Abstract
Trypanosomes of anurans and fish are grouped into the Aquatic Clade which includes species isolated from fish, amphibians, turtles and platypus, usually transmitted by leeches and phlebotomine sand flies. Trypanosomes from Brazilian frogs are grouped within the Aquatic Clade with other anuran trypanosome species, where there seems to be coevolutionary patterns with vertebrate hosts and association to Brazilian biomes (Atlantic Forest, Pantanal and Amazonia Rainforest). We characterised the anuran trypanosomes from two different areas of the Cerrado biome and examined their phylogenetic relationships based on the SSU rRNA gene. A total of 112 anurans of six species was analysed and trypanosome prevalence evaluated through haemoculture was found to be 7% (8 positive frogs). However, only three isolates (2.7%) from two anuran species were recovered and cryopreserved. Analysis including SSU rDNA sequences from previous studies segregated the anuran trypanosomes into six groups, the previously reported An01 to An04, and An05 and An06 reported herein. Clade An05 comprises the isolates from Leptodactylus latrans (Steffen) and Pristimantis sp. captured in the Cerrado biome and Trypanosoma chattoni Mathis & Leger, 1911. The inclusion of new isolates in the phylogenetic analyses provided evidence for a new group (An06) of parasites from phlebotomine hosts. Our results indicate that the diversity of trypanosome species is underestimated since studies conducted in Brazil and other regions of the world are still few.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trypanosomes are protozoan haemoparasitic flagellates that infect all classes of vertebrates (fish, amphibians, reptiles, birds and mammals) with life-cycles involving alternation between vertebrate and invertebrate hosts. Most species develop in haematophagous arthropods, which may belong to different orders and families, whereas amphibian and fish parasites are transmitted by blood-sucking leeches and insects (Gruby, 1843; Hoare 1972; Vickerman 1994; Stevens et al., 2001; Cavalier-Smith, 2004; Hamilton et al., 2005; Simpson et al., 2006; Hamilton et al., 2007). Trypanosoma rotatorium (Mayer, 1843) isolated from Pelophylax esculentus L. (=Rana esculenta) is the first species described in the genus Trypanosoma Gruby, 1843. Trypanosomes of anurans exhibit high morphological and genetic diversity and are generally more complex and polymorphic than mammalian trypanosomes (Bardsley & Harmsen, 1973; Ferreira et al., 2007).
Traditionally, the phylogenies of Trypanosoma spp. based on SSU rRNA and gGAPDH genes segregated species into different clades which exhibit associations with hosts, geographical origin or host biology (Stevens et al., 2001; Hamilton et al., 2004; Marcili et al., 2013). The Aquatic Clade includes trypanosome species isolated from fish, amphibians, turtles and platypus that are generally transmitted by leeches and phlebotomines (Stevens et al., 2001; Hamilton et al., 2004; Simpson et al., 2006; Ferreira et al., 2007, 2008). Trypanosomes from Brazilian frogs are grouped within the Aquatic Clade together with other anuran trypanosome species, where there seems to be coevolutionary patterns with vertebrate hosts, and association to Brazilian biomes (Atlantic Forest, Pantanal and Amazonia Rainforest), though sampling gaps are huge (Ferreira et al., 2007; 2008).
In this study, we have collected and isolated anuran trypanosomes from frogs in two different areas of the Brazilian Cerrado biome. The new isolates were characterised molecularly based on the rRNA gene and their relationships with the other anuran trypanosomes from Brazil were inferred from phylogenetic analyses.
Materials and methods
Study area and animal capture
Anurans were captured in two areas in the Cerrado biome: an indigenous reserve for Tapirapé ethnicity, municipality of Confresa (10°38′22″S, 51°34′08″W), Mato Grosso State (average annual temperature 27°C; annual precipitation higher than 1,800 mm), and an island formed by damming the River San Francisco for the construction of hydroelectric reservoir Três Marias and transformed into conservation area (Ecological Station of Pirapitinga), municipality of Morada Nova de Minas (18°20′S–18°23′S; 45°17′W–45°20′W), Minas Gerais State (average annual temperature 22°C; annual precipitation 1,300 mm). Pittfall traps and manual capture were used to capture the anurans in two field trips (ten days each) in October 2013 and March 2014. Anurans were identified using identification keys and original descriptions and anesthetised. Blood samples were collected by means of heart puncture. Some specimens were collected as vouchers and deposited in museum herpetological collections. These specimens were killed by anesthetic overdose, fixed in formalin 10%, preserved in alcohol 70%, and deposited at the Museum of Zoology, University of São Paulo. All animals were captured and manipulated in accordance to the recommendations of the Brazilian Institute for the Environment and Renewable Natural Resources Chico Mendes Institute of Biodiversity Conservation (IBAMA ICMbio) and approved by the Animal Research Committee of the Faculty of Veterinary Medicine, University of São Paulo (FMVZ-USP), Brazil.
Isolation of anuran trypanosomes
For trypanosome isolation, blood samples from anurans were inoculated into Vacutainer tubes containing a biphasic medium consisting of 15% sheep red blood cells as the solid phase (blood agar base), overlain by liquid LIT medium supplemented with 20% FBS at ambient temperature (25–30°C) (Marcili et al., 2013). Epimastigote forms from positive cultures were used to infect monolayers of insect cells (SF9) in 25 cm2 flasks kept in TC100 medium containing 10% bovine calf serum at 28°C. The isolates were cryopreserved in liquid nitrogen in the Brazilian Trypanosomatid Collection (Coleção Brasileira de Tripanossomatídeos, CBT), Department of Preventive Veterinary Medicine and Animal Health, FMVZ-USP. The primary samples (blood) were fixed in ethanol for molecular detection.
Molecular data and phylogenetic analysis
DNA was extracted from the trypanosome culture samples using the phenol-chloroform method and the primary samples (blood) were purified using the Wizard DNA Clean-Up System (Promega, Madison, USA). The DNA samples were subjected to polymerase chain reaction (PCR) amplification for trypanosome barcode (V7–V8 region of SSU rDNA) according Marcili et al. (2013). PCR products of the expected size were purified and sequenced in an automated sequencer (ABI Prism 310). The nucleotide sequences generated were deposited in GenBank (Table 1).
The newly-generated sequences were aligned with sequences previously determined for other trypanosome species available in GenBank (Table 1) using ClustalX (Thompson et al., 1997) and were adjusted manually using GeneDoc (Nicholas et al., 1997). The alignment was used to construct phylogenetic trees using maximum parsimony, as implemented in PAUP version 4.0b10 (Swofford, 2002) with 500 bootstrap replicates and Bayesian analysis performed using MrBayes v3.1.2 (Huelsenbeck & Ronquist, 2001) with 1,000,000 replicates. The first 25% of the trees represented ‘burn-in’, and the remaining trees were used to calculate Bayesian posterior probabilities.
Results
A total of 112 anurans belonging to six species was captured (Table 2). Trypanosome prevalence, as evaluated through haemoculture, was 7% (n = 8). Positive haemocultures were recovered from Rhinella schneideri (Wemer), Leptodactylus latrans (Steffen), Leptodactylus fuscus (Schneider) and Pristimanis sp., but only three isolates (ex L. latrans and Pristimanis sp.) were established and cryopreserved in the Coleção Brasileira de Tripanossomatídeos (CBT) (Table 1). Parasites were initially grown in biphasic isolation media, but epimastigote forms started dying probably by nutritional requirements not supplied in the liver infusion tryptose (LIT) medium. The transfer of parasites to the monolayer of SF9 cells led to the growth and isolation of trypanosomes. Primary samples (blood) were obtained from only 57 anurans when collection of sufficient quantities for molecular studies was possible, with priority to haemoculture for parasite isolation. All samples were negative for trypanosome DNA barcoding, including the blood from the positive animals.
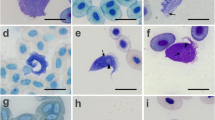
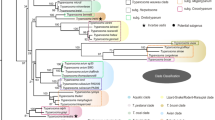
Sequences of the SSU rDNA (V7–V8 region) from three isolates recovered were aligned with sequences for different trypanosome species of the Aquatic Clade retrieved from GenBank (Table 1). The phylogenetic relationships based on SSU rDNA sequences inferred by maximum parsimony and Bayesian analysis corroborated the monophyly of the Aquatic Clade and the fish and anuran subclades in congruent topologies (Fig. 1). Species of Trypanosoma in the Anuran Clade (maximum support in both analyses) were segregated into six groups, An01 to An04, described previously (Ferreira et al., 2007), An06 comprising isolates from phlebotomines previously included in group An03 (Ferreira et al., 2008) and An05, a new clade, discovered in the present study. The latter comprises three isolates of anuran trypanosomes captured in the Cerrado biome (maximum node support, see Fig. 1); these exhibited intraspecific (An05) divergence of 0.46% and 3.62%, excluding and including T. chattoni, respectively. The divergence between An05 and the remaining groups within the Anuran Clade was 8.19%, 10.28%, 10.86%, 11.98% and 21.25% with An03, An06, An02, An01 and An04, respectively.
Maximum parsimony (MP) tree inferred from SSU rRNA gene sequences for 48 trypanosomes with Trypanosoma avium as an outgroup (827 characters; 296 parsimony-informative sites) with indication of the nodal support from Bayesian inference analysis. Numbers at nodes are the support values ordered as MP/BI. The sequences obtained in this study are indicated in bold. The scale-bar indicates the expected number of substitutions per site
Discussion
The determination of how many anuran trypanosomes represent valid species or lineages is a subject far from being solved. Different authors consider differently the validity of species and the morphological characters that should be used to classify anuran trypanosomes (Desser, 2001; Ferreira et al., 2007). Morphological polymorphism of blood forms is common, and anuran trypanosomes with distinct geographical origins or hosts can be morphologically similar (Desser, 2001; Martin et al., 2002; Lemos et al., 2008). More than 70 anuran trypanosomes are described with a worldwide distribution. However, world regions are far from being adequately sampled, isolates are difficult to obtain and parasite detection is difficult or imprecise because of the low levels of infection and the inadequacy of molecular markers for diversity exploration and phylogenetic studies.
Many phylogenetic studies have positioned the anuran trypanosomes into the Aquatic Clade together with fish, turtle and platypus trypanosomes transmitted by leeches (Stevens et al., 2001; Hamilton et al., 2004; Simpson et al., 2006; Ferreira et al., 2007; Bartlett-Healy et al., 2009; Paparini et al., 2014). However, anuran trypanosomes can also be transmitted by terrestrial leeches (Hamilton et al., 2005), phlebotomines (Ferreira et al., 2008) and mosquitoes (Bartlett-Healy et al., 2009). In Brazil, trypanosomes of anurans from different biomes are found and segregated into four subgroups (An01–An04) in relation to other species from North America, Europe and Africa based on phylogenetic analyses (Ferreira et al., 2007). Brazilian isolates from these groups seem to be related to host species and/or biomes (Atlantic Forest, Pantanal and Amazonia) (Ferreira et al., 2007). Samples originating from frogs captured at the Cerrado biome have never been included in a phylogenetic analysis; these were segregated into a new subgroup which we named An05. Isolates from phlebotomines previously included in An03 (Ferreira et al., 2008) were segregated in An06 subgroup. The groups An01 to An04 were recovered and their position within the anuran trypanosomes and inside the Aquatic Clade was confirmed.
The subgroups in the Anuran Clade do not exhibit clear association with different factors that can interfere with and modulate the evolution of the group. Previous studies conducted by Ferreira et al. (2007, 2008) suggest an association with the different biomes studied (Amazonia, Pantanal and Atlantic Forest) and/or host families. The results of our study also suggest an association with the biome of origin because of the differentiation of separate AN05 group from hosts captured in the Cerrado biome. However, the recovery of the new group An06 formed by trypanosomes isolated from phlebotomines suggests that evolutionary patterns in anuran trypanosomes can be associated with vectors/hosts.
In spite there appears a relation among the Brazilian anuran trypanosomes and their origin (Brazilian biomes), the correlations are still weak due to large sampling gaps. Amphibians have more restricted distributions compared to other vertebrates due to their restricted niches and limited dispersal (Buckley & Jetz, 2007; Smith & Green, 2005). The Neotropical region has the greatest diversity of frogs in the world and this diversity decreases from wet to dry areas, and with altitude (Buckley & Jetz, 2007). Roughly, these species can be split into two ecological groups, one associated with forests, and the other associated with open formations (Heyer, 1988). Yet, the variety of natural history and reproductive modes of the species is very high within these groups, and usually related to microhabitat specificity.
Anuran trypanosomes from Cerrado presented different nutritional requirements compared with the other already described groups, since they required insect cell monolayer for their development whereas the other groups have been cultured in LIT medium supplemented with bovine fetal serum. The LIT medium was originally described for the isolation of Trypanosoma cruzi (Chagas, 1909) and mimics the conditions found in the insect gut (Camargo, 1964); this medium has been effective in isolating various species of trypanosomes (Ferreira et al., 2007; Viola et al., 2009a, b). Haemocultures can select growth of the trypanosome species (Maia da Silva et al., 2009; Marcili et al., 2013).
The subgroup AN04 comprises species of trypanosomes described from Africa, Europe, Asia and North America, except for Trypanosoma chattoni (Mathis & Leger, 1911) that clustered within the An05 group. Trypanosoma chattoni was described from North America and has not been so far included in other phylogenetic clusters described (Ferreira et al., 2007, 2008). Despite the great morphological variability of blood trypomastigotes and cultured epimastigotes, parasites with morphology similar to T. chattoni have been described in other studies conducted in Brazil (Ferreira et al., 2007; Lemos et al., 2008).
The phylogenetic relationships and the position of anuran trypanosome subgroups are still provisional, since the inclusion of isolates from different geographical origins in the Americas and other continents, different anuran species with distinct microhabitats and vectors, may change the relationships and position of species in the phylogeny. The absence of clear evolutionary patterns reflects the small number of species or isolates of anuran trypanosomes described and included in phylogenetic studies compared to high diversity of frogs and phlebotomines and other possible vectors. The inclusion of new Brazilian isolates in phylogenetic analysis revealed greater than previously known diversity within the Anuran Clade. In addition to sampling effort, nutritional requirements represent another major factor for the isolation of frog trypanosomes. The use of new culture media in association with cell monolayers has increased the success of isolation and assessment of the diversity within the Anuran Clade. Further efforts should be concentrated on the isolation of trypanosomes from different species of frogs and vectors from already sampled or different locations but with culture media different from those used in previous studies.
References
Bardsley, J. E., & Harmsen, R. (1973). The trypanosomes of anura. Advances in Parasitology, 2, 1–73.
Bartlett-Healy, K., Crans, W., & Gaugler, R. (2009). Vertebrate Hosts and Phylogenetic Relationships of Amphibian Trypanosomes from a Potential Invertebrate Vector, Culex territans Walker (Diptera: Culicidae). Journal of Parasitology, 95, 381–387.
Buckley, L. B., & Jetz, W. (2007). Environmental and historical constraints on global patterns of amphibian richness. Proceedings of the Royal Society B: Biological Sciences, 274, 1167–1173.
Camargo, E. P. (1964). Growth and differentiation of Trypanosoma cruzi. I. Origin of metacyclic trypanosomes in liquid media. Revista do Instituto de Medicina Tropical de São Paulo, 6, 93–100.
Cavalier-Smith, T. (2004). Only six kingdoms of life. Proceedings of the Royal Society B: Biological Sciences, 271, 1251–1262.
Desser, S. S. (2001). The blood parasites of anurans from Costa Rica with reflections on the taxonomy of their trypanosomes. Journal of Parasitology, 87, 152–160.
Ferreira, R. C., Campaner, M., Viola, L. B., Takata, C. S. A., Takeda, G. F., & Teixeira, M. M. G. (2007). Morphological and molecular diversity and phylogenetic relationships among anuran trypanosomes from the Amazonia, Atlantic Forest and Pantanal biomes in Brazil. Parasitology, 134, 1623–1638.
Ferreira, R. C., Souza, A., Freitas, R. A., Campaner, M., Takata, C. S. A., Barretti, T. V., Shaw, J. J., & Teixeira, M. M. G. (2008). Phylogenetic lineage of closely related trypanosomes (Trypanosomatidae, Kinetoplastida) of anurans and sand flies (Psychodidae, Diptera) sharing the same ecotopes in Brazilian Amazonia. Journal of Eukaryotic Microbiology, 55(5), 427–445.
Gruby, M. (1843). Recherches et observations sur une nouvelle espèce d’hématozoaire, Trypanosoma sanguinis. Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences, 55, 1134–1136.
Hamilton, P. B., Stevens, J. R., Gaunt, M. W., Gidley, J., & Gibson, W. C. (2004). Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. International Journal for Parasitology, 34, 1393–1404.
Hamilton, P. B., Stevens, J. R., Gidley, J., Holz, P., & Gibson, W. C. (2005). A new lineage of trypanosomes from Australian vertebrates and terrestrial bloodsucking leeches (Haemadipsidae). International Journal for Parasitology, 35, 431–443.
Hamilton, P. B., Gibson, W. C., & Stevens, J. R. (2007). Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Molecular Phylogenetics and Evolution, 44, 15–25.
Heyer, W. R. (1988). On frog distribution patterns east of the Andes. In: Vanzolini, P. E. & Heyer, W. R. (Eds) Proceedings of a Workshop on Neotropical Distribution Patterns. Rio de Janeiro: Academia Brasileira de Ciências, pp. 245–273.
Hoare, C. A. (1972). The trypanosomes of mammals. A zoological monograph. Oxford, UK: Blackwell Scientific Publications, 748 pp.
Huelsenbeck, J. P., & Ronquist, F. (2001). MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Lemos, M., Morais, D. H., Carvalho, V. T., & D’Agosto, M. (2008). First record of Trypanosoma chattoni in Brazil and occurrence of other Trypanosoma species in Brazilian Frogs (Anura, Leptodactylidae). Journal of Parasitology, 94, 148–151.
Maia da Silva, F., Marcili, A., Lima, L., Cavazzana, M, Jr., Ortiz, P. A., Campaner, M., Takeda G. F., Paiva F., Nunes V. L., Camargo E. P., & Teixeira, M. M. G. (2009). Trypanosoma rangeli isolates of bats from Central Brazil: genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Tropica, 109, 199–207.
Marcili, A., Costa, A. P., Soares, H. S., Acosta, I. C. L., Lima, J. T. R., Minervino, A. H. H., Melo, A. T. L., Aguiar, D. M., Pacheco, R. C., & Gennari, S. M. (2013). Isolation and phylogenetic relationships of bat trypanosomes from different biomes in Mato Grosso, Brazil. Journal of Parasitology, 99, 1071–1076.
Martin, D. S., Wright, A. D. G., Barta, J. R., & Desser, S. S. (2002). Phylogenetic position of the giant trypanosomes Trypanosoma chanttoni, Trypanosoma fallisi, Trypanosoma mega, Trypanosoma neveulemairei, and Trypanosoma ranarum inferred from 18S rRNA gene sequences. Journal of Parasitology, 88, 566–571.
Nicholas, K. B., Nicholas, H. B, Jr., & Deerfield, D. W., II (1997). GeneDoc: Analysis and Visualization of Genetic Variation. Embnew News, 4, 14.
Paparini, A., Macgregor, J., Irwin, P. J., Warren, K., & Ryan, U. M. (2014). Novel genotypes of Trypanosoma binneyi from wild platypuses (Ornithorhynchus anatinus) and identification of a leech as a potential vector. Experimental Parasitology, 145, 42–50.
Simpson, A. G. B., Stevens, J. R., & Lukes, J. (2006). The evolution and diversity of kinetoplastid flagellates. Trends in Parasitology, 22, 168–174.
Smith, A. M., & Green, M. D. (2005). Dispersal and the metapopulation paradigm in amphibian ecology and conservation: are all amphibian populations metapopulations? Ecography, 28, 110–128.
Stevens, J. R., Noyes, H. A., Schofield, C. J., & Gibson, W. (2001). The molecular evolution of Trypanosomatidae. Advances in Parasitology, 48, 1–56.
Swofford, D. L. (2002). PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sunderland Massachusetts, USA: Sinauer Associates.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., & Higgins, D. G. (1997). The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882.
Vickerman, K. (1994). The evolutionary expansion of the trypanosomatid flagellates. International Journal for Parasitology, 24, 1317–1331.
Viola, L. B., Almeida, R. S., Ferreira, R. C., Campaner, M., Takata, C. S., Rodrigues, A. C., Paiva, F., Camargo, E. P., & Teixeira, M. M. (2009a). Evolutionary history of trypanosomes from South American caiman (Caiman yacare) and African crocodiles inferred by phylogenetic analyses using SSU rDNA and gGAPDH genes. Parasitology, 136, 55–65.
Viola, L. B., Attias, M., Takata, C. S., Campaner, M., De Souza, W., Camargo, E. P., & Teixeira, M. M. (2009b). Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake Trypanosomes: Trypanosoma serpentis n. sp. from Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. Journal of Eukaryotic Microbiology, 56, 594–602.
Acknowledgements
We are grateful to several people from the ESEC Pirapitinga for their invaluable help during frog collection. This research was financially supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). SMG and AHHM benefit from fellowships from CNPq, JIGS from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and AM and APC from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
da S. Ferreira, J.I.G., da Costa, A.P., Ramirez, D. et al. Anuran trypanosomes: phylogenetic evidence for new clades in Brazil. Syst Parasitol 91, 63–70 (2015). https://doi.org/10.1007/s11230-015-9558-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-015-9558-z