Abstract
Citations play a fundamental role in supporting authors’ contribution claims throughout a scientific paper. Labelling citation instances with different function labels is indispensable for understanding a scientific text. A single citation is the linkage between two scientific papers in the citation network. These citations encompass rich native information, including context of the citation, citation location, citing and cited paper titles, DOI, and the website’s URL. Nevertheless, previous studies have ignored such rich native information during the process of datasets’ accumulation, thereby resulting in a lack of comprehensive yet significantly valuable features for the citation function classification task. In this paper, we argue that such important information should not be ignored, and accordingly, we extract and integrate all of the native information features into different neural text representation models via trainable embeddings and free text. We first construct a new dataset entitled, NI-Cite, comprising a large number of labelled citations with five key native features (Citation Context, Section Name, Title, DOI, Web URL) against each dataset instance. In addition, we propose to exploit the recently developed text representation models integrated with such information to evaluate the performance of citation function classification task. The experimental results demonstrate that the native information features suggested in this paper enhance the overall classification performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A scientific paper does not stand alone. The reference is the acknowledgment that one document gives to another, whereas, a citation is the acknowledgment that one document receives from another (Narin, 1976). Citations in the online scientific publications reveal authors’ rationale about the cited article (Smith, 1981). Citations play an indispensable role in supporting authors’contributions throughout a scientific paper. Citation (and its context) primarily refers to the text encompassing a citation sign employed for referring to relevant scientific literature (Bornmann & Daniel, 2008). It presents a useful opportunity to ascertain the salient contributions of the referred scientific publication in a scientific paper and itself possesses rich semantic information (Abu-Jbara & Radev, 2012). Over the past decade or so, a number of research scholars have conducted thorough investigations in the promising paradigm of citation content from diverse perspectives which could be summarized into different areas—citation sentiment (Yousif et al., 2019), citation recommendation (Färber & Jatowt, 2020), citation function (Teufel et al., 2006), and citation summarization (Cohan & Goharian, 2018).
Among all the above tasks, citation function classification is an indispensable constituent of the citation context analysis. The citation function refers to the significance of cited literature in the citing literature (Moravcsik & Murugesan, 1975). Different citations provide different functions in a scientific publication. Citations may serve as:
-
1.
An introductory information for referring to a certain concept, e.g., in “... pre-trained sentence encoders such as ELMo [1] and BERT [2] have rapidly ...”, BERT is cited as a concept.
-
2.
An adaptation of newly proposed technical basis, e.g., in “... this work adopts BERT [10] as the base model as it achieves the state-of-the-art performance on MRC”, BERT is served as a technical basis.
Valenzuela and Etzioni (2015) and Hassan et al. (2017) address the problem of classifying cited work into important and non-important ones primarily based on the citation functions. They believe if a citing paper used or extended a cited paper’s work, then this particular citation is of higher significance. On the contrary, if a cited paper only employs a cited paper as a reference for related work, then the citation is regarded as less significant. It can be noticed from the second sentence that the cited BERT is playing a more important role than the former one, i.e., not all citations are equal in the scientific paper (Valenzuela & Etzioni, 2015). With different types of citation functions, more fine-gained research evaluation is implementable (Moed, 2006). Traditional scientometric indices, i.e., impact factor and H-index, treat all citations with equal importance and ignore their functions (Zhu et al., 2014). Once we are able to tell the important and meaningful citation amongst all the citations, it may help us in creating a novel and realistic evaluation index based on the important citations.
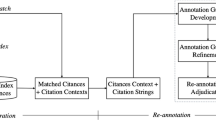
Given the large volume of online scientific publications proliferating with each passing day, it is impractical to perform manual labelling of citation functions. Although a number of researchers have carried out research on automatic classification of the citation function (Teufel et al., 2019; Jurgens et al., 2018; Cohan et al., 2019), we note that previous studies on automatic classification of the citation function primarily focused on the citation sentence, including but not limited to, sentence-level lexical, syntactic and semantic information (Zhao et al., 2019). Orthogonal to the sentence-level information, each citation additionally comes with rich associated native information from the online scientific paper. As depicted in Fig. 1, there is a huge amount of native information (i.e., citation context sentence (CCS), Section where the citation belongs to (Sec), cited and citing paper title (Title), Web URL of the citing paper (Web), and DOI of the citing paper (DOI)) around the citation in that respective paper’s website. Our research object is to find out whether native information around the citation can provide additional information to help with classifying citation function.
In this paper, we explored all the above native information and proposed to exploit the recently developed text representation models integrated with the native information to evaluate the classification performance. Our motivation comes from two aspects. Firstly, we note that existing datasets either possess relatively less native information or are small scale in terms of the number of instances as summarized in Table 1. To overcome this issue, we constructed a new dataset, entitled, NI-Cite, pertinent to online scientific publications in the best possible manner. The dataset encompasses a large number of labelled citations with five key native information, against each dataset instance (refer to the details in “Dataset construction and analysis” section). NI-Cite, owing to its rich native information, can be used for analysis of the citation function classification task at a relatively large scale. Additionally, after several years of research in the text representation (Yan, 2009), the research community has already developed a strong ability to employ the underlying techniques of text representation in text classification task. However how to make the best use of additional native information in citation function classification task remains unclear and demands further investigation. To the best of our knowledge, our envisaged models are the first to integrate native information with the state-of-the-art Long Short-Term Memory (LSTM) based model (Alikaniotis et al., 2016), SciBERT based model (Beltagy et al., 2019), and T5 based model (Raffel et al., 2019) to carry out the citation function classification task. The experimental results demonstrate that the native information suggested in this paper enhances the overall classification performance.
Figure 2 depicts the flowchart of our envisaged approach in this paper. We first discuss the related corpus that we have investigated in “Related work” section (Step 1). Then, we present the details on obtaining the citation context and the dataset construction with native information in “Dataset construction and analysis” section (Step 2–Step 6). Our proposed models and experiments are depicted in “Hybrid method” and “Experiment” sections (Step 7–Step 8) respectively. Furthermore, the results and discussions are delineated in “Experiment” and “Conclusion and future work” sections (Step 9). Our primary contributions in this paper are as follows:
-
We found that the proposed native information can enhance the performance of classification models in the citation function classification task.
-
We proposed different integration solutions for three popular neural text representation models including Long Short-Term Memory, Transformer, and Seq2seq (T5).
-
We built a new benchmark for using the native information in this task and further explored the state-of-the-art models and their structures that can best use those native information pertinent to a citation.Footnote 1
Related work
In our envisaged citation function classification task, datasets and text representation models are the two essential components. In the rest of this section, we review different datasets (Teufel et al., 2019; Jurgens et al., 2018; Cohan et al., 2019; Zhao et al., 2019; Agarwal et al., 2010; Dong & Schafer, 2011) and highlight their limitations as compared to our dataset, along with the computational models that have not integrated all the native information into the citation function classification task (Teufel et al., 2019; Jurgens et al., 2018; Cohan et al., 2019; Zhao et al., 2019; Jochim & Schiitz, 2012).
Existing datasets
The study of citation function classification can be traced as early as 1965 when Garfield (1965) proposed a fifteen categories of citation function, and a similar scheme was also introduced by Weinstock (1971) in 1971, while the data scale is too small. To the best of our knowledge, there are four open access state-of-the-art datasets in the context of citation classification as illustrated in Table 1. Teufel et al. (2019) comprises 2829 manually-tagged citations arising from 116 scientific papers randomly drawn out of 360 conference papers. Zhao et al. (2019) encompasses crawled scientific literature from numerous online databases and manually annotated 3088 citations. Whilst the full text of the scientific literature was extracted during the development process, nevertheless, only 5 sentences around each citation were opted to be incorporated into the above dataset. The ACL-ARC citation dataset (Jurgens et al., 2018) contains 1436 context citations annotated from the fully-labeled 52 papers and another 533 supplemental contexts from 133 papers. Although some contextual and native information could be observed in the ACL-ARC citation dataset (Jurgens et al., 2018), nevertheless, its scale is extremely small. In contrast to the above referred three datasets, SciCite (Cohan et al., 2019) is significantly larger in terms of its number of instances and contains papers from computer science and medicine-related domains. The pros of the said dataset are that they are still new and accessible, while the cons are that a lot of important native information is still missing for some of those instances, and therefore we reconstructed the datasets to fit the citation function classification task. Besides, we also found some relevant datasets in this filed. However, those datasets cannot be used because their instances either lack important native information (Zhao et al., 2019; Jochim & Schiitz, 2012) or are not accessible anymore (Agarwal et al., 2010; Dong & Schafer, 2011).
Sentence-level classification model
Pre-deep learning With the rapid development of the NLP technology and promising advances in machine learning, those techniques have been widely used in scientific text processing (Tuarob et al., 2015), citation context analysis (Hernández-Alvarez et al., 2016) and automatic classify citation function (Garzone & Mercer, 2000). In recent years, numerous researchers have carried out the citation function classification task in a wide variety of ways. Teufel et al. (2019) combined cue phrases with K-nearest neighbors algorithm to analyze the citation function classification result. Jochim and Schiitz (2012) trained the Stanford MaxEnt classifier with their annotated dataset and find out new useful features. Hassan et al. (2017) proposed a novel algorithm by integrating citation context and subsequently classifying them into 4 different categories for detecting the important citation from all citations in the research papers with five traditional machine learning models. Jurgens et al. (2018) trained citation function classification model using a Random Forest classifier and which proved to be robust to overfitting even with a large numbers of features. Pride and Knoth (2017) imported random forests classifier to detect important and incidental citations in the research paper. Tuarob et al. (2013) presented an initial effort in understanding the semantics of algorithms and consequently new classification scheme has been designed for cited algorithms’ functions. Tuarob et al. (2019) proposed a set of heterogeneous ensemble machine-learning methods with handcraft feature to classify the cited algorithm’s function in the citing paper.
Deep learning Kim (2014) proposed a series of experiments using a convolutional neural network (CNN) to train sentence-level categorization tasks on pretrained word vectors. Moreover, the CNN model discussed by the authors improved on four of the seven tasks, including but not limited to, sentiment analysis and question classification. Lai et al. (2015) introduced a recursive convolutional neural network (RNN) text classification method without any human-designed features. The authors used a recursive structure to obtain as much contextual information as possible and which caused less noise than the traditional window-based neural networks. Furthermore, maximum pool layer that automatically determines which words play a key role in text categorization has been proposed, thereby capturing key components in the text. Safder et al. (2020) proposed a bidirectional long short-term memory networks (BiLSTM) to classify citation contexts and extract algorithmic metadata.
Pretrained language model Cohan et al. (2019) used Embeddings from Language Models (ELMo) and attention-based LSTM model for citation function classification. They also introduced two additional scaffold tasks to further improve the performance. Similarly, the BERT model architecture (Devlin et al., 2018) is based on a multilayer bidirectional transformer (Vaswani et al., 2017). Roman et al. (2021) applied BERT to the citation intent classification task and achieved a better performance compared to the static Glove word embedding (Pennington et al., 2014). Moreover, Zhao et al. (2019) used a multi-task learning framework, SciResCLF, which was applied to jointly predict two-level citation roles and functions by sharing the BERT context representations. Recently, Beltagy et al. (2019) proposed SciBERT which shares a similar training scheme and architecture with BERT, but was trained on 1.14M scientific papers. In particular, SciBERT achieved the state-of-the-art on the SciCite and the ACL-ARC citation datasets (Beltagy et al., 2019). Joshi et al. (2020) proposed SpanBERT, which via changing the mask objective in BERT and obtain a better performance in different NLP tasks. T5 (Raffel et al., 2019) model is based on the transformer and shares the same encoder-decoder transformer architecture. It was pretrained on the “Colossal Clean Crawled Corpus” which consist a huge amount of clean English text. There are also two parallel cutting-edge works TDM-CFC (Zhang et al., 2021) and MULTICITE (Lauscher et al., 2021) both leveraged BERT in their multi-label citation function classification task.
It is worth mentioning that none of the previous works systematically researched how to integrate the proposed native information proposed in this paper with different kinds of text representation models to classify the citation function.
Dataset construction and analysis
In order to carry out the experiments, we require a dataset encompassing online scientific publications duly labeled with citation function and native information for each citation. As depicted in Fig. 1, we demonstrate a snapshot from our dataset with the following native information and delineate on the rationale as to why the native information may be helpful for the citation function classification task:
-
1.
Citation context sentences (CCS)—Sentences just before and after the citation sentence may provide additional information pertinent to citation function.
-
2.
The section to which it belongs (Sec)—Section names provide the structural information of the targeted citation.
-
3.
Citing and cited publication titles (Title)—The similarity between the two titles may indicate the purposes of a citation.
-
4.
Digital Object Identifier Number (DOI)—DOI helps trace a publication’s research domain and research focus as assigned by the publisher.
-
5.
Website URL of the online publications (Web)—The website addresses provide more information about publication sources.
In the dataset construction process, we selected raw instances from the SciCite (Cohan et al., 2019) dataset and the ACL-ARC citation dataset (Jurgens et al., 2018). We added native information against each instance in our dataset, and removed the instances that cannot be traced back to their original sources. In our dataset, there are three citation function labels: Background (depicting background information), Technical basis (implying a method, tool, approach or dataset), and Contrast (comparison of the paper’s results). Background corresponds to Background Information in SciCite and Background in ACL-ARC citation dataset. Technical Basis corresponds to Method in SciCite and Use in ACL-ARC citation dataset. Finally, Contrast corresponds to Result Comparison in SciCite and Comparison or Contrast in ACL-ARC citation dataset. Our dataset is a unique research contribution in the citation analysis research community. The details of the reconstruction include the following semi-automate stages:
-
Stage A: Obtaining the Full Text of the PDF Files We obtained PDF files from Semantic Scholar and other publishers’ websites corresponding to the titles and Semantic Scholar ID of each publication in the SciCite. Employing the license of Macquarie University, some of the PDFs were directly downloaded from Semantic Scholar, whereas, the other PDFs were found on the Publishers’ websites, i.e., Springer LinkFootnote 2 and IEEE XploreFootnote 3 and so on. In this process, we found that the source of online publication is complex. For example, there might be different versions or languages for one particular scientific publication. We also manually checked and downloaded 154 publications (approximately 2% of the total processed ones) which cannot be downloaded automatically.
-
Stage B: Converting PDF to Structured JSON We used SCIENCE-PARSE,Footnote 4 an open-source PDF structure analysis tool, to extract full text from the PDF and output it in the JSON format. We further extracted section names (Sec) and the sentence context (CCS) (ones before and after the citation sentence) from the JSON file. For the section names, we normalized them into a more fine-grained IMRaD structure (Bertin et al., 2016), i.e., abstract, introduction, related work, method, experiment, results and discussion, and conclusion, primarily since this structure covers most of section structure in our dataset. Normalization was implemented via lexical overlapping. For example, section names with word “result” were normalized into “results and discussion”. We note that the section titles containing multiple normalized names are rare in our dataset.
-
Stage C: Extracting DOI, Year, Title, Cited Paper Title, and Website Name information We retrieved DOI, year, title, and cited paper title from Semantic Scholar,Footnote 5 nevertheless, there was still some missing information in Semantic Scholar. In order to cater for the same, we automatically searched the titles on DBLPFootnote 6 and PubMedFootnote 7 and subsequently matched the title with each instance of the JSON file. We manually checked 215 instances for the DOI and website information. The DOI number was further used to trace the source of each publication, i.e., the corresponding website name for it. When using DOI and web address, we extracted the first 7 digits in DOI and the web root address as the representations for each publication. For example, in our dateset, one instance’s URL was ‘https://cancerres.aacrjournals.org/content/77/12/3194’, and we kept ‘cancerres.aacrjournals’ as the web root. From the URL, we could know that this instance discussed about cancer research and we believe that papers from similar subject sources should have the similar citing pattern. We also observed that some of the online scientific publications lacked the DOI information, e.g., PhD/Master’s theses, working papers, and reports. However, they are also considered as valuable online resources, and therefore, we assigned “empty” values for such DOIs. For reference, we came across 410 empty instances in total, i.e., the rate of such DOIs was 3.6%.
-
Stage D: Merging Instances from the ACL-ARC citation dataset We integrated some of the instances that are semantically similar to the ones in the SciCite dataset from the ACL-ARC citation dataset. To enrich the publication diversity, we first chose at least one instance from each paper in ACL-ARC citation dataset and then randomly sampled the other. We mapped some instances labelled with background in the ACL-ARC citation dataset to background information class in the SciCite dataset. In the same way, we integrated some instances in the ACL-ARC citation dataset labelled with use into the technically basis class. We integrated some instances labelled with comparison or contrast from the ACL-ARC citation dataset into the contrast class. We manually checked the instances from both datasets and ensured that they have the same function. In total, we replenished 300 background and 100 technical basis instances into our dataset and manually fixed 45 contrast instances.
We noted that some instances in the SciCite dataset cannot be traced back to the original papers or the original sources were corrections or notices which are not standard scientific papers. Those instances were therefore removed. As a result, we formulated a large scale annotated citation function dataset NI-Cite with richer native information. NI-Cite comprises 6468 online scientific publications with 11,195 instances with citation function and native information. The DOI is the identity of an online publication and differs for different publishers. The first 7 digits in a DOI signifies the publisher, and therefore, we chose the said digits from a DOI for each instance and ascertained its distribution. To prevent the long tail in the distribution, we merged those instances whose first 7 digits in a DOI appear less than 5 times into the others category. The number of instances, where the DOI appeared more than five times is around 96.46%. The section-wise proportion is the highest for Result and Discussion followed by Introduction and Method. We visualized the frequency of the top 30 frequent DOI and Web root address, as well as the section name distribution in Fig. 3. Both DOI and Web follow a long-tail distribution and Web is slightly sparser than the DOI. As for the section name, 88.8% of our citations are found in the Results and Discussion, Method, and Introduction sections.
We noticed that only 3.4% of citations are found in the Related Work section, and which in its essence, is a bit abnormal. However, our work is built on top of the sampled citation instances delineated in the existing literature. They selected a huge amount of instances from medical papers in PubMedFootnote 8 and did not pick instances in related work section. Therefore, their sampling strategies lead to this phenomenon. We also summarized the label distributions in different sections of Table 2. Not surprisingly, most of Method and Result citation instances can be found in Method and Result & Discussion Section. Yet, the Background instances distribute somewhat equally in Introduction and Result & Discussion.
Finally, as depicted in Table 3, we investigated the top frequent words in the title and context. Compared with context words (e.g., number, model), the words in title are more informative (e.g., Cell, Patients) and may provide additional information for the model.
Hybrid method
We formulate the proposed citation classification task with mathematical notations and discuss it. The input of the models is the native information features and citation sentence. The input citation sentence is a sequence of words \(C = [x^c_1, x^c_2, \dots ,x^c_n]\). Each C has five features, where \(f_{\mathrm{cc}}\) stands for the citation context, \(f_{\mathrm{sn}}\) for the section name, \(f_{tt}\) for the title, \(f_{\mathrm{d}}\) for the DOI, \(f_{\mathrm{w}}\) for the Web URL. The feature \(f_{\mathrm{cc}} = \left\{ s^p,s^n\right\}\) has two sentences, where \(s^p = [x^p_1, x^p_2, \dots , x^p_{l_p}]\) and \(s^n = [x^n_1, x^n_2, \dots , x^n_{l_n}]\) are two sequence of words of citation context sentence. The \(f_{\mathrm{sn}} \in\) {Result and Discussion, Method, Introduction, Experiment, Abstract, Related Work, Conclusion} is a label of section name. The feature \(f_{tt} = \left\{ t^{o}, t^{t}\right\}\) has two titles, where \(t^{o} = [t^{o}_1, t^{o}_2, \dots , t^{o}_{l_o}]\) and \(t^t = [t^{t}_1, t^{t}_2, \dots ,t^{t}_{l_t}]\) are two sequence of words of citing paper title and cited paper title. The features \(f_{\mathrm{d}}\) and \(f_{\mathrm{w}}\) are two symbols of numbers and website address. We use the context of each instance’s citation and feature highlighted above to denote the input. The expected output is one specific citation function label \(Y \in\) {Background, Technical basis, Contrast} that describe a function of each C. The citation function classification task is to learn a function \(f : C \rightarrow Y\), i.e., to predict label Y for the input C. For discrete native information, i.e., section name, DOI and Web, we used randomly initialized and trainable embeddings \(e^{\mathrm{DOI}}\), \(e^{\mathrm{WEB}}\), \(e^{\mathrm{SN}}\) to represent the discrete values in section name, DOI and Web. For textual native information, i.e., title and citation context sentences, we used word embedding to encode textual information. At this piont, the t-th word in the citation sentence C text can be represented as \(e^c_t \in R^d\) (d is dimention of the word embedding vector). When the length of text is \(T_c\), the input text can be represented as:
We also represented \(f_{\mathrm{cc}}\) and \(f_{tt}\) in the above referred way as follows:
The t-th word in the title \(f_{tt}\) and citation context \(f_{\mathrm{cc}}\) can be represented as \(e^{f_{\mathrm{cc}}}_t \in R^d\) and \(e^{f_{tt}}_t \in R^d\) separately and d is dimention of the word embedding vector.
LSTM-based model
The LSTM-based model (Alikaniotis et al., 2016) inputs word sequences through the multi-layered neural network shown in Fig. 4 to predicts scores. We processed citation sentence with LSTM-based model as followed:
To obtain the long-distance word dependencies, in the LSTM network, at each timestep, the input vector will be converted into an output vector. In the memory cell, we have the forget gate for controlling the deletion of the historical information, the input gate for controlling the update of the current cell state of the LSTM unit, and the output gate for getting the hidden state for next cell. When the \(e^c_t\) is d-dimensional word embedding vector, the number of LSTM hidden layer nodes is H. The three gates (input \(i_t\), output \(o_t\), and forget \(f_t\)) at the time step t is updated as follows:
The state of the memory cell at the time step t is \(c_t\), which is determined by the input gate, input vector, and forget gate. \(h^c_t\) is the final output of the LSTM and its dimension is equal to the number of hidden layer nodes H. \(\sigma\) is the sigmoid function. \(W_i, W_o, W_f, W_c \in R^{H \times E}, U_i, U_o, U_f, U_c\in R^{H \times H}, b_i, b_o, b_f, b_c \in R^{H \times 1}\) are the trainable network parameters. The model process the title \(f_t\) and the citation context \(f_{\mathrm{cc}}\) in the same way:
For discrete native information, i.e., DOI, Web, section name, we used randomly initialized and trainable embeddings as discussed before, i.e., \(e^{\mathrm{DOI}}\), \(e^{\mathrm{WEB}}\), and \(e^{\mathrm{SN}}\) to represent the discrete values in DOI, Web and section name. We concatenated feature representations with the citation sentence representation, \(H^c_{\mathrm{cls}}\) (from LSTM), before the \({\mathrm {softmax}}\) layer. we then encoded different features into fixed size vectors and concatenated them together to classify the citation functions:
We will use the combination of \(h^c_{\mathrm{last}}\) and the other native information’s feature’s dense vector to compute the model.
SciBERT-based model
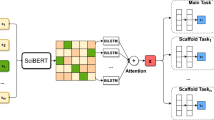
In this section, we opted for the state-of-the-art text encoder, SciBERT, as our baseline model. The baseline model only accepts the citation sentence as its input. We then incorporated each of our proposed native feature into the SciBERT baseline to verify its usefulness. In contrast to BERT, the SciBERT model was trained on 1.14M biomedical and computer science papers and is better at understanding scientific writings. Therefore, we used SciBERT, as depicted in Fig. 5, similar to Devlin et al. (2018), we used \(h_{\mathrm{cls}}\) as the aggregated representation for the whole input sentenceas as our baseline model, and incorporated the model with all of the native information. We processed citation sentence with SciBERT-based model as follows:
As the basic building block of SciBERT, a self-attentive layer takes a citation sentence sequence of token embeddings, \(E^c=[e^c_{1}, e^c_{2}, \dots , e^c_{T_c}]\), as inputs and uses k attention heads \(H^i\) to model \(E^c\):
In H, each word embedding \(e^c_i\) is projected into Query, Key, and Value vectors. Subsequently, the inter-word attention scores are computed using Query and Key vectors. The value vectors are aggregated accordingly to obtain the attention scores:
Finally, the position-wise feed-forward network is computed, followed by a LayerNorm layer:
Given input citation sentence C, SciBERT adds the special tokens [CLS] and [SEP] at the beginning and at the end of the sequence to represent the starting and the end of the sequence respectively. The corresponding output vector of [CLS] is used in one of SciBERT’s pre-training objective to judge if the given sentence pair is next to each other. We used SciBERT to encode textual native information, i.e., title and citation context sentences encode textual information into dense vectors in the same way as follows:
Integrating additional information into neural networks has been studied intensively (Wang et al., 2019). As discussed before we discrete native information, i.e., DOI, Web, section name, we used randomly initialized and trainable embeddings, i.e., \(e^{\mathrm{DOI}}\), \(e^{\mathrm{WEB}}\), and \(e^{\mathrm{SN}}\) to represent the discrete values in DOI, Web and section name. Finally, we concatenated feature representations with the citation sentence representation, \(H^c_{\mathrm{cls}}\) (from SciBERT), before the \({\mathrm {softmax}}\) layer, and concatenated them together to classify the citation functions:
In this paper, we will always use \(H^c_{\mathrm{cls}}\), but for the other features we will drop it when we don’t use it. Yet, for simplicity, we encoded diverse types of native information with dense vectors, whereas, the search for optimal neural architecture is left as the future work.
T5-based model
In this section, we adopt T5 as our computational model. T5 is an encoder and decoder structure. The encoder model is the same as the SciBERT as discussed in “SciBERT-based model” section. Let \(P=[p_1, \dots ,p_n]\) be the prefix sequence in the T5 model. The input sequence of citation sentence with prefix sequence \(P^c\) (i.e., citation sentence) is \(C^p = [p^c_1, \dots , p^c_k, x^c_1, \dots ,x^c_n]\). In the same method, we have \(f^p_{\mathrm{cc}}=[p^{f{\mathrm{cc}}}_1, \dots , p^{f{\mathrm{cc}}}_k, x^p_1, x^p_2, \dots , x^p_{l_p}, x^n_1, \dots , x^n_{l_n}]\), where \(P^{\mathrm{cc}}\) is the prefix sequence (i.e., this is the citation context), \(f^p_{tt}=[p^{f{tt}}_1, \dots , p^{f{tt}}_k, t^{o}_1, \dots , t^{o}_{l_o}, t^{t}_1, \dots ,t^{t}_{l_t}]\), where \(P^{tt}\) is the prefix sequence (i.e., this is the title), and \(f^p_{\mathrm{sn}} = [p^{f{\mathrm{sn}}}_1, \dots ,p^{f{\mathrm{sn}}}_k, f_{\mathrm{sn}} ]\), where \(P^{\mathrm{sn}}\) is the prefix sequence (i.e., this is the section name). In the T5 encoder model, we combine the \(C^p\) and \(f^p_{\mathrm{cc}}\), \(f^p_{tt}\), \(f^p_{\mathrm{sn}}\) separately:
Different from the SciBERT, T5 changed the formal task in a text to text manner, since T5 basically changed all the NLP task into “text-to-text” implying that T5 introduces a unified framework that converts every language problem into a text-to-text format. Let T5 decoder input be \(H^i_{t5}\in \left\{ H^{c;{f^p_{\mathrm{cc}}}}_{t5}, H^{c;{f^p_{tt}}}_{t5}, H^{c;{f^p_{\mathrm{sn}}}}_{t5}\right\}\), and predict the citation function classification label Y as:
The T5 model is trained to predict missing, or otherwise, corrupted tokens in the input which is offen referred to as the denoising objective (Devlin et al., 2018; Taylor, 1953). In our task, as depicted in Fig. 6, we amalgamate native information text and citation sentence text together as the input of the T5 model. We trained T5 model to predict the function of each input instance, i.e., we took the data depict in the Fig. 1 to train the model with the native information-(Title), where the input to the T5 model is “citation sentence: Further, we establish that our models have fewer parameters (almost as few as native network) and so are easier to train than methods like [23]; This is the title: Scale-recurrent Netword for Deep Image Debulrring”. We give the prefix (i.e., this is the title:...) to the native information text, then we let the model to predict the citation function label.
Experiment
In this section, we first describe our experimental setup and then report the performance for each of our proposed native information with different computational models.
Experimental setup
Previous datasets (Zhao et al., 2019; Cohan et al., 2019) split annotated instances in a random manner without the considering paper-level and the temporal-level information leak. In the real-world scenario, when we build a citation function prediction system on a particular date, we can only use paper already published as the training data. For the newly published paper, we will use them to test. Therefore, we split NI-Cite in a temporal order (1968–2015 as training, 2016 as dev., 2017–2021 as test), and each split has a similar distribution of the three citation function labels (see Table 4). Our temporal-based splitting scheme gives about the same function distributions for three splits. For discrete DOI and Web features, we represent values that appear at least 5 times in the training data with dense embedding vectors and merge all other minor values into the “other” category. We used trainable dense vectors with size \(k=256\) to represent them. This results in 3.54% “other” in DOI and 6.61% “other” in Web. We simply selected \(k=256\) for both features.
Baseline models
To highlight the improvement of our proposed models, we first compared them with several competitive baselines as follows:
-
Majority Baseline: this baseline model always predicts the Background label.
-
Random Baseline: this baseline model always randomly predicts a label.
-
Section Guess Baseline: this baseline model always predicts based on the Section Name.
-
SVM and Naive Bayes: these two baselines use the bag-of-words feature.
Proposed models
LSTM We used a word-level LSTM in our experiment, which contains embedding layer, hidden layer and single fully connected. We first used the embedding layer to carry out the word embedding, then fed the output into the hidden layer with 64 hidden nodes. Subsequently, the last hidden state of the LSTM will be fed to a single-layer fully connected network with 128 neurons for the citation function classification. we evaluate our model we Macro-F1 score, averaged F1 score across all three labels.
SciBERT According to Beltagy et al. (2019), SciBERT uses the same architecture as \({\mathrm{BERT}}_{\mathrm{BASE}}\) model and includes 12 self-attentive layers with a hidden size of 768 and 12 heads. We trained our model for 5 epochs and fine-tuned the SciBERT model parameters during the training. To evaluate our model, we used Macro-F1 score and averaged F1 score across all three labels in
NI-Cite to treat three labels equally. We picked the model with the best development performance and reported the corresponding performance.
T5 According to Raffel et al. (2019), we used T5-Base whose encoder and decoder are both sized similarly to \({\mathrm{BERT}}_{\mathrm{BASE}}\). Specifically, both the encoder and decoder consist of 12 blocks (each block comprising self-attention, optional encoder-decoder attention, and a feed-forward network). T5 predicted the label of the each inputting instance and we evaluated our model via Macro-F1 score, and averaged F1 score across all three labels. We picked the model with the best development performance and reported the corresponding performance.
Experimental evaluation
To evaluate our model, we used Macro-F1 score, averaged F1 score across all three labels in NI-Cite. This is because the distribution of three labels is unbalanced and each function label has its own practical application in the downstream tasks. They should be treated equally in the evaluation. Micro-F1 or classification accuracy metrics would be largely dominated by the Background label. The functions of instances in each split are shown in Table 4. As observed, the ratio of function distribution in the three sets is about the same. Each dataset function distribution can be found in Table 4 (test data: contrast 162, technical basis 375, background 731; development data: contrast 127, background 521, and technical basis 272; train data: contrast 1191, background 5117, and technical basis 2699). We trained our LSTM, SciBERT, and T5 model for 5 epochs and fine-tuned the SciBERT and T5 model parameters during the training. We picked the model with the best development performance and reported the corresponding performance.
Results
In our experiment, we conducted experiments (i.e., a baseline experiment and experiments injected with different types of native information) for 5 times and reported their averaged F1 score for each function label and Macro-F1 score in Table 5. With T5, we only conducted four experiments since it is a text-to-text model, and therefore we only injected text native information in it. We first observed that all of our models achieved noticeable improvements over the baselines. Although we have seen the high correction between ground labels and section names, using section names in the prediction only achieves 53.5 Macro-F1 score, much lower than our SciBERT Baselines implying that the section names alone cannot improve the performance. As for the traditional machine learning algorithm SVM and NaiveBayes, SVM outperforms NaiveBayes by 23.6 F1. This could because n-grams play an important role in citation function classification and NaiveBayes is based on the word independent assumption (i.e., each uni-gram is independent with each other.)
In LSTM-based model we observed that some of the proposed native information features demonstrate a better F1 score as compared with the baseline. Title reached the most significant improvement of 2.1%, same as CCS which also improved 2.1%. The Sec shows a simailar improvement of 2.0% compared with the baseline. For the DOI and Web the improvement is tiny but still we can see the improvement. As depicted in Table 5 Finally, we aggregated the four native information features with LSTM (Comb) and the resulintg model achieved 78.9 Marco-F1 score, outperforming the LSTM model by 2.2%. We also re-implemented the SOTA model ‘bi-lstm Attn w / Elmo + both scaffolds’ (Bil-Scaf) from Cohan et al. (2019). We observed that, their model achieved lower F1 score than the SciBert model. After integrating the native information, we could observe a 1% F1 score improvement, which proves our native information is useful. We also note that the improvement gap of the Bil-Scaf (Comb) is not as high as SciBERT (Comb) since Bil-Scaf was trained in the multitask learning process which has already used section information.
In SciBERT-based model, as depicted in Table 5 we observed that all of the proposed native information features demonstrated a better F1 score in contrast to the baseline except for the Web. Specifically, Sec achieves the largest improvement of 1.4%. It attained the best performance in the Background and Technical basis categories. The model with citing and cited paper titles obtained the best performance in the Contrast category. As for the DOI, we also observed 0.5% improvement in the F1 score. Finally, we explored the potential of these native information features and aggregated the four positive native information features with SciBERT (Comb). The resulting model achieved 84.7 Macro-F1 score, thereby outperforming the SciBERT model by 1.6% Macro-F1 score. For the individual feature, we found that Sec is the most informative one which improves the performance of our SciBERT baseline by 1.4% F1 score.
In T5-based model we observed that all of the three native information obtained a better F1 score than the T5 model baseline. For the Background and Contrast categories, CCS demonstrated a highest performance, i.e., it improved by 0.6 F1 score and by 0.8 F1 score respectively as compared to the baseline. For the technical basis categories, the model with the Sec improved 1.7% F1 score, i.e., the highest in the three native information features. Overall, the model integrated with Sec obtained the best performance of 84.5% F1 score. As for the Title, compared with the T5 baseline, the performance of each label has been considerably improved.
We also conducted t-student test for the baseline models and models with all features Comb, and all p values are less than 0.01.
Discussion and implication
It can be observed from the experimental results that all of our proposed models achieved a higher Marco-F1 score in contrast to all the baseline models. This manifests that we have constructed a high quality dataset for this research task and our evaluation metric is reasonable.
We further demonstrated why the native information we proposed is useful in this citation function classification task. Title improves the classification performance implying that the model can quickly learn the information from the representation. For Title, it is likely that papers with comparable results often include same or similar keywords in their titles (which are encoded in their representations), and titles that share similar lexical items are likely to be referred for result comparison purposes. As for the Sec, it is a very efficient native information feature in all the models. This may be due to the reason that each function label has its own positional contraction and Sec provides valuable prior information to the model. The CCS which is connected with the citation can provide a rich semantic information for the citation itself. This means that model can better understand the citation sentence with citation context. Different DOI implies different publishers which may have different publishing strategy and which further leads to different citation distributions.
Overall, in all of the three labels, we can see the improvement of Marco-F1 score. Among those three function labels, the Background label has the highest performance, since in NI-cite dataset, the Background label has the largest number. We note the data imbalance in our dataset. However, this scenario is commonly seen in different benchmarks of the citation function classification task. In addition, in the real world scenario, it is the truth that majority of citations are used for introducing background knowledge. This also reflects that the citation function classification task is no-trivial and worth research effort. When class imbalance exists within a training data, the deep learning based models will typically over-classify the majority group due to its increased prior probability (Johnson & Khoshgoftaar, 2019).
The citation function classification allows researchers to have a better understanding of a citation function, which can facilitate us to (a) create better informative citation indexers (Teufel et al., 2019), (b) identify the significant references (Valenzuela & Etzioni, 2015), and (c) search the target online scientific paper in a database (Cohan et al., 2019). For example, AI based scientific literature database such as semantic scholar Footnote 9 leverage the citation function of each scientific literature to help the reader better understand and search the references. While adopting our proposed method, there are two main points worth to be considered. Firstly, our model cannot directly handle citation context in other languages like Spanish or Chinese, since the model we used is pretrained with English corpus. We note that multilingual pretrained language model (Pires et al., 2019) is possible to handle other languages. In addition, since the data used in this paper mainly comes from computer science and medical domains, our model may achieve better performance if it is adopted to the related domains. Although a pretrained language model is robust to handle data from different areas, domain adaptation is still a challenging problem which is out of scope of this paper.
Conclusion and future work
In this paper, we proposed to integrate various native information with the state-of-the-art text representation models in the citation function classification task. We proposed different integration solutions for three popular neural text representation models including LSTM, transformer, and seq2seq. Our experimental results suggest that the native information of citations does contribute to the citation function classification performance to a significant extent. We also constructed a novel large scale dataset, NI-Cite, with rich native information. We further built a new benchmark for using this information in citation function classification task and explored the state-of-the-art model and its structure that can best use those native information around a citation. In the near future, we intent to further improve our model by integrating native information into the language model’s pretraining stage and will enhance our dataset by collecting high-quality scientific paper corpus with more native information. The other limitation of our work is that there are only three citation function labels in our dataset. In the future, we plan to extent the dataset into a more fine-grained citation function scheme.
Change history
18 July 2022
A Correction to this paper has been published: https://doi.org/10.1007/s11192-022-04451-1
Notes
References
Abu-Jbara, A., & Radev, D. (2012). Reference scope identification in citing sentences. In Proceedings of the 2012 conference of the North American Chapter of the Association for Computational Linguistics: Human language technologies (pp. 80–90).
Agarwal, S., Choubey, L., & Yu, H. (2010). Automatically classifying the role of citations in biomedical articles. In Proceedings of American Medical Informatics Association fall symposium (pp. 11–15).
Alikaniotis, D., Yannakoudakis, H., & Rei, M. (2016). Automatic text scoring using neural networks. In Proceedings of the 54th annual meeting of the Association for Computational Linguistics (pp. 715–725).
Beltagy, I., Lo, K., & Cohan, A. (2019). SciBERT: A pretrained language model for scientific text. Retrieved from arXiv:1903.10676
Bertin, M., Atanassova, I., Gingras, Y., & Lariviere, V. (2016). The invariant distribution of references in scientific articles. Journal of the American Society for Information Science and Technology, 67(1), 164–177.
Bornmann, L., & Daniel, H. (2008). What do citation counts measure? A review of studies on citing behavior. Journal of Documentation, 64(1), 45–80.
Cohan, A., Ammar, W., van Zuylen, M., & Cady, F. (2019). Structural scaffolds for citation intent classification in scientific publications. In Proceedings of 2019 conference of the North American Chapter of the Association for Computational Linguistics (pp. 3586–3596).
Cohan, A., & Goharian, N. (2018). Scientific document summarization via citation contextualization and scientific discourse. International Journal on Digital Libraries, 19(2), 287–303.
Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2018). BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805
Dong, C., & Schafer, U. (2011). Ensemble-style self-training on citation classification. In Proceedings of the 5th international joint conference on natural language processing (pp. 623–631).
Färber, M., & Jatowt, A. (2020). Citation recommendation: Approaches and datasets. International Journal on Digital Libraries, 21(1), 375–405.
Garfield, E. (1965). Can citation indexing be automated? In M. E. Stevens, V. E. Giuliano, & L. B. Heilprin (Eds.), Statistical association methods for mechanical documentation. National Bureau of Standards.
Garzone, M., & Mercer, R. E. (2000). Towards an automated citation classifier. In Proceedings the conference of the Canadian society for computational studies of intelligence (pp. 337–346). Springer.
Hassan, S., Akram, A., & Haddawy, P. (2017). Identifying important citations using contextual information from full text. In Proceedings of 2017 ACM/IEEE joint conference on digital libraries (pp. 1–8).
Hernández-Alvarez, M., & Gomez, M. J. (2016). Survey about citation context analysis: Tasks, techniques, and resources. Natural Language Engineering, 22(3), 327–349.
Jochim, C., & Schiitz, H. (2012). Towards a generic and flexible citation classifier based on a faceted classification scheme. In Proceedings of the 2012 international conference on computational linguistics (pp. 1343–1358).
Johnson, J. M., & Khoshgoftaar, T. M. (2019). Survey on deep learning with class imbalance. Journal of Big Data, 6(27), 1–54. https://doi.org/10.1186/s40537-019-0192-5.
Joshi, M., Chen, D., Liu, Y., Weld, D. S., Zettlemoyer, L., & Levy, O. (2020). Spanbert: Improving pre-training by representing and predicting spans. Transactions of the Association for Computational Linguistics, 8, 64–77.
Jurgens, D., Kumar, S., Hoover, R., McFarland, D., & Jurafsky, D. (2018). Measuring the evolution of a scientific field through citation frame. Transactions of the Association for Computational Linguistics, 6, 391–406.
Kim, Y. (2014). Convolutional neural networks for sentence classification. In Proceedings of the 2014 conference on empirical methods in natural language processing (pp. 1746–1751).
Lai, S., Xu, L., Liu, K., & Zhao, J. (2015). Recurrent convolutional neural networks for text classification. In Proceedings of twenty-ninth AAAI conference on artificial intelligence (pp. 2267–2273).
Lauscher, A., Ko, B., Kuehl, B., Johnson, S., Jurgens, D., Cohan, A., & Lo, K. (2021). MultiCite: Modeling realistic citations requires moving beyond the single-sentence single-label setting. arXiv preprint arXiv:2107.00414
Moed, H. F. (2006). Citation analysis in research evaluation (Vol. 9). Springer.
Moravcsik, M. J., & Murugesan, P. (1975). Some results of the function and quality of citations. Social Studies of Science, 5(1), 86–92.
Narin, F. (1976). Evaluative bibliometrics: The use of publication and citation analysis in the evaluation of scientific activity (pp. 334–337). Computer Horizons.
Pennington, J., Socher, R., & Manning, C. D. (2014). Glove: Global vectors for word representation. In Proceedings of the 2014 conference on empirical methods in natural language processing (pp. 1532–1543).
Pires, T., Schlinger, E., & Garrette, D. (2019). How multilingual is multilingual BERT? In Proceedings of the 57th annual meeting of the Association for Computational Linguistics (pp. 4996–5001).
Pride, D., & Knoth, P. (2017). Incidental or influential?—Challenges in automatically detecting citation importance using publication full texts. In Research and advanced technology for digital libraries (pp. 572–578). https://doi.org/10.1007/978-3-319-67008-9_48
Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S., & Matena, M., Zhou, Y., Li, W., & Liu, P. J. (2019). Exploring the limits of transfer learning with a unified text-to-text transformer. arXiv:1910.10683
Roman, M., Shahid, A., Khan, S., Koubaa, A., & Yu, L. (2021). Citation intent classification using word embedding. IEEE Access, 9, 9982–9995.
Safder, I., Hassan, S. U., Visvizi, A., Noraset, T., Nawaz, R., & Tuarob, S. (2020). Deep learning-based extraction of algorithmic metadata in full-text scholarly documents. Information Processing & Management, 57, 102269.
Smith, L. C. (1981). Citation analysis. Library Trends, 30(1), 83–106.
Taylor, W. L. (1953). Cloze procedure: A new tool for measuring readability. Journalism Quarterly, 30, 415–433.
Teufel, S., Siddharthan, A., & Tidhar, D. (2006). An annotation scheme for citation function. In Proceedings of the 7th SIGdial workshop on discourse and dialogue (pp. 80–87).
Teufel, S., Siddharthan, A., & Tidhar, D. (2019). Automatic classification of citation function. In Proceedings of 2006 conference on empirical methods in natural language processing (pp. 103–110).
Tuarob, S., Kang, S. W., Wettayakorn, P., Pornprasit, C., Sachati, T., Hassan, S. U., & Haddawy, P. (2019). Automatic classification of algorithm citation functions in scientific literature. IEEE Transactions on Knowledge and Data Engineering, 32(10), 1881–1896.
Tuarob, S., Mitra, P., & Giles, C. L. (2013). A classification scheme for algorithm citation function in scholarly works. In Proceedings of the 13th ACM/IEEE-CS joint conference on digital libraries (pp. 367–368).
Tuarob, S., Mitra, P., & Giles, L. C. (2015). A hybrid approach to discover semantic hierarchical sections in scholarly documents. In Proceedings of the 13th international conference on document analysis and recognition (pp. 1081–1085).
Valenzuela, M., Ha, V., & Etzioni, O. (2015). Identifying meaningful citations. In Proceedings of AAAI workshop: Scholarly big data (pp. 13–18).
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., Kaiser, L., & Polosukhin, I. (2017). Attention is all you need. In Proceedings of the 31st international conference advances in neural information processing systems (pp. 5998–6008).
Wang, Y., Johnson, M., Wan, S., Sun, Y., & Wang, W. (2019). How to best use syntax in semantic role labelling. In Proceedings of the 57th annual meeting of the Association for Computational Linguistics (pp. 5338–5343).
Weinstock, M. (1971). Citation indexes. In M. Drake (Ed.), Encyclopedia of library and information science (Vol. 5). Dekker.
Yan, J. (2009). Text representation. In L. Liu & M. T. Özsu (Eds.), Encyclopedia of database systems (pp. 3069–3072). Springer.
Yousif, A., Niu, Z., Tarus, J. K., & Ahmad, A. (2019). A survey on sentiment analysis of scientific citations. Artificial Intelligence Review, 52(1), 1805–1838. https://doi.org/10.1007/s10462-017-9597-8.
Zhang, Y., Wang, Y., Sheng, Q. Z., Mahmood, A., Emma Zhang, W., & Zhao, R. (2021). TDM-CFC: Towards document-level multi-label citation function classification. In Proceedings of international conference on web information systems engineering (pp. 363–376).
Zhao, H., Luo, Z., Feng, C., Zheng, A., & Liu, X. (2019). A context-based framework for modeling the role and function of on-line resource citations in scientific literature. In Proceedings of 2019 conference on empirical methods in natural language processing and the 9th international joint conference on natural language processing (pp. 5209–5218).
Zhu, X., Turney, P., Lemire, D., & Vellino, A. (2014). Measuring academic influence. Journal of the Association for Information Science and Technology, 66, 408–427.
Acknowledgements
This research is funded by Australian Research Council (ARC) Discovery Project DP200102298 and the National Social Science Fund of China (No. 18ZDA325).
Author information
Authors and Affiliations
Corresponding author
Additional information
The original online version of this article was revised: In the original version the first affiliation was incorrectly linked to the author name, Adnan Mahmood.
Rights and permissions
About this article
Cite this article
Zhang, Y., Zhao, R., Wang, Y. et al. Towards employing native information in citation function classification. Scientometrics 127, 6557–6577 (2022). https://doi.org/10.1007/s11192-021-04242-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-021-04242-0