Abstract
In higher plants, ferredoxin (Fd) and ferredoxin-NADP+ reductase (FNR) are each present as distinct isoproteins of photosynthetic type (leaf type) and non-photosynthetic type (root type). Root-type Fd and FNR are considered to facilitate the electron transfer from NADPH to Fd in the direction opposite to that occurring in the photosynthetic processes. We previously reported the crystal structure of the electron transfer complex between maize leaf FNR and Fd (leaf FNR:Fd complex), providing insights into the molecular interactions of the two proteins. Here we show the 2.49 Å crystal structure of the maize root FNR:Fd complex, which reveals that the orientation of FNR and Fd remarkably varies from that of the leaf FNR:Fd complex, giving a structural basis for reversing the redox path. Root FNR was previously shown to interact preferentially with root Fd over leaf Fd, while leaf FNR retains similar affinity for these two types of Fds. The structural basis for such differential interaction was investigated using site-directed mutagenesis of the isotype-specific amino acid residues on the interface of Fd and FNR, based on the crystal structures of the FNR:Fd complexes from maize leaves and roots. Kinetic and physical binding analyses of the resulting mutants lead to the conclusion that the rearrangement of the charged amino acid residues on the Fd-binding surface of FNR confers isotype-specific interaction with Fd, which brings about the evolutional switch between photosynthetic and heterotrophic redox cascades.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The photosynthetic electron transport chain reduces Fd, donating reducing equivalents to FNR and other Fd-dependent enzymes, such as nitrite reductase, sulfite reductase, and glutamate synthase, involved in the assimilation of inorganic nitrogen and sulfur into amino acids (Knaff 1996; Hanke and Mulo 2013; Goss and Hanke 2014). In non-photosynthetic tissues such as roots, these Fd-dependent enzymes are also functioning, but the reduction of Fd is catalyzed by FNR, using NADPH generated through the oxidative pentose phosphate cycle (Bowsher et al. 1993). These discrete redox cascades are conducted by the combination of genetically distinct isoforms of FNR and Fd present differentially in photosynthetic and non-photosynthetic tissues (Emes and Neuhaus 1997). Cyanobacterial FNRs contain structural elements of both types of higher plant FNRs, while FNRs from unicellular green algae are more similar to root type than to leaf type (Arakaki et al. 1997). The plant-type FNRs were also found in bacteria in the genus Leptospira, of which some are entirely parasites of vertebrates (Catalano-Dupuy et al. 2011), and their FNRs are thought to be segregated with FNRs found in the apicomplexan eukaryotic parasites such as Toxoplasma gondii and Plasmodium falciparum. These apicomplexan FNRs display some sequence similarity with root-type FNRs (Vollmer et al. 2001), while Leptospira FNR contains some structural features from both types (Catalano-Dupuy et al. 2011). These information have prompted speculation that root and leaf FNRs have evolved through gene duplication and differentiation following the endosymbiotic origin of plastids. To date, X-ray crystal structure-based discussions of molecular interaction and electron transfer between FNR and Fd have been confined to maize leaf (Kurisu et al. 2001) and cyanobacteria (Morales et al. 2000). Consequently, elucidation of the interactions between root FNR and Fd at atomic resolution is fundamental to understand the non-photosynthetic redox cascade. Maize root FNR is functionally distinct from maize leaf FNR, having a higher affinity for root Fd than for leaf Fd (Onda et al. 2000). The 3D structure of maize root FNR shows several structural variations from that of spinach leaf FNR (Aliverti et al. 2001). Here we show the crystal structure of the maize root FNR:Fd electron transfer complex, which provides the details of the interaction between the Fd and FNR (preliminarily reviewed by Hanke et al. 2004). Despite the similarity in the overall structure of the two types of Fd and FNR isoforms in the free state, the orientation of Fd relative to FNR is largely different between the leaf and root complexes. In order to clarify the structural basis for the differential recognition of leaf and root Fds by the FNR isoforms, a series of site-directed mutagenesis of the isotype-specific residues on the interface of Fd and FNR were performed, and their effects on the protein–protein interaction were evaluated.
Experimental section
Preparation of wild-type and site-directed mutants of Fd and FNR
Recombinant maize L-FNR(I) and FdI, and R-FNR and FdIII (Hase et al. 1991; Onda et al. 2000), representative isoforms of leaf- and root-type FNR and Fd, respectively, were used in this study. Site-specific mutants of leaf Fd and root- and leaf FNRs were prepared using a Quikchange mutagenesis kit (Agilent Technologies). The wild-type and mutant FNR and Fd proteins were expressed in Escherichia coli and purified according to the methods described previously (Matsumura et al. 1999; Onda et al. 2000). Absorption spectra of the mutant proteins in the UV–visible region were essentially the same as those of the parental FNRs and Fd.
X-ray crystallographic studies of maize root FNR:Fd complex and free-state forms
A 1:2 molar mixture of R-FNR and FdIII in 50 mM Tris–HCl, pH 7.5 was subjected to microfiltration to remove excess Fd. Crystals were obtained at 4 °C from equal volumes of the concentrated proteins (10 mg/mL) and reservoir solution [20% (w/v) PEG6000, 100 mM sodium cacodylate, pH 6.5] by the hanging drop vapor diffusion method. Crystals of free-state R-FNR and FdIII were also prepared. The reservoir solution for R-FNR was 50 mM Tris–HCl, pH 7.5 containing 1.2 M sodium citrate, and that for FdIII was 50 mM Tris–HCl, pH 8.5 containing 3.6 M ammonium sulfate, and 1% [v/v] ethanol. X-ray intensity data of the complex were collected at a wavelength of 1.0 Å at beamline 41XU on SPring-8 synchrotron radiation (Hyogo, Japan), and those of R-FNR and FdIII at beamline BL18B, Photon Factory (Tsukuba, Japan) and Rigaku RU-200 X-ray generator equipped with R-axis IV imaging plate system. Diffraction data were collected at 100 K and processed with the program DPS/MOSFLM (Rossman and van Beek 1999). The structures were determined by molecular replacement with the program AMORE in the CCP4 program package (CCP4 1994) using maize leaf FNR (Kurisu et al. 2001) and spinach FdI (Binda et al. 1998) as search models. One R-FNR:FdIII and one free R-FNR molecule for the complex crystal, one R-FNR molecule and six FdIII molecules for each free-state crystal were refined in the asymmetric unit with the program CNS (Brunger et al. 1998). Crystal data and refinement statistics are given in Table 1.
Enzymatic analysis
Measurements of NADPH-dependent Fd reduction by FNR were performed as described previously (Onda et al. 2000) by monitoring the increase of reduced cytochrome c at 500 nm in the reaction mixture containing 50 µM NADPH, 200 µM cytochrome c, 40 nM FNR, 50 mM Tris–HCl, pH 7.5, and 100 mM NaCl in the presence of the NADPH- generating system at 25 °C. The reaction was initiated by the addition of Fd at the final concentrations from 0 to 40 µM. Kinetic parameters were extracted from Fd saturation curves, and K m and k cat were determined from a double-reciprocal plot. The measurements were repeated at least three times.
Affinity chromatography
Maize FNRs were immobilized on CNBr-activated Sepharose 4B (GE Healthcare Bioscience) following the manufacturer’s directions. FNR-conjugated resin (approx. 2 ml) was packed into a column, and chromatography was performed as described previously (Sakakibara et al. 2012; Kimata-Ariga et al. 2007). Wild-type and mutant Fds (2–4 nmol) were loaded on the column, and a linear gradient of NaCl from 0 to 300 mM in 50 mM Tris–HCl, pH 7.5 was applied to elute proteins at a flow rate of 0.5 mL/min, monitoring the conductivity and the absorbance at 422 nm for Fd.
Results
Crystal structure of the maize root FNR:Fd complex
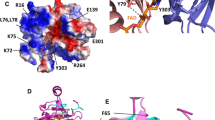
The root electron transfer complex, as shown in Fig. 1a, consists of one molecule each of FNR and Fd, with their redox centers (the [2Fe-2S] cluster in Fd, and FAD in FNR) located face to face. The relative arrangement of the two proteins is similar to the leaf counterpart (Fig. 1b), but the orientation of Fd is different; compared to the leaf complex, Fd in the root complex is rotated approximately 60 degrees along an axis connecting the two prosthetic groups. Within the interface of the intermolecular contact region of the root complex, 3 ionic bridges and 11 hydrogen bonds (8 of which are through water molecules) occur (Fig. 1c), in contrast to 5 ionic bridges in the leaf complex (Kurisu et al. 2001), making a major contribution for determining protein orientation and stabilizing the complex. The buried accessible surface area between root Fd and FNR is relatively narrow (589 and 602 Å2 respectively) compared to those of leaf Fd and FNR (812 and 765 Å2 respectively) resulting in a vacant space at the boundary, unique to the root complex (Fig. 1a).
Structures of the FNR:Fd complexes from maize roots and leaves. Stereo views of a the maize root FNR:Fd complex (this study, PDB code:5H5J) and b the maize leaf FNR:Fd complex (PDB code:1GAW) in the same orientation with respect to FNR. Ribbon diagrams show FNRs colored blue to yellow from the N to C-terminus, and Fds yellow to red. The prosthetic groups, FAD and the [2Fe-2S] cluster, are drawn as ball-and-stick models. c Stereo view of electrostatic and hydrogen bonding interactions at the interface. Interacting amino acids are shown as ball-and-stick models with residue numbers, and water molecules as red balls with numbers. Salt-bridged residue pairs are as follows: Glu35OE2-Lys310NZ; Glu30OE1-Lys156NZ; Asp67OD2-Lys84NZ. Hydrogen bonding residue pairs are as follows: Arg41O-Wat707-Gly155N; Ser39O-Wat554-Ile157N; Ser39O-Wat554-Glu315OE1; Ser39OG-Wat610-Val316N; Arg41O-Wat707-Gly155N; Asp27OD2-Wat530-Asn27OD1; Tyr24OH-Lys30NZ; Asp22O-Wat484-Lys30NZ; Tyr81OH-Lys30NZ; Asp61OD1-Wat415-Lys32N; Ser60OG-Lys32NZ. All figures were generated by Bobscript (Esnouf 1997) and RENDER from the Raster 3D package (Merritt and Murphy 1994)
The close proximity of the prosthetic groups of Fd and FNR in the root complex leads us to postulate that electron transfer can occur through space, as in the leaf complex (Kurisu et al. 2001). The C8 carbon atom of FAD is the closest to the sulfur atom of Fd Cys 40 ligating the FE1 atom of the [2Fe-2S] cluster (Fig. 2a). In the leaf complex, the closest sulfur atom is that of the other cysteine ligand (Cys 44; Fig. 2b), suggesting that the root and leaf complexes utilize the different sides of the [2Fe-2S] cluster for the intermolecular electron transfer. There are no significant conformational changes in root FNR upon complex formation with Fd, whereas several changes were observed in the case of leaf complex (Kurisu et al. 2001). At the time of the leaf complex formation, an intramolecular salt bridge between Arg40 and Glu29 of Fd is broken, and a new intermolecular salt bridge is formed between Arg40 of Fd and Glu154 of FNR (Kurisu et al. 2001). Structural comparison of the root complex with free-state Fd and FNR (determined at 2.49 and 1.65 Å resolution, respectively, structures not shown) reveals that the equivalent intramolecular salt bridge between Arg41 and Glu30 of root Fd remains in the complex.
Comparison of the relative position of the [2Fe-2S] cluster and FAD in root and leaf Fd:FNR complexes. Stereo view of the final σ-weighted 2Fo-Fc electron density omit map around the prosthetic groups in the root (a) and leaf (b) complexes. Distances (Å) from the C8M carbon atom of FAD to the FE1, and SG atoms of cysteines ligating FE1 are shown
We previously reported that maize root FNR exhibits a higher affinity for maize root Fd (K m: 3.3 µM) than for leaf-type Fds (K m: 32.5 µM for maize leaf Fd, and 28.8 µM for spinach leaf Fd) during NADPH oxidation (Onda et al. 2000). Consistently, the K d of root FNR for root Fd (2.5 µM) was less than one-tenth of that for leaf Fd (32.6 µM) from maize (Onda et al. 2000). Therefore, root FNR preferentially interacts with root Fd over leaf Fd, as depicted in Fig. 3a. In contrast, leaf FNR shows similar high affinity for both types of Fds (Onda et al. 2000). The differential interaction mode shown in the crystal structures of leaf and root FNR:Fd complexes could be responsible for this phenomenon. The amino acid residues salt-bridging to FNR in the crystal structures (Fig. 3b, c) are largely conserved between leaf and root Fds. Thus, the structural basis for this differential recognition was investigated using site-directed mutagenesis of the isotype-specific residues on the interface of Fd and FNR, based on the crystal structures of the FNR:Fd complexes from maize leaves and roots.
Differential interaction of Fd and FNR from leaves and roots. a Root FNR preferentially interacts with root Fd while leaf FNR shows similar affinity for both types of Fds. b, c Ribbon drawing of Fd:FNR complexes from maize leaves (PDB code:1GAW) and roots (this study, PDB code:5H5J). Their intermolecular salt bridges are shown as ball-and-stick model. The prosthetic group, FAD and the [2Fe-2S] cluster are drawn as ball-and-stick model and spheres, respectively
Fd mutagenesis on the interface with FNR
In the first place, structural components on the Fd side, responsible for the differential recognition of leaf and root Fds by root FNR, were investigated. For this purpose, isotype-specific amino acid residues of Fd located on the interface with FNR were searched using maize leaf Fd (FdI) and root Fd (FdIII) (Fig. 4a), based on the crystal structure of the root-type complex. There are four pairs of isotype-specific residues on the core region around the [2Fe-2S] cluster, D34/E35, S43/A44, S46/T47, and Q61/G62 of leaf/root type (root Fd has one insertion relative to leaf Fd at the N-terminal region) (Fig. 4b). In addition, maize leaf Fd is longer at the C-terminus (T96G97A98) than root Fd (Y97), where the interaction with FNR is indicated in the leaf-type complex (Fig. 4c). Accordingly, three mutants of leaf Fd containing root-type substitutions were prepared: (1) FdIcon1 with quadruple mutations of D34E/S43A/S46T/Q61G at the core region, (2) FdIctm with one substitution and two deletions of T96Y/∆G97A98 at the C-terminus, and (3) FdIcon1ctm containing both of the above two sets of mutations. The affinity of these mutants for root FNR was investigated, using kinetic analysis (Table 2) and affinity chromatography (Fig. 5). During NADPH oxidation, root FNR has K m values about 5 times higher for leaf Fd (FdI) than for root Fd (FdIII) (Table 2) as reported previously (Onda et al. 2000). The K m values for FdIcon1 mutant was about a half of that of leaf Fd (FdI), and for FdIctm mutant, decreased to about one-third. Moreover, the combination of these mutations, FdIcon1ctm, decreased the K m value as low as that of root Fd (FdIII). The physical interaction between FNR and Fd was further examined using a root FNR-immobilized column (Fig. 5). Both leaf Fd (FdI) and root Fd (FdIII) bound to the FNR-affinity column, but root Fd eluted slower by developing them with a linear gradient of NaCl, indicating that root Fd interacts with root FNR more strongly than leaf Fd, as expected. FdIcon1 eluted at the intermediate volume of leaf and root Fds, and FdIctm and FdIcon1ctm eluted even slower than root Fd, indicating that physical interaction with root FNR became stronger by these mutations. Therefore, the substitutions of the four residues at the core region and three residues at the C-terminus of leaf Fd for corresponding root-type residues were sufficient to attain the higher affinity observed in root Fd/FNR combination.
Isotype-specific amino acid residues of Fds located on the interface with FNR, colored in red on the primary structures of maize leaf Fd (ZmFdI) and root Fd (ZmFdIII) (a), and shown as ball-and-stick model on the three-dimensional structures of the maize root Fd:FNR complex at the core region (b) and the leaf complex at the C-terminus region (c)
Affinity chromatography of wild-type and mutant Fds on a root FNR-immobilized column. Conditions for the chromatography are described under “Experimental section”
FNR mutagenesis on the interface with Fd
The results of Fd mutagenesis indicate that root FNR discriminates the isotype-specific Fd residues shown in Fig. 4, and thus lowers the affinity for leaf Fd. In contrast, leaf FNR retains similar and relatively high affinity for both Fds irrespective of such structural difference. Therefore, next we investigated the structural components on the FNR side, responsible for such a differential recognition for leaf and root Fds. As with the case of Fd mutagenesis, isotype-specific amino acid residues of leaf and root FNRs from maize (Fig. 6a) located on the interface with Fd were searched based on the crystal structure of the leaf-type complex. Nine residues were found (Fig. 6b) and grouped into six pairs; root/leaf-type pairs of P85A87N90/N86K88K91, S154I157/V151E154, M281/E278, D300/F297, A304/K301, and K310/D307 (leaf FNR has several insertions relative to root FNR). Root FNR mutants containing leaf-type substitutions at these sites were prepared, and the resulting six mutants showed variable effect on their affinity for leaf and root Fds (Table 3); (1) P85N/A87K/N90K, D300F, and A304K largely decreased the K m values for both leaf and root Fds, while (2) S154V/I157E, M281E, and K310D largely increased the K m values for both Fds. Because the group (1) mutants confer the increase in the positive charge (or decrease in the negative charge), and group (2) mutants confer the increase in the negative charge, these effect on the affinity can be at least partly explained by the changes in the extent of electrostatic interaction with Fd, of which FNR-interacting surface is largely negatively charged. Notably, group 1) mutants exhibited the affinity for root Fd even higher than that of wild-type root FNR, with about 1 µM of K m values. Irrespective of the large changes in the K m values, both of the two groups of the FNR mutants still retained preferred affinity for root Fd over leaf Fd, with 3–7 time differences in the K m values. However, the combination of each one of the group (1) and group (2) mutations, resulting in P85N/A87K/N90K/S154V/I157E, slightly reduced this difference down to 2.5 times. Further addition of group 2) K310D mutation yielding P85N/A87K/N90K/S154V/I157E/K310D almost diminished this difference, although the K m values became higher than 40 µM for both Fds. Nevertheless, the more addition of D300F mutation of group 1), resulting in seven-residue substitutions, lead to the moderate and similar affinity for both Fds (13.0 ± 3.5 µM for leaf Fd and 11.0 ± 2.0 µM for root Fd; in the last line of Table 3). Therefore, these seven residues on the Fd-binding interface of FNR were shown to be involved in the conversion of isotype-specific recognition mode. Complemental mutagenesis of leaf FNR, which conversely replace the eight isotype-specific residues described above, caused the reverse effects observed in the root FNR mutagenesis (Table 4). The K m values increased with N86P/K88A/K90N, F297D, and K310A mutants, and decreased with V151S/E154I and E278M mutants, although the effects were not so significant except for N86P/K88A/K90N mutant. All these leaf FNR mutants did not show significant difference between the K m values for leaf and root Fds.
Discussion
Comparison of the 3D structures of the two maize FNR:Fd complexes revealed a remarkable difference in the orientation of FNR and Fd between leaf and root types (Fig. 1). Accordingly, the orientation of the prosthetic groups and the electron transfer route are altered between them (Fig. 2), which may relate to the opposite direction of electron transfer occurring in the photosynthetic and non-photosynthetic processes. Notably, relatively narrow buried accessible surface area between root Fd and FNR leaves a vacant space at the boundary (Fig. 1a). This situation is similar to those observed in the crystal structures of FNR-related redox enzymes containing Fe–S domain, phthalate dioxygenase reductase of Pseudomonas cepacia (Correl et al. 1993) and Benzoate dioxygenase reductase of Acinetobacter sp. (Karlsson et al. 2002). These enzymes which deliver electrons from their own Fe–S cluster to a downstream enzyme have an open space between the Fe–S and flavin/NAD(P)H domains, possibly used for the site of protein–protein interactions. Therefore, the vacant space observed in the root Fd:FNR complex may be used for the formation of a ternary complex with downstream enzymes. An interesting hypothesis is that the vacant space could allow the access of a third molecule of Fd-dependent enzyme, such as nitrite reductase and sulfite reductase (Akashi et al. 1999), forming a ternary complex for efficient electron channeling from NADPH to those assimilatory metabolisms. The possibility for the formation of a ternary complex with sulfite reductase was investigated by (1) gel filtration and (2) pull down analysis using R-FNR-immobilized resin, but no evidence for such a complex formation has been obtained so far (data not shown).
Whereas the Fd internal salt bridge between Arg40 and Glu29 is disrupted in the leaf complex (Kurisu et al. 2001), the equivalent salt bridge is retained in the root complex. This structural alteration in leaf Fd may cause a negative redox potential shift of the [2Fe-2S] cluster, which favors the electron flow from Fd to FNR, as discussed previously (Kurisu et al. 2001). In the root complex, on the other hand, such a negative redox potential shift would be detrimental to electron flow from FNR to Fd, and therefore, these differences in the Fd structural alteration between the two complexes may be physiologically relevant. Another difference between leaf and root complexes revealed by the structure comparison with their free-state FNRs is the structural perturbation of FNR, such as the large movement of E312 and slight movement of the entire NADPH domain, which is prominent only in the case of leaf complex (Kurisu et al. 2001). Such structural changes are not observed in the cyanobacterial complex, and therefore may relate to leaf-specific modulation of enzymatic property of FNR, such as induced-fit like changes in the active site upon complex formation, which optimize the orientation of NADP+ for the productive interaction with the FAD (Kurisu et al. 2001).
Because the amino acid residues salt-bridging to FNR are largely conserved between leaf and root Fds (Fig. 3b, c), the mechanism of how root FNR discriminates the two types of Fds was intriguing. In this study, several isotype-specific residues of Fd on the interface with FNR (Fig. 4) were shown to be responsible for the differential affinity for root FNR. With regard to the residues in the core region (Fig. 4b), the substitutions do not seem to confer large changes in the charge and/or size except Q61/G62. In addition, the backbone structure of this region is very similar between leaf and root Fds. Thus, differences in the minute structures and/or polarity in this region of Fd may be important for the strict recognition by root FNR. Also, differences in the length of D34/E35 may be crucial for the affinity so that E35 forms a salt bridge with K310 of root FNR in the root complex (Fig. 3c). On the other hand, larger differences in the structure were introduced by substitutions at the C-terminal region (Fig. 4c). The extra two residues of leaf Fd may hinder the stable interaction with root FNR, and/or the C-terminal Tyr residue of root Fd may be important for the interaction with root FNR, although this Tyr residue appears to be freely exposed to the solvent in the crystal structure of the root complex. More extensive single mutagenesis of leaf Fd and experimental structure analysis of the cross complex between leaf Fd and root FNR will be required to verify these considerations.
Unlike the case of Fd, three (K88, K91, and E154) out of five leaf FNR residues and one (K310) of the three root FNR residues involving salt bridges with Fd (Fig. 3b, c) are unique to each isoform. Seven isotype-specific residues of FNR including these four salt-bridging residues were shown to be responsible for the differential interaction with Fds. Additions of other two isotype-specific substitutions which were not included into the above septuplet mutant, M281E and A304K, may further optimize the leaf-type recognition. Substitutions of the seven residues result in considerable changes in the charge distribution on the Fd-binding surface of FNR as shown in Fig. 7. Thus, these results lead to the mechanism that the changes in the charge distribution on the Fd-interface of FNR induce isotype-specific binding mode of Fd relative to FNR. Among the seven residues changed in this study, P85N/A87K/N90K and S154V/I159E appear to induce leaf-type-specific salt bridges while K310D breaks root-type-specific salt bridge. The remaining D300F may stabilize the hydrophobic interaction with the core region of Fds in leaf-type orientation. Because the D300 in root FNR is freely exposed to solvent in root Fd:FNR complex, the function of this isotype-specific residue is not clear, but may be important for the function other than the recognition of Fd, such as binding with a third molecule. Alternatively, this D300 may be important for root FNR not to assume the leaf-type Fd:FNR interaction mode. In this connection, it is intriguing that group 1) root FNR mutants, which shows higher affinity (as low as approx. 1 µM of K m for root Fd) than wild-type root FNR, were obtained. Root FNR may have acquired root-type Fd specificity at the expense of the decrease in the affinity, and/or specific distribution of negative and positive charges would be important for adequate affinity for Fd (around 3 µM of K m ) which may be suitable for the physiological function. In conclusion, root FNR discriminates leaf and root Fds by recognizing the differences in their precise structure on the FNR interface, which would not be recognized by leaf FNR. Such differential recognition of leaf and root FNRs could be attained through differential distribution of electrostatic-interaction sites, which is induced by the replacement of negative and positive charges on the Fd-binding surface of leaf and root FNR molecules. In higher plants, isotype-specific amino acid residues of FNR, shown in Fig. 7, are mostly conserved among the same group of leaf type and root type (Hanke et al. 2004). However, the residues of the corresponding sites varied among those FNRs from cyanobacteria, unicellular green algae, Leptospira, and apicomplexan parasites (Catalano-Dupuy et al. 2011). In fact, the orientation and interaction mode of Fd and FNR in the cyanobacterial complex are largely different from either of leaf and root complexes (Morales et al. 2000). The similarity of the Fd-binding mode between root FNR and P. falciparum FNR which drives heterotrophic metabolism has been presented using NMR analysis (Kimata-Ariga et al. 2007). Therefore, the strategy used for the interaction between Fd and FNR seems to be variable, and would be developed according to the roles of this redox pair in each organism.
Surface charge distribution on the Fd-binding side of maize leaf and root FNRs. Positive potential is shown in blue and negative potential in red. Seven isotype-specific residues shown to be responsible for the differential interaction with Fds in this study are indicated (yellow circles). These residues at least partly involve the differences in the surface charge distribution between the two types of FNRs. The figure was produced with PyMol (DeLano 2002)
Physiological significance of the differential recognition of leaf and root Fd by root FNR is not clear at present. Non-photosynthetic-type FNR and Fd are constitutively expressed and present in photosynthetic tissues also (Kimata and Hase 1989; Hase et al. 1991; Onda et al. 2000). Thus, root-type FNR may need to deliver electrons from NADPH preferentially to root-type Fd over leaf-type Fd in photosynthetic tissues under certain circumstances, possibly by forming a ternary complex with downstream redox enzymes.
References
Akashi T, Matsumura T, Ideguchi T, Iwakiri K, Kawakatsu T, Taniguchi I, Hase T (1999) Comparison of the electrostatic binding sites on the surface of ferredoxin for two ferredoxin-dependent enzymes, ferredoxin-NADP-reductase and sulfite reductase. J Biol Chem 274:29399–29405
Aliverti A, Faber R, Finnerty CM, Ferioli C, Pandini V, Negri A, Karplus PA, Zanetti G (2001) Biochemical and crystallographic characterization of ferredoxin-NADP+ reductase from nonphotosynthetic tissues. BioChemistry 40:14501–14508
Arakaki AK, Ceccarelli EA, Carrillo N (1997) Plant-type ferredoxin-NADP+ reductases: a basal structural framework and a multiplicity of functions. FASEB J 11:133–140
Binda C, Coda A, Aliverti A, Zanetti G, Mattevi A (1998) Structure of the mutant E92K of [2Fe-2S] ferredoxin I from Spinacia oleracea at 1.7 A resolution. Acta Crystallogr D 54:1353–1358
Bowsher CG, Hucklesby DP Emes MJ (1993) Induction of ferredoxin-NADP+ oxidoreductase and ferredoxin synthesis in pea root plastids during nitrate assimilation. Plant J 3:463–467
Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54:905–921
Catalano-Dupuy DL, Musumeci MA, López-Rivero A, Ceccarelli EA (2011) A highly stable plastidic-type ferredoxin-NADP(H) reductase in the pathogenic bacterium Leptospira interrogans. PLoS One 6(10):e26736
Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D 50:760–763
Correll CC, Ludwig ML, Bruns CM, Karplus PA (1993) Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Protein Sci 2:2112–2133
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos
Emes MJ, Neuhaus HE (1997) Metabolism and transport in non-photosynthetic plastids. J Exp Bot 48:1995–2005
Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph 15:132–134
Goss T, Hanke G (2014) The end of the line: can ferredoxin and ferredoxin NADP(H) oxidoreductase determine the fate of photosynthetic electrons? Curr Protein Pept Sci 15(4):385–393
Hanke G, Mulo P (2013) Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ 36(6):1071–1084
Hanke GT, Kurisu G, Kusunoki M, Hase T (2004) Fd:FNR electron transfer complexes: evolutionary refinement of structural interactions. Photosynth Res 81:317–327
Hase T, Kimata Y, Yonekura K, Matsumura T, Sakakibara H (1991) Molecular-cloning and differential expression of the maize ferredoxin gene family. Plant Physiol 96:77–83
Karlsson A, Beharry ZM, Eby DM et al (2002) X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. Strain ADP1. J Mol Biol 318:261–272
Kimata Y, Hase T (1989) Localization of ferredoxin isoproteins in mesophyll and bundle sheath cells in maize leaf. Plant Physiol 89:1193–1197
Kimata-Ariga Y, Saitoh T, Ikegami T, Horii T, Hase T (2007) Molecular interaction of ferredoxin and ferredoxin-NADP+ reductase from human malaria parasite. J Biochem 142:715–720
Knaff DB (1996) Ferredoxin and ferredoxin-dependent enzymes. In: Ort DR, Yocum CF (eds) Oxygenic photosynthesis: the light reactions. Advances in photosynthesis and respiration, vol 4. Springer, Dordrecht, pp 333–361
Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimata-Ariga Y, Hase T (2001) Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP+ reductase. Nat Struct Biol 8:117–121
Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Murata H, Takao T, Shimonishi Y, Hase T (1999) Complementary DNA cloning and characterization of ferredoxin localized in bundle-sheath cells of maize leaves. Plant Physiol 119:481–488
Merritt EA, Murphy MEP (1994) Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D 50:869–873
Morales R, Charon MH, Kachalova G, Serre L, Medina M, Gómez-Moreno C, Frey M (2000) A redox-dependent interaction between two electron-transfer partners involved in photosynthesis. EMBO Rep 1:271–276
Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T (2000) Differential interaction of maize root ferredoxin:NADP+ oxidoreductase with photosynthetic and nonphotosynthetic ferredoxin isoproteins. Plant Physiol 123:1037–1045
Rossmann MG, van Beek CG (1999) Data processing. Acta Cryst D 55:1631–1653
Sakakibara Y, Kimura H, Iwamura A, Saitoh T, Ikegami T et al (2012) A new structural insight into differential interaction of cyanobacterial and plant ferredoxins with nitrite reductase as revealed by NMR and X-ray crystallographic studies. J Biochem 151(5):483–492
Vollmer M, Thomsen N, Wiek S, Seeber F (2001) Apicomplexan parasites possess distinct nuclear-encoded, but apicoplast-localized, plant-type ferredoxin-NADP+ reductase and ferredoxin. J Biol Chem 276:5483–5490
Acknowledgements
The authors thank Prof. Michael J. Emes (University of Guelph), Prof. Masami Kusunoki (University of Yamanashi), Prof. T. Tsukihara (IPR, Osaka University), Drs. N. Kamiya (Riken), M. Kawamoto (JASRI), N. Igarashi, M. Suzuki, N. Watanabe, and N. Sakabe (PF, KEK) for their helpful discussions, and Mses. R. Igarashi and S. Ujita for initial crystallization trials. This work was supported in part by grants-in-aids from the Ministry of Culture, Education, Science and Sports and Technology of Japan and from Japan Science and Technology Agency (JST)–CREST.
Author information
Authors and Affiliations
Corresponding author
Additional information
Fumio Shinohara, Genji Kurisu and Toshiharu Hase have contributed equally.
Coordinates: Atomic coordinates have been deposited in the Protein Data Bank under ID codes of 5H57 for FdIII, 5H59 for R-FNR and 5H5J for FdIII:R-FNR.
Rights and permissions
About this article
Cite this article
Shinohara, F., Kurisu, G., Hanke, G. et al. Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP+ oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation. Photosynth Res 134, 281–289 (2017). https://doi.org/10.1007/s11120-016-0331-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-016-0331-1