Abstract
The reactions of N-alkyliminodiacetamide derivatives, namely N-ethyliminodiacetamide (CH3CH2N(CH2CONH2)2; Etimda) and N-isopropyliminodiacetamide (CH3)2CHN(CH2CONH2)2; i-Primda), with sodium tetrachloropalladate(II) in aqueous solutions were investigated. Three new palladium(II) complexes, [Pd(Etimda−H)2]∙2H2O (1), [Pd(i-Primda−H)2]∙2H2O (2) and [PdCl2(i-Primda)] (3), were obtained and characterized by X-ray structural analysis, infrared spectroscopy and thermal analysis (TGA). The square planar coordination environments around the palladium(II) ions in complexes 1 and 2 consist of two N,N′-bidentate N-alkyliminodiacetamidato ligands, with imino N atoms in trans-position. The complex 3 also exhibits a square planar coordination environment around Pd(II), but with two chloride ions and one neutral N-isopropyliminodiacetamide ligand bound in an N,O′-bidentate coordination mode. The described coordination modes, as well as the presence of deprotonated amide groups in ligands in 1 and 2, are found for the first time in palladium(II) complexes with iminodiacetamide type ligands. The molecular geometries and infrared spectra of these three complexes were also modelled using DFT calculations, at the BP86-D3/def2-TZVPP/PCM level of theory. The RMSD values suggest a good agreement of the calculated and experimental geometries. A QTAIM analysis suggests a qualitative correlation between bond lengths and energy densities, also supported by an NBO analysis. The dimer interaction energy between complex units was estimated at about −15 kcal/mol for all complexes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An iminodiacetamide, HN(CH2CONH2)2 (imda), is an amide derivative of the well-known complexone iminodiacetic acid, HN(CH2COOH)2 (idaH2) [1]. Being a neutral ligand, iminodiacetamide forms cationic complexes with copper(II) and nickel(II), [M(imda)2]2+ (M = Cu(II) of Ni(II)) [2, 3], but neutral one with palladium(II), [PdBr2(imda)2] [4]. The last one is, at the same time, the only example of a palladium(II) complex with iminodiacetamide type ligand that has been structurally described until now. This is surprising, since palladium(II) complexes are known to be highly efficient catalysts in modern synthetic organic chemistry [5, 6] and many of them also show remarkable antitumor potential [7]. Transition metal complexes with N-alkyliminodiacetamides and structurally related ligands [8,9,10,11,12,13,14,15,16,17,18,19,20] are generally scarce in the literature.
In order to get more insight into coordination chemistry of the palladium(II) complexes with iminodiacetamide type ligands, we have prepared three new palladium(II) complexes with N-ethyliminodiacetamide and N-isopropyliminodiacetamide (Scheme 1): [Pd(Etimda−H)2]∙2H2O (1), [Pd(i-Primda−H)2]∙2H2O (2) and [PdCl2(i-Primda)] (3). These complexes significantly differ from [PdBr2(imda)2], the only palladium(II) complex with iminodiacetamide type ligand reported by now [4], which contains a neutral ligand bound in an N-monodentate fashion via imino N atom of imda ligand. For the first time, the palladium(II) complexes with deprotonated amide groups of iminodiacetamide type ligands (Etimda in 1 and i-Primda in 2) were reported, exhibiting an N,N′-bidentate coordination mode of iminodiacetamide type ligands. The complex 3 also differs from [PdBr2(imda)2] and both 1 and 2, as it contains neutral i-Primda ligand bound in an N,O′-bidentate coordination mode. The studies based on comparison of experimental and calculated molecular structures and infrared spectra of the palladium(II) complexes with iminodiacetamide type ligands have not been reported in literature until now.
Experimental
Materials and physical measurements
All chemicals for the syntheses were purchased from commercial sources (Sigma-Aldrich, Acros Organics or Alfa Aesar) and used as received without further purification. The ligands were prepared as previously reported [11]. The CHN analyses were performed on a Perkin-Elmer 2400 Series II CHNS analyzer in the Analytical Services Laboratories of the Ruđer Bošković Institute, Zagreb, Croatia. The FTIR spectra were obtained in the range 4000–400 cm−1 on a Perkin-Elmer Spectrum Two™ FTIR-spectrometer in the ATR mode. The TGA measurements were performed at a heating rate of 10 °C min−1 in the temperature range of 25–600 °C, under oxygen flow of 150 mL min−1 on a Mettler-Toledo TG/SDTA 851e instrument. Approximately 10 mg of sample was placed in a standard aluminium crucible (40 μL).
Synthesis of the complexes [Pd(Etimda−H)2]∙2H2O (1) and [Pd(i-Primda−H)2] . 2H2O (2)
The ligand (1 mmol) was added to an aqueous solution of Na2PdCl4∙xH2O (x ≈ 3) (0.17 g; 0.5 mmol in 40 mL) and the mixture was heated to 80 °C, after which a clear red-orange solution was obtained. Sodium hydrogencarbonate (0.08 g; 1 mmol) was then added with stirring and the solution became yellow. Yellow crystals of 1 and 2, suitable for X-ray structural analysis, were obtained by standing of the above-described reaction mixtures for 1 day at room temperature. The complexes were filtered off, washed with water (10 mL) and dried in air.
[Pd(Etimda−H)2]∙2H2O (1): From 0.16 g Etimda. Yield: 0.17 g (74%). Anal. Calc. for C12H28N6O6Pd (458.80): C 31.41, H 6.15, N 18.32%. Found: C 31.51, H 6.28, N 18.19%. IR data (cm−1): 3381(s), 3326(s, br), 3274(s), 3077(w), 2985(w), 2952(w), 1688(s), 1634(m), 1589(vs), 1465(w), 1441(m), 1426(m), 1410(m), 1385(w), 1372(w), 1340(w), 1319(w), 1300(m), 1272(w), 1164(w), 1132(w), 1103(w), 1066(w), 1016(w), 976(w), 954(w), 920(w), 892(w), 848(w), 793(w), 750(w), 702(w), 677(w), 581(w), 519(w), 465(w).
[Pd(i-Primda−H)2]∙2H2O (2): From 0.17 g i-Primda. Yield: 0.19 g (77%). Anal. Calc. for C14H32N6O6Pd (486.85): C 34.54, H 6.62, N 17.27%. Found: C 34.76, H 6.88, N 17.09%. IR data (cm−1): 3422(m, br), 3334(s), 3217(s, br), 3094(s, br), 3005(w), 2988(m), 2954(w), 1686(vs), 1650(m), 1589(vs), 1463(w), 1439(m), 1427(m), 1409(m), 1395(m), 1372(w), 1340(w), 1315(m), 1276(m), 1265(m), 1177(w), 1152(m), 1133(m), 1072(w), 1030(w), 996(w), 949(w), 917(w), 902(w), 800(m), 749(w), 720(m), 688(w), 665(m), 597(m), 561(w), 529(w), 482(w).
Synthesis of the complex [PdCl2(i-Primda)] (3)
The ligand (1 mmol; 0.17 g) was added to an aqueous solution of Na2PdCl4∙xH2O (x ≈ 3) (0.34 g; 1 mmol in 40 mL) and the mixture was heated for few minutes, until a clear red-brown solution was obtained. Red-brown crystals of the product, suitable for X-ray structural analysis, were obtained by the slow evaporation of the reaction mixture for 1 week. The product was filtered off and washed with water (10 mL). Yield: 0.25 g (71%). Anal. Calc. for C7H15N3O2Cl2Pd (350.52): C 23.98, H 4.31, N 11.99%. Found: C 24.16, H 4.40, N 12.11%. IR data (cm−1): 3371(s, br), 3307(m), 3231(m), 3131(m), 2993(w), 2977(w), 2965(w), 1676(vs), 1652(vs), 1603(m), 1572(s), 1482(w), 1461(w), 1436(w), 1416(m), 1396(w), 1376(w), 1362(w), 1332(w), 1314(m), 1260(w), 1175(w), 1148(w), 1132(w), 1061(w), 1035(w), 983(w), 951(w), 935(w), 906(w), 895(w), 879(w), 807(w), 754(w), 704(w), 630(w), 608(w), 578(m), 508(w).
X-ray crystallographic analysis
The suitable single crystal of 1–3 was selected and mounted in Paratone-N oil onto a thin glass fibre. The data collection was carried out on an Oxford Diffraction Xcalibur four-circle kappa geometry diffractometer with Xcalibur Sapphire 3 CCD detector, using a graphite monochromated MoKα (λ = 0.71073 Å) radiation at room temperature (296(2) K) and by applying the CrysAlisPro Software system, Version 1.171.37.35 [21]. The data reduction and cell refinement were performed by the same program. The structures were solved by SHELXS-2014 and refined by SHELXL-2014 [22]. The refinement procedure was done by full-matrix least-squares methods based on F 2 values against all reflections. The figures were made with MERCURY (Version 3.5.1) [23]. The crystallographic data for 1–3 are summarized in Table 1.
Computational details
In our previous work, the BP86-D3/6-311 + G(d,p)/SDD level of theory was used to model IR spectra of similar compounds [24]. For complexes modelled in this work, the ECP basis set did not prove useful, as too much mixing between the ECP orbitals and valence orbitals was detected, which resulted in problems with SCF convergence. Instead, we used Alrichs’ balanced def2 basis sets [25, 26]. First, a well-behaved wave function was constructed using a single-point BP86-D3/def2-TZVP calculation. This result was then used as a guess wave function for further calculations. The functionals BP86-D3, B3LYP-D3 and mPW1PW91 were tested, along with def2-TZVP and def2-TZVPP basis sets [27,28,29,30,31]. In cases where the conventional SCF procedure failed, the quadratically convergent SCF procedure was used [32].
The starting geometries for all modelled complexes were obtained from crystallographic data. As complexes 1 and 2 are hydrates, water molecules present in the crystal structure were added to the geometries. In case of 3, only a single monomer unit was considered. The optimizations were carried out both in vacuo and using the PCM model [33] and water as an implicit solvent, and were followed by frequency calculations carried out at the same level of theory. All calculations were performed using the Gaussian 09 program package [34].
QTAIM analysis was done on optimized geometries using AIMAll [35], while the single NBO calculations were performed with the NBO 6 program [36] at the BP86-D3/def2-TZVPP/PCM level of theory. Energies of interaction between two complex units were estimated at the same model chemistry by optimizing dimers of complexes, with the starting geometries taken from crystallographic data.
Results and discussion
Synthesis and general properties of the complexes
N-alkyliminodiacetamide derivatives contain three potential donor atoms, imino-nitrogen and two amide nitrogen or oxygen atoms. However, their reactions with sodium tetrachloropalladate(II) lead to the formation of complexes with different chemical composition and molecular geometry, depending on the reaction conditions. The reactions of sodium(II) tetrachloropalladate(II) with these ligands and NaHCO3 in molar ratio 1:2:2 (Scheme 2, a) resulted in formation of the complexes of the formula [Pd(Rimda−H)2]∙2H2O (R = Et or i-Pr). In the absence of any additional base, the reaction of Na2PdCl4 with i-Primda gave the complex [PdCl2(i-Primda)] (Scheme 2, b), but no such complex could be isolated (possibly due to greater solubility) from the analogous reaction mixture containing Etimda instead of i-Primda.
All three complexes are stable substances which are practically insoluble in water. Thermal decomposition of 1 and 2 is characterized by dehydration above 100 °C and is finished around 150–170 °C. Complex 3, as well as 1 and 2 after their dehydration, decompose in one step which corresponds to the loss of the coordinated ligands. The amount of undecomposed residue in oxygen atmosphere is in good accordance with the calculated amount of PdO. Full thermal analysis data for these complexes are given in Supplementary material.
Crystal structures
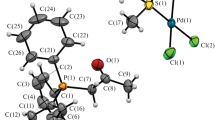
The palladium(II) ion is placed at the inversion centre and coordinated by two deprotonated N,N′-bidentate Etimda−H ligands in 1 or i-Primda−H ligands in 2, resulting with the square planar coordination and the formation of trans isomers in both cases (∠(N1–Pd1–N1i), 180° in 1 and 2; symmetry code ('): −x + 1, −y + 1, −z + 1 (1) or −x + 1, −y + 1, −z (2)). The Etimda−H and i-Primda−H ligands are coordinated to palladium(II) ion via imino N1 and amide N2 atoms (Fig. 1a, b, Table 2). The τ4 index [37] amounts 0 for both 1 and 2. The palladium(II) ion in 3 is coordinated in a square planar fashion by a neutral N,O′-bidentate i-Primda ligand and two chloride atoms Cl1 and Cl2 in cis position (∠(Cl1–Pd1–Cl2), 90.52(3)°). The i-Primda ligand is coordinated via imino N1 and amide O1 atoms (Fig. 1c, Table 2). The τ4 index for 3 amounts 0.04 indicating almost perfect square planar geometry.
The crystal packings of 1 and 2 are very similar. The molecules of [Pd(Etimda−H)2] (in 1) or [Pd(i-Primda−H)2] (in 2) and water molecules of crystallization in the respective crystal structures are assembled together into an infinite 3D framework by intermolecular N–H⋅⋅⋅O and O–H⋅⋅⋅O hydrogen bonds (Fig. 2a, b, Table 3). Each [Pd(Etimda−H)2] molecule is hydrogen-bonded to four neighbouring [Pd(Etimda−H)2] molecules and to eight neighbouring water molecules, while each water molecule is hydrogen-bonded to four neighbouring [Pd(Etimda−H)2] molecules in the crystal structure of 1. In the crystal structure of 2, each [Pd(i-Primda−H)2] molecule is hydrogen-bonded to four neighbouring [Pd(i-Primda−H)2] molecules and to six neighbouring water molecules, while each water molecule is hydrogen-bonded to three neighbouring [Pd(i-Primda−H)2] molecules. The [PdCl2(i-Primda)] molecules in the crystal structure of 3 are assembled into an infinite 2D network in the (1 0 0) plane by intermolecular N–H⋅⋅⋅O and N–H⋅⋅⋅Cl hydrogen bonds (Fig. 3, Table 3). Each [PdCl2(i-Primda)] molecule is hydrogen-bonded to seven neighbouring molecules.
The packing diagrams of [Pd(Etimda−H)2]∙2H2O (1) (a) and [Pd(i-Primda−H)2]∙2H2O (2) (b) (a view in the (1 0 0) plane). The molecules of [Pd(Etimda−H)2] (a) or [Pd(i-Primda−H)2] (b) and water molecules of crystallization are assembled together into an infinite 3D framework by intermolecular N–H⋅⋅⋅O and O–H⋅⋅⋅O hydrogen bonds (shown by the dotted lines)
Analysis and comparison of the experimental and calculated infrared spectra and structures
Infrared spectra of all three complexes in the solid state (Fig. 4) are characterized by the presence of very strong and sharp bands of carbonyl group stretching, ν(C=O) [38, 39]. Two bands were found in similar spectral regions in the spectra of 1 (1688, 1634 cm−1), 2 (1686, 1650 cm−1) and 3 (1676, 1652 cm−1), indicating the presence of two non-equivalent carbonyl groups (coordinated and non-coordinated) in the molecules of these complexes. Two pairs of characteristic bands of medium intensity, assigned as antisymmetric (ν a) and symmetric stretching (ν s) of the amide NH2 groups, were observed in the range of 3100–3400 cm−1 for complex 3. However, only three such bands appeared in the spectra of 1 and 2 since one of the four amide hydrogen atoms is replaced by palladium atom upon complexation. Clearly, two types of spectra, belonging to one of two kinds of the complexes, can be distinguished. The indication of the water molecules of crystallization in the hydrates 1 and 2 is due to the appearance of a broad band of medium intensity, observed at 3381 cm−1 (1) and 3422 cm−1 (2), which was assigned as O−H stretching, ν(OH, H2O). The band is absent in the spectra of anhydrous complex 3.
Of all tested methods, BP86-D3/def2-TZVPP in implicit solvent showed best correlation with experimental spectral data. This is not surprising, as the BP86 functional in combination with Alrichs’ basis sets and empirical dispersion corrections has been often used in recent publications [29, 40, 41]. The comparison of experimental and calculated spectra was done by analysing the signals between 800 and 3100 cm−1 from experimental spectra. The calculated harmonic vibrational frequencies were matched with experimental values using linear regression. In this way, a relationship υ exp = (1.004 ± 0,002)υ calc + (9 ± 3)cm−1 between experimental and calculated frequencies was obtained. This method produced a mean unsigned error of 8.8 cm−1 between experimental and predicted frequencies below 3100 cm−1, as shown in Table 4. For frequencies above 3100 cm−1, describing the vibrations of H bonded groups, the discrepancies were larger since only a single molecule of the modelled complex was included in the calculation. In case of compound 1, the frequencies of peaks at 3381 (ν(OH)) and 3274 cm−1 (ν s (NH2), ν(OH)) were underestimated in silico by about 100 cm−1, corresponding to 3230 and 3188 cm−1. On the other hand, good agreement was found in case of 2, where the experimental peaks at 3422 (ν(OH)), 3334 (ν s (NH2)) and 3217 cm−1 (ν(OH)) corresponded to 3426, 3318 and 3195 cm−1, respectively. In case of anhydrous 3, the errors were largest—the ν(NH) and ν(NH2) frequencies were not dampened by any intramolecular interactions and thus were grossly overestimated.
Under 3100 cm−1, the agreement was much better. All peaks corresponding to carbonyl stretching compounds were found in calculated spectra, with their intensities roughly corresponding to experimental ones (see Fig. 4). This trend remained in most of the other considered peaks describing the C–H, C–C and C–N bends. The small errors there can be attributed to the lack of anharmonic corrections [42,43,44].
For further validation, structures obtained by optimization were compared to experimental ones. Their overlay is shown in Fig. 5, and the comparison between geometric parameters in Table 2. The obtained RMSD values show acceptable agreement, suggesting that a single molecule of the complex can be used in DFT calculations [45, 46].
QTAIM and NBO analysis
Using Natural Bond Orbital (NBO) analysis, the donor-acceptor relationship between the metal and the neighbouring ligands was further studied at the BP86-D3/def2-TZVPP/PCM level of theory. The relevant localized orbitals are shown in Fig. 6. Interaction energies between NBOs were calculated using second-order perturbative analysis of donor-acceptor interactions, E2PERT [47]. It suggests strong interactions between orbitals where N, O and Cl are the donors, while the acceptors are the vacant orbitals on Pd. For complexes 1 and 2, the best donors are the nitrogen lone pair orbitals (150–100 kcal/mol for 1 and 50–100 kcal/mol for 2), followed by Pd–N bonding orbitals (25–40 kcal/mol). In case of complex 3, nitrogen and oxygen are much better donors (55–60 kcal/mol) than chlorine (10–15 kcal/mol).
Bader’s quantum theory of atoms-in-molecules (QTAIM) was used for a topological analysis of electronic density. In QTAIM, the electron density ρ values in bond critical points (BCPs) can be used to estimate the strength of the bond—ρ BCP values on the order of 0.1 indicate a covalent (or similar strong) interaction, while weaker interactions (such as hydrogen bonds) typically have ρ BCP values above 0.001 [48, 49].
Figure 7 and Table 5 show the results of the QTAIM analysis, done at the BP86-D3/TZVPP/PCM level of theory. Generally, all of the ρ BCP values shown in Table 5 are consistent with strong coordinative bonds. Both in complexes 1 and 2, the N2 and N2’ atoms are more strongly bound to Pd than N1 and N1’. In complex 2, the ρ BCP values are larger than the corresponding ones in complex 1, which is in agreement with the donor-acceptor stabilization energy trends obtained with E2PERT analysis. This is also observed in complex 3, where the ρ BCP values for N and O bonds are larger than for Cl1 and Cl2. Also, the ρ BCP values are qualitatively correlated to bond lengths (shorter bonds correspond to larger ρ BCP values), which was reported previously [50]. When considering hydrogen bonds between complexes and water molecules (present in 1 and 2), all of the ρ BCP values are between 0.0200 and 0.0300 and all the ∇2 ρ BCP values are positive, suggesting typical closed-shell interactions.
Complex dimerization energy estimation
As it can be seen from the above discussion, the modelling of a single unit of the complex can provide a lot of insight into its vibrational and electronic structure. However, the crystal structure is largely influenced by the interactions between neighbouring complex units of the investigated molecule. In order to estimate the energy of interaction between two units of the complex, the energy of the reaction 2 Complex → (Complex)2 was calculated using the energies obtained by optimization at the BP86-D3/def2-TZVPP level of theory with water as the implicit (PCM) solvent. In case of 1 and 2, monomeric units with 6 water molecules (previously used for vibrational analysis) were fused, resulting in products with 12 water molecules, one of which bridges the two molecules of the complex. The dimer products are shown in Fig. 8. Obtained dimerization energies are similar: −17.26, −14.63 and −16.93 kcal/mol for compounds 1, 2 and 3, respectively. This is consistent with the formation of three new hydrogen bonds, as indicated in Fig. 8.
Conclusions
In the presence of a mild base, complexes [Pd(Etimda−H)2]∙2H2O (1) and [Pd(i-Primda−H)2]∙2H2O (2) were obtained, containing two deprotonated amide ligands bound in an N,N′-bidentate coordination mode. In the absence of a mild base, complex [PdCl2(i-Primda)] (3) was produced, containing a neutral amide ligand bound in an N,O′-bidentate coordination mode. These coordination modes were found for the first time in palladium(II) complexes with iminodiacetamide type ligands, as the only known coordination mode in such type of complexes was N-monodentate via imino N atom [4]. Also, complexes 1 and 2 are the first palladium(II) complexes containing deprotonated amide groups of iminodiacetamide type ligands.
The substituents at imino N atom in the ligands (ethyl vs. i-propyl) do not have any influence on the kind of complexes prepared, as the type of complexes depends on the basicity of the reaction solution. Moreover, the substituents at imino N atom in the ligands do not have a significant influence on the crystal packings, apparent from very similar crystal structures of 1 and 2. This is not very surprising, as ethyl and i-propyl groups are weak proton donors and participate only in weak hydrogen bonding, thus not affecting significantly the respective crystal packings of 1 and 2. The complex molecules and water molecules of crystallization of 1 and 2 are assembled into 3D framework by a complex network of N–H⋅⋅⋅O and O–H⋅⋅⋅O hydrogen bonds, very similar to the crystal structures of [Pd(HBnida)2]∙2H2O [51] and [Pd(i-PridaH)2]∙2H2O [52]. However, in the case of 3, molecules are connected into 2D network. This difference can be hardly attributed to the presence of weaker N–H⋅⋅⋅Cl hydrogen bonds in the crystal structure of 3, but rather to the absence of water molecules of crystallization, leading to a less complicated structure due to the decrease of potential proton donors and acceptors.
The structure and spectra of the complexes were additionally validated using BP86-D3/def2-TZVPP/PCM level of theory. A very good agreement between experimental and DFT calculated results for both geometries and harmonic frequencies was achieved in the case of all three complexes.
References
Bell CF (1977) Principles and applications of metal chelation. Oxford Chemistry Series, Clarendon Press, Oxford University Press, Oxford
Sekizaki M (1974). Bull Chem Soc Jpn 47:1447–1450
Sekizaki M (1976). Acta Crystallogr B 32:1568–1570
Sekizaki M (1981). Bull Chem Soc Jpn 54:3861–3862
Clayden J, Greeves N, Warren S, Wothers P (2001) Organic chemistry. Oxford University Press, Oxford,
Tsuji J (2004) Palladium reagents and catalysts. Wiley, Chichester,
Abu-Surrah AS, Al-Sa’doni HH, Abdalla MY (2008). Cancer Therapy 6:1–10
Smrečki N, Kukovec B-M, Đaković M, Popović Z (2015). Polyhedron 93:106–117
Smrečki N, Stilinović V, Merkaš M, Lučić A, Kukovec B-M, Popović Z (2016). Aust J Chem 69:896–904
Smrečki N, Stilinović V, Jović O, Kukovec B-M, Popović Z (2017). Inorg Chim Acta 462:57–63
Smrečki N, Jović O, Molčanov K, Kukovec B-M, Kekez I, Matković-Čalogović D, Popović Z (2017). Polyhedron 130:115–126
Ran J-W, Pei J (2013). Acta Crystallogr E69:m325–m325
Deng X-H, Nie Q-J, Zhu F-J (2013). Acta Crystallogr E69:m89–m89
Clapp LA, Siddons CJ, Van Deerver DG, Reibenspies JH, Jones SB, Hancock RD (2006) Dalton Trans 2001–2007
Smith DA, Sucheck S, Pinkerton A (1992) J Chem Soc Chem Commun 367–368
Claudio ES, Horst MA, Forde CE, Stern CL, Zart MK, Godwin HA (2000). Inorg Chem 39:1391–1397
Clapp LA, Siddons CJ, Whitehead JR, Van Derveer DG, Rogers RD, Griffin ST, Jones SB, Hancock RD (2005). Inorg Chem 44:8495–8502
Burdinski D, Pikkemaat JA, Lub J, de Peinder P, Garrido LN, Weyhemuller T (2009). Inorg Chem 48:6692–6712
Krot K, Liamas-Saiz AL, Vembu N, Nollan KB (2007). Z Anorg Allg Chem 633:1900–1910
Skrzypczak-Jankun E, Smith DA (1994). Acta Crystallogr C50:1585–1588
CrysAlisPro (2014) Agilent Technologies, Yarnton, Oxfordshire, England. Version 1.171.37.35
Sheldrick GM (2008). Acta Crystallogr A64:112–122
Macrae CF, Bruno IJ, Chisholm JA, Edgington PR, McCabe P, Pidcock E, Rodriguez-Monge L, Taylor R, van de Streek J, Wood PA (2008). J Appl Crystallogr 41:466–470
Smrečki N, Rončević I, Popović Z (2016). Aust J Chem 69:1285–1291
Weigend F, Ahlrichs R (2005). Phys Chem Chem Phys 7:3297–2305
Weigend F (2006). Phys Chem Chem Phys 8:1057–1065
Cramer CJ, Truhlar DG (2009). Phys Chem Chem Phys 11:10757–10816
Steinmetz M, Grimme S (2013). ChemistryOpen 2:115–124
Raoufmoghaddam S, Mannathan S, Minnaard AJ, de Vries JG, Reek JN (2015). Chemistry 21:18811–18820
Goerigk L, Grimme S (2011). Phys Chem Chem Phys 13:6670–6688
Tsipis AC (2014). Coord Chem Rev 272:1–29
Schlegel HB, McDouall JJ (1991) In: Ögretir C, Csizmadia IG (eds) Computational advances in organic chemistry. Kluwer Academic, The Netherlands, pp. 167–185
Tomasi J, Mennucci B, Cammi R (2005). Chem Rev 105:2999–3093
Gaussian 09, Revision A.02, Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich A, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov AF, Sonnenberg JL, Williams-Young D, Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Keith T, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, Fox DJ (2016) Gaussian, Inc., Wallingford
AIMAll (Version 17.01.25), Todd A. Keith (2017) TK Gristmill Software, Overland Park KS, USA, (aim.tkgristmill.com)
NBO 6.0., Glendening ED, Badenhoop JK, Reed AE, Carpenter JE, Bohmann JA, Morales CM, Landis CR, Weinhold F (2013) Theoretical Chemistry Institute, University of Wisconsin, Madison
Yang L, Powell DR, Houser RH (2007) Dalton Trans 955–964
Silverstein RM, Webster FX, Kiemle D (2005) Spectrometric identification of organic compounds, 7th edn. Wiley, New York,
Nakamoto K (2009) Infrared and Raman spectra of inorganic and coordination compounds, part B, 6th edn. Wiley, Hoboken,
Pandey KK, Patidar P, Vishwakarma R (2014). Eur J Inorg Chem 18:2916–2923
Weymuth T, Couzijn EPA, Chen P, Reiher M (2014). J Chem Theory Comput 10:3092–3103
Pirc G, Stare J, Mavri J (2010). J Chem Phys 132:224506–224513
Stare J, Mavri J, Grdadolnik J, Zidar J, Maksić ZB, Vianello R (2011). J Phys Chem B 115:5999–6010
Brela M, Stare J, Pirc G, Sollner-Dolenc M, Wojcik MJ, Mavri J (2012). J Phys Chem B 116:4510–4518
Smrečki N, Jović O, Stilinović V, Kukovec B-M, Đaković M, Popović Z (2016). Inorg Chim Acta 453:95–103
Kukovec B-M, Kodrin I, Mihalić Z, Popović Z (2011). Inorg Chim Acta 378:154–162
Weinhold F (1998) Natural Bond Orbital Methods. In: Schleyer P v R (ed) Encyclopedia of Computational Chemistry, vol 3, pp. 1792–1811
Bader RFW (1990) Atoms in molecules: a quantum theory. Oxford University Press, Oxford,
Koch U, Popelier PLA (1995). J Phys Chem 99:9747–9754
Dimić D, Petković M (2015). Int J Quantum Chem 116:27–34
Smrečki N, Kukovec B-M, Jazwinski J, Liu Y, Zhang J, Mikecin A-M, Popović Z (2014). J Organomet Chem 760:224–230
Smrečki N, Kukovec B-M, Rotim K, Oršolić D, Jović O, Rončević T, Režić Mužinić N, Vinković M, Popović Z (2017). Inorg Chim Acta 462:64–74
Acknowledgements
This research was supported by the Foundation of the Croatian Academy of Sciences and Arts (year 2015/2016.) and the Croatian Science Foundation (Grant No. 7444).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
GA curves for the complexes (recorded in the oxygen). Mol2 files containing energies and geometries of the modelled complexes 1–3.
CCDC 1548186-1,548,188 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif (or from the Cambridge Crystallographic Data Centre (CCDC), 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44(0)1223-336,033; email: deposit@ccdc.cam.ac.uk).
ESM 1
(DOC 100 kb)
Rights and permissions
About this article
Cite this article
Smrečki, N., Kukovec, BM., Rončević, I. et al. New coordination modes of iminodiacetamide type ligands in palladium(II) complexes: crystallographic and DFT studies. Struct Chem 29, 195–206 (2018). https://doi.org/10.1007/s11224-017-1018-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-017-1018-8