents high percentage identity (69.52 %) with AtPIN5 gene of A. thaliana, also CpPIN6 gene was grouped with its ortholog AtPIN8 gene, these proteins, identified as SD transporters have a hydrophobic conserved domain as in their counterparts for A. thaliana (Fig. 5).
Neighbor-joining phylogenetic tree of AUX1/LAX amino acid sequences from A. thaliana and C. papaya cv. Sun Up showing the formed subclades. AUX1/LAX orthologs sequences from C. papaya and A. thaliana are highlighted with black and grey circles, respectively. The evolutionary history was inferred by using the Maximum Likelihood method based on the Whelan and Goldman model (Whelan and Goldman 2001). The bootstrap consensus tree inferred from 1000 replicates (Saitou and Nei 1987) is taken to represent the evolutionary history of the taxa analyzed (Jones et al. 1992). When the number of common sites was <1000 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise BIONJ method with MCL distance matrix was used. Phylogeny was generated with MEGA5 (Tamura et al. 2011)
Neighbor-joining phylogenetic tree of PIN amino acid sequences from A. thaliana and C. papaya cv. Sun Up showing the formed subclades. PIN orthologs sequences from C. papaya and A. thaliana are highlighted with black and grey circles, respectively. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein 1985). The evolutionary distances were computed using the JTT matrix-based method (Jones et al. 1992) and are in the units of the number of amino acid substitutions per site. Phylogeny was generated with MEGA5 (Tamura et al. 2011)
As mentioned above, auxin molecules can passively enter the cells; however, they could also be transported into cells through the activity of H+ of the AUX1/LAX family of PM permeases (Kerr and Bennett 2007). The need for such active auxin uptake processes in the cell causes a high and rapid influx of auxin, as in lateral root cap, where AUX1 plays an important role in the redirection of polar auxin flow (Kramer and Bennett 2006). The A. thaliana genome encodes one AUX1 and three AUX1-like (LAX1, LAX2 and LAX3) proteins, that share approximately 80 % similarity in their AA sequences. Therefore, our results agree with those reported by Parry et al. (2001a) and Swarup et al. (2008). Similarly, the gene CpAUX1 and their three homologous CpLAX genes presented between 88 and 95 % similarity at the AA level, suggesting a conserved function in auxin absorption. In Zea mays, all four proteins contained a highly conserved core region, which was composed of ten transmembrane helices, the AA composition analysis indicated that C-terminus of the ZmLAX was proline rich and the N-terminus was acidic AA-rich (Yue et al. 2015).
Regarding the PIN proteins, Zažímalová et al. (2007) have found in homologous sequences to those from the model plant A. thaliana, relatively high similarity values ranging between 32 and 82 % compared to other higher plants, this suggests that the evolution the PIN arose from a single ancestral sequence (Forestan et al. 2012). From our results, CpPIN proteins showed high homology percentages (values between 66 and 91 %) similarity to those of A. thaliana at the AA level, that agrees with those previously published. The more significant characteristics of theses CpPIN proteins was the presence of a hydrophilic loop that partially modulates the intracellular auxin homeostasis, depending on cell type and developmental stage, as described by Ganguly et al. (2014). As mentioned by Wabnik et al. (2011), the SD proteins from A. thaliana (AtPIN5 and AtPIN8 proteins) showed high homology, these SD proteins appear to be localized at the endoplasmic reticulum (ER), suggesting a possible role in regulating intracellular auxin homeostasis (Wu et al. 2007). The absence of the hydrophilic loop on SD proteins, suggested that there has been no selective advantage to keep this region, whose purpose is crucial to maintain polarity. In our study, we were unable to identify an ortholog for AtPIN6 protein in C. papaya. The classification of AtPIN6 protein as LD or SD is rather controversial, since the hydrophilic loop is only partially deduced, while the transmembrane regions show high sequence similarity with LD PINs proteins, as reported by Křeček et al. (2009) and Mravec et al. (2009). The proteins encoded by these genes can be classified in two groups: (1) PIN LD type proteins (PIN1, 2, 3, 4, and 7) located in the plasma membrane (PM) and involved in processes such as gravitropism, phototropism, embryo development, regulation of root meristem and formation of the apical hook, and (2) PIN SD type proteins (PIN5, 6 and 8) which lack the large hydrophilic loop, these proteins are found in the cytoplasm and the ER (Paponov et al. 2005; Mravec et al. 2009). LD PIN have a hydrophobic region with five to six transmembrane segments (residues 1–163), a predominantly hydrophilic core extending from residue 164–482, and other hydrophobic region with four to five transmembrane segments between residues 483 and 647 (Müller et al. 1998). The hydrophilic loop of PIN1 has three evolutionarily conserved TPRXS motifs that have central phosphorylated Ser residues. These residues are phosphorylated by PINOID (PID), a protein Ser/Thr kinase, whose purpose is to control the polarity of location of PIN genes by direct phosphorylation (Friml et al. 2004; Michniewicz et al. 2007; Fang et al. 2010). Studies in other plant species have revealed the biological functions of auxin transporters (Xu et al. 2014). The evolutionary relationships among C. papaya with A. thaliana should help us to understand the possible roles of these auxin transporter in C. papaya. Another interesting point is that in A. thaliana, the AtPIN3, 4 and 7 are closely related to each other, from our results, we also found that CpPIN3 and CpPIN4 showed a high level of similarity and they were grouped in the same clade I (Fig. 5). As it was mentioned by Dal Cin et al. (2009), there are a different organization of the PIN members in different species; this may be due to duplication and specialization of the PIN members in plants. Indeed, as reported by Wang et al. (2009) in rice, the LD OsPIN proteins were grouped with LD proteins AtPIN (AtPIN3, AtPIN4 and AtPIN7) in the same clade. A previous phylogenetic analysis suggested a close evolutionary relationship between these three A. thaliana transporters: AtPIN3, AtPIN4 and AtPIN7, that are located in the same clade. The reduction in the number of members for gene families in C. papaya in comparison with those in A. thaliana, has been previously reported for other genes families (Ming et al. 2008; Idrovo et al. 2012; Peraza-Echeverría et al. 2012), and it could be attributed to evolutive differences between C. papaya and A. thaliana (Ming et al. 2008).
IBA induces changes in the expression of CpAUX1/LAX and CpPIN genes in C. papaya cv. Maradol
In stem-base tissues
In the case of SB tissues, in general, except for in vitro plantlets without IBA, AET genes (dark bars) showed higher relative expression levels (REL) than those of the AIT genes (light bars). Within the auxin AIT genes, CpLAX3 was in general the one showing the highest REL. For the AET genes, CpPIN1 and CpPIN2 were the genes with the highest REL, while CpPIN5 and CpPIN6, had the lowest REL (Fig. 6).
qRT-PCR analysis of CpAUX1/LAXs and CpPIN genes expression from SB tissues from intact seedlings, IBA treated de-rooted seedlings, and in vitro plantlets of C. papaya cv. Maradol. The histograms show the relative expression level of CpAUX1/LAXs and CpPIN genes from seedlings at day 0 (a) and at day 21 (e), IBA treated de-rooted seedling at day 0 (b) and at day 21 (f), in vitro plantlets without IBA at day 0 (c) and day 21 (g) and in vitro plantlets treated with 2 mg L−1 IBA at day 0 (d) and at day 21 (h). The relative mRNA level of individual genes was normalized with respect to that of CpEF1α gene. The data are mean ± SD from three independent experiments each with three replicates (n = 3). Treatments with the same letters are not significantly different (one-way ANOVA followed by Tukey post-test)
In intact seedlings at day 0, all AIT genes were expressed but only the LD AET (CpPIN1, CpPIN2, CpPIN3, CpPIN4) genes were expressed, whereas the SD AET (CpPIN5 and CpPIN6) genes showed very low REL (Fig. 6a). At day 21 however, all AIT and AET genes (including CpPIN5 and CpPIN6) increased their expression, relative to that found at day 0, but they maintained the same expression pattern (Fig. 6e).
In de-rooted seedlings, at day 0 (taken when the new adventitious roots started to develop, i.e. 10 days after IBA application; Fig. 6b), they showed a similar AIT and AET expression patterns to those observed for intact seedlings. At day 21, however, all AIT and AET genes (including CpPIN5 and CpPIN6) increased their REL relative to that found at day 0 (Fig. 6f). Again, the expression patterns of those genes found at day 21 in this treatment, were similar to those found at the intact seedlings at day 21.
Interestingly, in the case of in vitro plantlets not treated with IBA (that did not produce roots), they showed a very low REL of all AIT and AET genes at day 0 (Fig. 6c), which remained low at day 21 (Fig. 6g).
On the contrary, in vitro plantlets treated with IBA, at day 0, again the REL of all auxin AIT and AET genes were as low as those found in in vitro plantlets without IBA (Fig. 6d vs. c), however, at day 21 a marked increase in the REL of those genes occurred (in all except CpPIN 5 and CpPIN 6; Fig. 6h). In fact, the expression patterns of those genes for IBA-treated in vitro plantlets at day 21 (that did generate roots; Fig. 6h), were similar and the REL were as high as those observed at day 21, for both intact seedlings (Fig. 6e) and IBA-treated de-rooted seedlings (Fig. 6f).
In root tissues
In R tissues, in general for those treatments that did generate adventitious R, AET genes (dark bars) showed higher REL than those of the AIT genes (light bars), similar to the pattern observed for SB tissues. Within the AIT genes, CpLAX3 was in general the one showing the highest REL. For the AET genes, CpPIN2 was the gene with the highest REL, while CpPIN5 and CpPIN6, were the genes with the lowest REL (Fig. 7). In R tissues from intact seedlings at day 0, all AIT genes were relatively highly expressed. In the case of the LD AET genes (CpPIN1, CpPIN2, CpPIN3, CpPIN4) they were highly expressed, whereas SD AET (CpPIN5 and CpPIN6) showed very low REL (Fig. 7a). At day 21, all AIT and AET genes (including CpPIN5 and CpPIN6) increased their REL, relative to that found at day 0 (Fig. 7e).
qRT-PCR analysis of CpAUX1/LAXs and CpPIN genes expression from R tissues from seedlings, IBA treated de-rooted seedlings and in vitro plantlets of C. papaya cv. Maradol. The histograms show the relative expression level of CpAUX1/LAXs and CpPIN genes from seedlings at day 0 (a) and at day 21 (e), IBA treated de-rooted seedling at day 0 (b) and at day 21 (f), in vitro plantlets without IBA at day 0 (c) and day 21 (g) and in vitro plantlets treated with 2 mg L−1 IBA at day 0 (d) and at day 21 (h). The relative mRNA level of individual genes was normalized with respect to that of CpEF1α gene. The data are mean ± SD from three independent experiments each with three replicates (n = 3). Treatments with the same letters are not significantly different (one-way ANOVA followed by Tukey post-test)
In de-rooted seedlings, at day 0 (taken when the new adventitious roots started to develop, i.e. 10 days after IBA application; Fig. 7b), a similar expression pattern of the AIT and AET genes to that observed for intact seedlings occurred. At day 21, again all AIT and AET genes (including CpPIN5 and CpPIN6) increased their REL relative to that found at day 0 (Fig. 7f). Again, the expression pattern found at day 21 in this treatment, was similar to that found at R tissues from intact seedlings at day 21. In the case of in vitro plantlets not treated with IBA, the REL are not available, as this treatment did not produce roots at any time (Fig. 7c, g).
The IBA-treated in vitro plantlets, at day 0 had no roots (Fig. 7d), however, at day 21, this treatment had already generated adventitious roots and interestingly, the expression patterns for both AIT and AET genes (Fig. 7h) were in general equivalent to those observed at the same day, for both intact seedlings (Fig. 7e) and IBA-treated de-rooted seedlings (Fig. 7f). In this treatment, CpLAX3, CpPIN2 and CpPIN4 were the genes with the highest REL (Fig. 7h). It is interesting that the REL of CpAUX1 were lower, in this treatment, than those found for this gene in seedlings (Fig. 7e) and IBA-treated de-rooted seedlings (Fig. 7f).
Auxin transport depends largely on specific auxin transporters, namely the AUX1/LAX and PIN-FORMED (PIN) proteins (Petrásek et al. 2006). Therefore, the expression patterns of AUX1/LAX and PIN genes in seedlings, de-rooted seedlings and in vitro plantlets of C. papaya cv. Maradol is crucial for understanding the role of IBA in the rhizogenesis processes of this difficult-to-root cultivar.
The large differences in the REL of CpAUX1/LAX and CpPIN genes in both SB and R tissues, between plantlets not treated with IBA and those treated with IBA (that induced high rooting), might indicate that those genes (particularly CpAux3 and CpPIN 1–4) were actively involved in regulating rhizogenesis in C. papaya. In our experiments, IBA-treated plants (both in vitro plantlets or de-rooted seedlings) CpLAX3 gene had a greater REL compared to the other members of this family. These data are consistent with those reported previously by Swarup et al. (2008), who confirmed that the expression of AtAUX1 and AtLAX3 genes in roots, is a requirement for lateral root development and apical hook formation in A. thaliana (Vandenbussche et al. 2010). In addition, it has been observed that defects in AtAUX1 and AtLAX1 genes, reduced initiation and/or elongation of lateral roots due a reduced movement of auxin (Marchant et al. 2002; Benková et al. 2003; Wu et al. 2007; Swarup et al. 2008). For instance, aux1 mutants are agravitropic and they have a decreased number of lateral roots. Similar data were observed in loss-of-function mutants for AtLAX3 gen, that results in a delayed lateral root emergence (Swarup et al. 2008).
The phenotypes of mutants for the first two PIN genes, indicated that PIN-driven PAT is crucial for processes as diverse as aboveground organogenesis and the root gravitropic response (Adamowski and Friml 2015). In seedling, PIN genes maintains activity of the root apical meristem, the local auxin maximum in the root tip is established by directional auxin transport driven by PIN genes and the action of PIN long-distance (LD) establishes a local reflux-loop of auxin (Friml and Palme 2002; Blilou et al. 2005). The process in which the root system is formed begins with the emergence of pericycle layer of the primary root, these events are caused in response to high concentrations of auxin involving changes in the distribution of PIN-driven auxin distribution, gradual concentration of auxin at the apical end of the growing lateral root organ (Benková et al. 2003; Dubrovsky et al. 2008; Adamowski and Friml 2015). In Zea mays roots, IAA treatment up-regulated the ZmPIN genes expression levels more than five-fold (Yue et al. 2015). From our results, it was observed that the LD CpPIN1, CpPIN2, CpPIN3 and CpPIN4 genes, increased their expression after 21 days of treatment in all seedlings and de-rooted seedlings in both SB and R tissue. In in vitro plantlets, no reports exist on the expression of these auxin genes transporters. However, clearly the in vitro plantlets of C. papaya need a stimulus for developing roots, this stimulus was achieved by adding IBA to the culture medium, with this condition the in vitro plantlets showed a marked increase in the REL of LD CpPIN genes. The increased expression of the CpPIN1, CpPIN2, CpPIN3 and CpPIN4 genes in response to IBA in C. papaya, is in line with reports of A. thaliana orthologs showing that they are involved in the induction and adventitious root formation (Zažímalová et al. 2010). Regarding the expression of these LD CpPIN genes in other species, Raven (1975), Benková et al. (2003) and Petrásëk et al. (2006), mentioned that AtPIN1, AtPIN2, AtPIN3, AtPIN4, and AtPIN7 genes were detected in the root tip, where they mediate tropism and root patterning, by modulating the maximum auxin concentration and auxin redistribution for root gravitropism.
In our R tissues from seedlings, IBA-treated de-rooted seedlings or IBA-treated in vitro plantlets, CpPIN2 gene showed high REL, what is consistent with the fact that in A. thaliana, the AtPIN2 transporter has been localized at the apical side in root epidermal cells and at the basal side in young cortex cells, contributing to the reflux of auxin towards to root tip, which is crucial in the control of root meristem size (Blilou et al. 2005). Our results also coincide with the work performed by Křeček et al. (2009), where this gene was found in radicular zones of A. thaliana, with absence or low expression in shoot apical areas. Our data confirmed that CpPIN2 gene was highly expressed in both SB and R of those treatments that were able to produce roots, which suggested that this gene may take part in C. papaya root architecture. The SD genes, CpPIN5 and CpPIN6 showed low expression in SB and R in all plants evaluated; what might be consistent with the fact that AtPIN8 and AtPIN5 genes of A. thaliana had not been reported to influence rhizogenesis, instead they have been expressed in male gametophyte and have a crucial role in pollen development and functionality, their ectopic expression in sporophytic tissues establishes a role in regulating auxin homoeostasis and metabolism (Dal Bosco et al. 2012; Ding et al. 2012).
Interestingly, it seems that CpPIN2 and CpLAX3 genes are directly involved in root formation in C. papaya, because they showed higher REL during root formation in seedlings and in de-rooted seedlings, as well as in in vitro plantlets after being treated with IBA and formed roots. Other authors have reported that PIN2 and LAX3 for A. thaliana (Blilou et al. 2005; Abas et al. 2006; Křeček et al. 2009) and Medicago truncatula (Schnabel and Frugoli 2004) are the best candidates for the maintenance of initial PAT in the cambial zone of the roots, therefore is feasible that CpPIN1 and CpPIN2 genes participate in different development processes and show functional redundancy in root zones in plants of C. papaya. Most likely, some of these members are also putative regulators and the comparative study of their function and activity will help to clarify their role in the regulation of the direction of PAT of in vitro plantlets of C. papaya. To understand the biological meaning behind this redundancy, it is necessary to further investigate the different endogenous and exogenous stimuli able to regulate CpAUX1/LAX and CpPIN genes in vitro plantlets of C. papaya cv. Maradol, to uncover how these auxin transporter genes might participate in PAT.
IBA induced histological changes in SB and R tissues of in vitro plantlets, equivalent to those naturally occurring in intact seedlings
From SB tissues
Sections from SB from seedlings of C. papaya cv. Maradol, showed a mono-stratified epidermis (ep), cortex (co), cambium and strands of xylem (xy) and phloem (ph). In those areas where the root primordia was naturally developing, an irregular arrangement of vv and meristematic zones (mz) can be observed (Fig. 8a). In SB sections from IBA-treated de-rooted seedlings, a similar irregular arrangement of vv and mz were observed, in areas where adventitious roots were developing (Fig. 8b). On the contrary, SB tissues from the non IBA-treated in vitro plantlets did show unaltered arrangement of tissues, and no signs of root primordia were visible (Fig. 8c). This unaltered SB histological arrangement, is consistent with the fact that those plantlets were unable to produce roots. However, 21 days after IBA treatment, in vitro plantlets suffered marked histological modifications, including the presence of adventitious root primordia (where vv and mz were visible; Fig. 8d). SB tissues from those IBA-treated in vitro plantlets were histologically equivalent to those from intact seedlings, bearing naturally formed roots (Fig. 8a), and to those IBA-treated de-rooted seedlings developing adventitious roots (Fig. 8b).
Histological studies from SB and R tissues from seedlings, IBA treated de-rooted seedlings, and in vitro plantlets of C. papaya cv. Maradol with IBA (2 mg L−1) or without IBA. Sections in SB (a) and R (e) from seedlings, Section in SB (b) and R (f) from de-rooted seedlings, and cross-sections from SB (c) from in vitro plantlets without IBA, plantlets in this treatment did not generate roots (g). Sections from SB (d) and R (h) from in vitro plantlets treated with 2 mg L−1 IBA. Sections were taken 21 days after each treatment was imposed. e Epidermis, ex exodermis, co cortex, en endodermis, pe pericycle, ph phloem, x xylem, mz meristematic zone, vv vascular vessels
From R tissues
Sections from R tissues from intact seedlings of C. papaya cv. Maradol, showed epidermis (e), cortex (co), endodermis (en), perycicle (pe) and strands of ph and xy (Fig. 8e). In R sections from IBA-treated de-rooted seedlings, a similar histological arrangement to those described for intact seedlings was observed in the new IBA-induced adventitious R (Fig. 8f). On the contrary, non-IBA treated in vitro plantlets, did not generate R (Fig. 8g, empty space). However, the addition of IBA to in vitro plantlets resulted in IBA-induced adventitious R, that showed root anatomical features (Fig. 8h) that were similar to those observed in R from IBA-treated de-rooted seedlings (Fig. 8f), and more importantly to those observed in naturally occurring R from intact seedlings (Fig. 8e).
As described by Iliev et al. (2001), auxins might increase the cambial activity and adventitious root formation, therefore tracheid-like cells are formed, providing vascular connection between the root-shoot systems. Our histological observations for C. papaya are similar to those reported for Manihot esculenta by Medina et al. (2007), where in vitro roots were highly similar to roots from their seedlings (both, in vitro roots and those from seedlings lacked starch granules and had an identical composition and organization). Ballester et al. (1999), also reported the same normal organization of epidermis, cortex and collateral vascular bundles forming a ring around the pith, in roots from in vitro plants, and in those from seedlings of Castanea sativa Mill.
Conclusion
We report here the identification, and in silico characterization, of the complete family members of AUX1/LAX genes involved in the influx transporters of auxin and of the complete family members of PIN genes involved in the efflux transport of auxins in C. papaya. Both families showed high homology with AUX1/LAX and PIN genes from A. thaliana. We identified in C. papaya the same number of AUX1/LAX genes (four) that those reported for A. thaliana, however, in the case of the PIN family members, we identified only six genes from the eight PIN genes reported in A. thaliana. Under normal in vitro culture conditions, non IBA-treated C. papaya plantlets are unable to generate adventitious roots, and the expression levels of CpAUX1/LAX and CpPIN genes from their stem base tissues from those plants, are extremely low. On the contrary, when exogenous IBA are applied on in vitro C. papaya plantlets, they generated adventitious roots (histologically equivalent to those from seedlings), and the expression levels of CpAUX1/LAX and CpPIN genes (in particular CpLAX3 and CpPIN2) from SB and R tissues of those plantlets were reestablished. Both the expression patterns and the REL of those genes at both SB and at the generated R tissues, reached equivalent values to those observed in SB and R tissues from intact seedlings. However, as it has been documented elsewhere that auxin transporters might experience further post-translational regulation (phosphorylation, ubiquitination, and maturation processes, changes in the plasma membrane), that might play an important role in regulating PAT, as suggested by Titapiwatanakun and Murphy (2009). Therefore, the apparent association reported here between the exogenous IBA-induced expression of those auxin transport genes, with the concomitant exogenous IBA-induced rooting generation, on in vitro cultured plantlets of C. papaya, merits to be further explored.
Abbreviations
- AET:
-
Auxin efflux transporters
- AIT:
-
Auxin influx transporters
- AUX1/LAX:
-
AIT genes
- IAA:
-
Indole-acetic acid
- IBA:
-
Indole-3-butyric acid
- L:
-
Leaf
- LD:
-
Long-distance
- NAA:
-
1-Naphthaleneacetic acid
- PAT:
-
Polar auxin transport
- PIN:
-
AET genes
- PPFD:
-
Photosynthetic photon flux density
- REL:
-
Relative expression levels
- RH:
-
Relative humidity
- R:
-
Root
- SB:
-
Stem-base
- SD:
-
Short-distance
- T:
-
Temperature
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
References
Abas L, Benjamins R, Malenica N, Paciorek T, Wišniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C (2006) Intracellular trafficking and proteolysis of the auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8:249–256. doi:10.1038/ncb1369
Adamowski M, Friml J (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27:20–32. doi:10.1105/tpc.114.134874
Bainbridge K, Guyomarc’h S, Bayer E, Swarup R, Bennett M, Mandel T, Kuhlemeier C (2008) Auxin influx carriers stabilize phyllotactic patterning. Genes Dev 22:810–823. doi:10.1101/gad.462608
Ballester A, San-José MC, Vidal N, Fernández-Lorenzo JL, Vieitez AM (1999) Anatomical and biochemical events during in vitro rooting of microcuttings from juvenile and mature phases of chestnut. Ann Bot 83:619–629. doi:10.1006/anbo.1999.0865
Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602. doi:10.1016/S0092-8674(03)00924-3
Berlin GP, Miksche JP (1976) Botanical microtechnique and cytochemistry, 3rd edn. Iowa State University Press, Ames, p 254
Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Mitsuhiro A, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44. doi:10.1038/nature03184
Chen Y, Hao X, Cao J (2014) Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J Integr Plant Biol 56:133–150. doi:10.1111/jipb.12127
Dall Bosco C, Dovzhenko A, Palme K (2012) Intracellular auxin transport in pollen. Plant Signal Behav 7:1504–1505. doi:10.4161/psb.21953
Dal Cin V, Barbaro E, Danesin M, Murayama H, Velasco R, Ramina A (2009) Fruitlet abscission: a cDNA-AFLP approach to study genes differentially expressed during shedding of immature fruits reveals the involvement of a putative auxin hydrogen symporter in apple (Malus domestica L. Borkh). Gene 442:26–36. doi:10.1016/j.gene.2009.04.009
De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, Inzé D, Bennett MJ, Beeckman T (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134:681–690. doi:10.1242/dev.02753
Ding Z, Wang B, Moreno I, Dupláková N, Simon S, Carraro N, Reemmer J, Pěnčík A, Chen X, Tejos R, Skůpa P, Pollmann S, Mravec J, Petrášek J, Zažímalova E, Honys D, Rolčík J, Murphy A, Orellana A, Geisler M, Friml J (2012) ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun 3:941–947. doi:10.1038/ncomms1941
Drew RA (1987) The effects of medium composition and cultural conditions on in vitro root initiation and growth of papaya (Carica papaya L.). J Hortic Sci Biotechnol 62:551–556. http://www.jhortscib.org/Vol62/62_4/20.htm. Accessed 6 Jun 2015
Drew RA, Miller RM (1993) Nutritional and cultural factors affecting rooting of papaya (Carica papaya L.) in vitro. Hortic Sci 64:767–773. doi:10.1080/14620316.1989.11516019
Drew RA, McComb JA, Considine JA (1993) Rhizogenesis and root growth of Carica papaya L. in vitro in relation to auxin sensitive phase and use of riboflavin. Plant Cell Tissue Organ Cult 33:1–7. doi:10.1007/BF01997591
Dubrovsky JG, Gambetta GA, Hernandez-Barrera A, Shishkova S, Gonzalez I (2006) Lateral root initiation in Arabidopsis: developmental window, spatial patterning, density and predictability. Ann Bot 97:903–915. doi:10.1093/aob/mcj604
Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105:8790–8794. doi:10.1073/pnas.0712307105
Fang H, Zago MK, Abas L, Van Marion A, Galván-Ampudia CS, Offringa R (2010) Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22:1129–1142. doi:10.1105/tpc.109.072678
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.1016/j.bse.2010.10.005
Forestan C, Farinati S, Varotto S (2012) The maize PIN gene family of auxin transporters. Front Plant Sci 3:1–16. doi:10.3389/fpls.2012.00016
Friml J, Palme K (2002) Polar auxin transport—old questions and new concepts? Plant Mol Biol 49:273–284. doi:10.1023/A:1015248926412
Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk P, Ljung K, Sandberg G, Hooykaas P, Palme K, Offringa R (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306:862–865. doi:10.1126/science.1100618
Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282:2226–2230. doi:10.1126/science.282.5397.2226
Ganguly A, Park M, Kesawat MS, Cho HT (2014) Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN-FORMED proteins. Plant Cell 26:1570–1585. doi:10.3410/f.718335431.793494308
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series No. 41. Oxford University Press. pp. 95–98. http://brownlab.mbio.ncsu.edu/JWB/papers/1999Hall1.pdf
Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14:2339–2351. doi:10.1105/tpc.004960
Hoffmann M, Hentrich M, Pollmann S (2011) Auxin–oxylipin crosstalk: relationship of antagonists. J Integr Plant Biol 53:429–445. doi:10.1111/j.1744-7909.2011.01053.x
Idrovo EFM, Peraza-Echeverria S, Fuentes G, Santamaría JM (2012) In silico cloning and characterization of the TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiol Biochem 54:113–122. doi:10.1016/j.plaphy.2012.02.011
Iliev I, Kitin P, Funada R (2001) Morphological and anatomical study on in vitro root formation of silver birch (Betula pendula Roth.). Prop Orn Plants 1:10–19
Islam R, Rahman SM, Hossain M, Joarder OI (1993) In vitro clonal propagation of papaya (Carica papaya L.). Pak J Bot 25:189–192. http://www.pakbs.org/pjbot/PDFs/25(2)/PJB25(2)13.pdf
Ivanchenko MG, Zhu J, Wang B, Medvecká E, Du Y, Azzarello E, Mancuso S, Megraw M, Filichkin S, Dubrovsky JG, Friml J, Geisler M (2015) The cyclophilin A DIAGEOTROPICA gene affects auxin transport in both root and shoot to control lateral root formation. Development 142:712–721. doi:10.1242/dev.113225
Jiménez VM, Mora-Newcomer E, Gutiérrez-Soto MV (2014) Biology of the papaya plant. In: Ming R, Moore PH (eds) Genetics and genomics of papaya, plant genetics and genomics: crops and models 10. Springer Science + Business Media, New York, pp 17–33
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi:10.1093/bioinformatics/8.3
Kerr ID, Bennett MJ (2007) New insight into the biochemical mechanisms regulating auxin transport in plants. Biochem J 401:613–622. doi:10.1042/BJ20061411
Kramer EM, Bennett MJ (2006) Auxin transport: a field in flux. Trends Plant Sci 11:382–386. doi:10.1016/j.tplants.2006.06.002
Křeček P, Skůpa P, Libus J, Naramoto S, Tejos R, Friml J, Zažímalová E (2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10:1–11. doi:10.1186/gb-2009-10-12-249
Laskowski M, Grieneisen VA, Hofhuis H, Ten Hove CA, Hogeweg P, Marée AF, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6:e307. doi:10.1371/journal.pbio.0060307
Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18:450–458. doi:10.1016/j.tplants.2013.04.006
Magdalita PM, Persley DM, Godwin ID, Drew RA, Adkins SW (1997) Screening Carica papaya × C. cauliflora hybrids for resistance to papaya ringspot virus-type P. Plant Pathol 46:837–841. doi:10.1046/j.1365-3059.1997.d01-90.x
Malabadi RB, Vijayakumar S, Mulgund GS, Nataraja K (2011) Induction of somatic embryogenesis in papaya (Carica papaya L). Res Biotechnol 2:40–55
Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14:589–597. doi:10.1105/tpc.010354
McManus JFA (1961) Periodate oxidation techniques. General cytochemical methods. Academic Press, New York, pp 171–201
Medina R, Faloci MM, Gonzalez AM, Mroginski LA (2007) In vitro cultured primary roots derived from stem segments of cassava (Manihot esculenta) can behave like storage organs. Ann Bot 99:409–423. doi:10.1093/aob/mcl272
Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130:1044–1056. doi:10.1016/j.cell.2007.07.033
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–996. doi:10.1038/nature06856
Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329:1306–1311. doi:10.1126/science.1191937
Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof YD, Dobrev PI, Schwarzerová K, Rolčík J, Seifertová D, Luschnig C, Benková E, Zažímalová E, Geisler M, Friml J (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459:1136–1140. doi:10.1038/nature08066
Muday GK, DeLong A (2001) Polar auxin transport: controlling where and how much. Trends Plant Sci 6:535–542. doi:10.1016/S1360-1385(01)02101-X
Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17:6903–6911. doi:10.1093/emboj/17.23.6903
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Paponov IA, Teale WD, Trebar M, Blilou I, Palme K (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10:170–177. doi:10.1016/j.tplants.2005.02.009
Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ (2001a) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J 25:399–406. doi:10.1046/j.1365-313x.2001.00970.x
Parry G, Marchant A, May S, Swarup R, Swarup K, James N, Graham N, Allen T, Martucci T, Yemm A, Napier R, Manning K, King G, Bennett M (2001b) Quick on the uptake: characterization of a family of plant auxin influx carriers. J Plant Growth Regul 20:217–225. doi:10.1007/s003440010030
Peraza-Echeverría S, Santamaria JM, Gabriela Fuentes, Mariana Menéndez, Angel Vallejo M, Virginia Herrera (2012) The NPR1 family of transcription cofactors in papaya: insights into its structure, phylogeny and expression. Genes Genom 34:379–390. doi:10.1007/s13258-011-0218-7
Petrásëk J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubeš M, Čovanová M, Dhonukshe P, Skůpa P, Benková E, Perry L, Křeček P, Lee OR, Fink GR, Geisler M, Murphy AS, Luschnig C, Zažímalová E, Friml J (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312:914–918. doi:10.1126/science.1123542
Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9:448–453. doi:10.1016/j.pbi.2006.07.006
Raven JA (1975) Transport of indoleacetic acid in plant cells in relation to pH and electrical potential gradients, and its significance for polar IAA transport. New Phytol 74:163–172. doi:10.1111/j.1469-8137.1975.tb02602.x
Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426:255–260. doi:10.1038/nature02081
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sawchuk MG, Scarpella E (2013) Control of vein patterning by intracellular auxin transport. Plant Signal Behav 8:e27205. doi:10.4161/psb.27205
Schnabel EL, Frugoli J (2004) The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol Genet Genom 272:420–432. doi:10.1007/s00438-004-1057-x
Shen C, Bai YH, Wang S, Zhang S, Wu YR, Chen M, Jiang DA, Qi YH (2010) Expression profile of PIN, AUX/LAX and PGP auxin transporters gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J 277:2954–2969. doi:10.1111/j.1742-4658.2010.07706.x
Shen C, Yue R, Youhuang B, Feng R, Sun T, Wang Y, Tie S, Wang H (2015) Identification and analysis of Medicago truncatula auxin transporter gene families uncover their roles in responses to Sinorhizobium meliloti infection. Plant Cell Physiol 56:1930–1943. doi:10.1093/pcp/pcv113
Swarup R, Kramer EM, Perry P, Knox K, Ottoline LH, Haseloff J, Beemster G, Bhalerao R, Bennett MJ (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7:1057–1065. doi:10.1038/ncb1316
Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, Levesque MP, Carrier D, James N, Calvo V, Ljung K, Kramer E, Roberts R, Graham N, Marillonnet S, Patel K, Jones J, Taylor C, Schachtman D, May S, Sandberg G, Benfey P, Friml J, Kerr I, Beeckman T, Laplaze L, Bennett MJ (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10:946–954. doi:10.1038/ncb1754
Talavera C, Espadas F, Contreras F, Fuentes G, Santamaría JM (2007) Acclimatization, rooting and field establishment of micropropagated papaya plants. Acta Hortic 812:373–378. doi:10.17660/ActaHortic.2009.812.52
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Teale WD, Ditengou FA, Dovzhenko AD, Li X, Molendijk AM, Ruperti B, Paponov I, Palme K (2008) Auxin as a model for the integration of hormonal signal processing and transduction. Mol Plant 1:229–237. doi:10.1093/mp/ssn006
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Titapiwatanakun B, Murphy AS (2009) Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60:1093–1107. doi:10.1093/jxb/ern240
Ugartechea-Chirino Y, Swarup R, Swarup K, Péret B, Whitworth M, Bennett M, Bougourd S (2010) The AUX1 LAX family of auxin influx carriers is required for establishment of embryonic root cell organization in Arabidopsis thaliana. Ann Bot 105:277–289. doi:10.1093/aob/mcp287
Vandenbussche F, Petrášek J, Žádníková P, Hoyorevá K, Pešek B, Raz V, Swarup R, Bennett M, Zažímalová E, Benková E, Van Der Straeten D (2010) The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137:597–606. doi:10.1242/dev.040790
Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136:1005–1016. doi:10.1016/j.cell.2009.03.001
Visser EJW, Heijink CJ, Van Hout KJGM, Voesenek LACJ, Barendse GWM, Blom CWPM (1995) Regulatory role of auxin in adventitious root formation in two species of Rumex, differing in their sensitivity to waterlogging. Physiol Plant 93:116–122. doi:10.1034/j.1399-3054.1995.930117.x
Wabnik K, Kleine-Vehn J, Govaerts W, Friml J (2011) Prototype cell-to-cell auxin transport mechanism by intracellular auxin compartmentalization. Trends Plant Sci 16:468–475. doi:10.1016/jplants.2011.05.002
Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P (2009) Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2:823–831. doi:10.1093/mp/ssp023
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi:10.1093/oxfordjournals.molbev.a003851
Wu G, Lewis DR, Spalding EP (2007) Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19:1826–1837. doi:10.1105/tpc.106.048777
Xu Y, Zhang S, Guo H, Wang S, Xu L, Li C et al (2014) OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J 79:106–117. doi:10.1111/tpj.12544
Yu TA, Yeh SD, Cheng YH, Yang JS (2000) Efficient rooting for establishment of papaya plantlets by micropropagation. Plant Cell Tissue Organ Cult 61:29–35. doi:10.1023/A:10064-759-0143-9
Yue R, Tie S, Sun T, Zhang L, Yang Y, Qi J, Yan S, Han X, Wang H, Shen C (2015) Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 10:1–23. doi:10.1371/journal.pone.0118751
Zažímalová E, Křeček P, Skůpa P, Hoyerová K, Petrášek J (2007) Polar transport of the plant hormone auxin—the role of PIN-FORMED (PIN) proteins. Cell Mol Life Sci 64:1621–1637. doi:10.1007/s00018-007-6566-4
Zažímalová E, Murphy AS, Yang H, Hoyerová K, Hošek P (2010) Auxin transporters—why so many? Cold Spring Harb Perspect Biol 2:1–14. doi:10.1101/cshperspect.a001552
Acknowledgments
This work was funded by CONACYT, México (Project No. CB155356). H.E.M. acknowledges a scholarship (254647) granted by CONACYT.
Author contribution
E.M.H., First author, PhD student, performed the gene expression studies and the bioinformatic analysis; E.M.H., F.O.G., C.L.A., I.E.F. and R.Z.L. contributed to the writing of the paper; F.O.G. and I.E.F., supervision on the expression and bioinformatics analysis; C.L.A., Assisted in the gene expression studies; C.T.M. and E.G.F., Assisted in the tissue culture work, including the experimental set up for root induction; B.P.F., Responsible of the anatomical studies; S.J.M., Corresponding author. General conception of the project. Design of the experimental strategy, responsible for the writing of the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest and the presented work is compliant with ethical standards of PCTOC. All the authors read and approved the manuscript in its final form.
Rights and permissions
About this article
Cite this article
Estrella-Maldonado, H., Fuentes Ortíz, G., Chan León, A.C. et al. The papaya CpAUX1/LAX and CpPIN genes: structure, phylogeny and expression analysis related to root formation on in vitro plantlets. Plant Cell Tiss Organ Cult 126, 187–204 (2016). https://doi.org/10.1007/s11240-016-0989-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-0989-2