Abstract
Chlorophyll biosynthesis plays a crucial role in the greening process and survival of etiolated seedlings and yet the mechanism underlying the regulation of this process is poorly understood. Upon light stimulation, NADPH: protochlorophyllide oxidoreductase (POR) catalyzes the reduction of protochlorophyllide (Pchlide) to chlorophyllide. Whereas this represents a key step in the chlorophyll biosynthetic pathway, the regulation of POR remains largely unknown. Three POR isoforms exist in Arabidopsis thaliana, i.e., PORA, PORB, and PORC. In this study, we identified a transcription factor, REVEILLE1 (RVE1), that binds directly to the PORA promoter through the EE-box cis-regulatory element. Analysis of PORA expression in RVE1 loss-of-function (rve1) and overexpression (RVE1-OX) Arabidopsis plants showed that RVE1 positively regulates the transcription of PORA. We found that Pchlide levels were reduced in RVE1-OX seedlings. Furthermore, rve1 etiolated seedlings had lower greening rates than the wild type when exposed to light, whereas RVE1-OX seedlings had higher greening rates. In addition, when etiolated seedlings were exposed to light, RVE1-OX plants had less reactive oxygen species (ROS) accumulation and cell death than the wild type, and had reduced levels of ROS-responsive gene expression. Taken together, our study reveals an important role for RVE1 in regulating chlorophyll biosynthesis and promoting seedling greening during early plant growth and development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorophylls are the major pigments that harvest light energy and drive electron transfer during photosynthesis in plants. Abnormal chlorophyll formation affects chloroplast biogenesis (Frick et al. 2003; Pogson and Albrecht 2011). Chlorophylls are biosynthesized in chloroplasts via a branch of the tetrapyrrole metabolic pathway that involves sequential enzymatic conversion by more than a dozen nucleus-encoded plastid-localized enzymes (Tanaka and Tanaka 2007; Tanaka et al. 2011), and chlorophyll biosynthesis is regulated at the transcriptional, post-translational, and redox levels (Tanaka and Tanaka 2007). Miniarray profile analysis demonstrated that light and circadian signals broadly affect the expression of genes involved in the chlorophyll biosynthetic pathway (Matsumoto et al. 2004). Two transcription factors of the light signaling pathway, FAR-RED ELONGATED HYPOCOTYLS3 and FAR-RED-IMPAIRED RESPONSE1, bind to the promoter of HEMB1, which encodes 5-aminolevulinic acid dehydratase, and positively regulate its expression (Tang et al. 2012). A regulatory protein, FLU, binds to glutamyl-tRNA reductase and strongly represses 5-aminolevulinic acid synthesis (Meskauskiene et al. 2001). Furthermore, thioredoxin has been reported to regulate CHLI1 at the redox state level, with the ATPase activity of CHLI1 being reversibly inactivated by oxidation (Ikegami 2007). Misregulation of metabolism can lead to severe photooxidative stress.
Under dark conditions, the chlorophyll biosynthetic pathway is paused at the intermediate protochlorophyllide (Pchlide). Light activates NADPH: protochlorophyllide oxidoreductase (POR) which catalyzes the reduction of Pchlide to produce chlorophyllide and then chlorophyll a and b (Runge et al. 1996; Buhr et al. 2008; Heyes and Hunter 2005). POR has a photoprotective role during plant seedling greening (Sperling et al. 1997; Buhr et al. 2008). Over-accumulation of Pchlide in darkness and/or impairment of POR activity may result in the production of reactive oxygen species (ROS) upon light stimulation and even cause cell death in the cotyledons (op den Camp et al. 2003). Therefore, chlorophyll biosynthesis is critical for plant growth and survival and must be properly regulated.
POR is an important target for transcriptional regulation in chlorophyll biosynthesis. The Arabidopsis genome encodes three POR homologs, designated as PORA, PORB, and PORC, which are differentially expressed during plant development (Masuda et al. 2003; Frick et al. 2003). PORA is highly expressed in etiolated seedlings and its transcript levels decline rapidly after exposure to light, whereas PORB and PORC are up-regulated in response to light (Su et al. 2001). Several transcription regulators have been shown to mediate the expression of the POR genes. PHYTOCHROME-INTERACTING FACTOR1 (PIF1), a bHLH transcription factor of the light signaling pathway, regulates chlorophyll biosynthesis by binding to the promoter of PORC and activating its expression (Moon et al. 2008). ETHYLENE INSENSITIVE3 (EIN3), a central component of ethylene signaling, directly activates the expression of PORA and PORB, and consequently the ein3 loss-of-function mutant accumulates excess Pchlide in etiolated seedlings and becomes photobleached after light exposure (Zhong et al. 2009). In addition, DELLA proteins up-regulate POR expression and limit ROS accumulation and photooxidative damage during seedling de-etiolation (Cheminant et al. 2011). However, the level of POR mRNA was moderately reduced in the pif1, ein3, and della mutants, suggesting that other regulator(s) might be involved in regulating the transcription of POR genes.
In this study, we focused on the regulation of PORA and identified REVEILLE1 (RVE1) as its direct regulator by yeast one-hybrid screening. We demonstrate that RVE1 binds to the promoter region of PORA in vitro and in vivo. Moreover, we showed that RVE1 regulates PORA expression and etiolated seedling greening. Our results demonstrate that the RVE1 transcription factor is a novel positive regulator of chlorophyll biosynthesis and seedling de-etiolation.
Materials and methods
Plant materials and growth conditions
The rve1-1 (Salk_057420) mutant and RVE1-OX transgenic plants are in the Arabidopsis thaliana Columbia (Col) background (Rawat et al. 2009). After sterilization, seeds were sown on MS medium containing 1 % Suc and 0.8 % agar, incubated at 4 °C in darkness for 3 days, and then exposed to white light for 9 h to promote germination. White light was supplied by cool-white fluorescent lamps.
Plasmid construction
To amplify the open reading frame (ORF) of RVE1, total RNA was isolated from Col wild-type seedlings, and first-strand cDNA was reverse transcribed using oligo(dT)18 primer. The ORF of RVE1 was amplified by high-fidelity Pfu DNA polymerase (Invitrogen) and cloned into the pEASY vector (Transgen) to generate pEASY-RVE1. The pEASY-RVE1 plasmid was digested with EcoRI and SalI and the RVE1 ORF was ligated into the EcoRI/XhoI sites of the JG4-5 vector (Clontech), resulting in pAD-RVE1. The same fragment was cloned into the EcoRI/XhoI sites of pET28a (Novagen), to generate pET-HIS-RVE1.
To generate 35S: Myc-RVE1 transgenic plants, the pEASY-RVE1 plasmid was digested with EcoRI and SalI to release the RVE1 ORF, which was then ligated into the EcoRI/XhoI sites of pRI101AN-6Myc (Takara), resulting in 35S:Myc-RVE1.
The promoter sequence (~1.2-kb fragment upstream of the start codon) of PORA was amplified from Col genomic DNA and ligated into the pEASY vector, giving rise to pEASY-PORAp. This plasmid was then digested with EcoRI and XhoI to release the RVE1 fragment, which was then inserted into the EcoRI/XhoI sites of pLacZi2u (Lin et al. 2007) to produce pPORA:LacZ. pEASY-PORAp was digested with EcoRI and XbaI, and the PORA fragment was ligated into the EcoRI/XbaI sites of the pHISi-1 vector (Clontech), resulting in pPORA: HIS.
Plant transformation
The 35S: Myc-RVE1 binary vector was electroporated into the Agrobacterium tumefaciens strain GV3101 and then introduced into the rve1 mutant via the floral dip method (Clough and Bent 1998). Transgenic plants were selected on MS plates containing 50 mg/L hygromycin.
Yeast one-hybrid assay
Yeast one-hybrid screening was performed as described in the Yeast Protocol Handbook (Clontech). Briefly, the pPORA: HIS target reporter construct was first integrated into the genome of yeast strain YM4271, and the background expression was tested in the presence of a series of concentrations of 3-amino-1,2,4-triazole (3-AT). The plasmid DNAs were isolated from the Arabidopsis cDNA Library (CD4-30, TAIR, ligated in the pAD-GAL4-2.1 vector), which contains about 1 × 106 clones. These plasmid DNAs were then transformed into the yeast strain containing the pPORA: HIS reporter gene, and the transformants were selected on dropout plates supplemented with synthetic dextrose (SD)/-His-Leu plus 45 mM 3-AT. The largest colonies were picked and restreaked on the same medium, and positive colonies were sequenced.
pAD-RVE1 (or pJG4-5 control) and pPORA: LacZ plasmids were co-transformed into yeast strain EGY48 using the method described in the Yeast Protocols Handbook. Transformants were grown on SD/-Trp-Ura dropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for color development. Protein-DNA interactions resulted in blue yeast colonies.
EMSA
Electrophoresis mobility shift assay (EMSA) was performed as previously described (Tang et al. 2012). HIS-RVE1 recombinant fusion proteins were expressed in the Escherichia coli BL21 (DE3) strain. The proteins were then purified using Ni-NTA Agarose (Qiagen), according to the manufacturer’s instructions. The core evening element sequence, AAAATATCT, was mutagenized to AAACTGCAG. The DNA–protein interaction signal was detected on X-ray film.
Chromatin immunoprecipitation assay
The ChIP experiment was carried out as previously described (Tang et al. 2012). The chromatin complexes were isolated from the 35S: Myc-RVE1 and Col wild-type seedlings and precipitated with anti-Myc antibody (Abcam). The precipitated DNA fragments were recovered and quantified by quantitative PCR. The primer sequences are shown in Supplemental Table 1.
Gene expression analysis
Plant total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen), and first-strand cDNA was synthesized using reverse transcriptase (Invitrogen). Quantitative PCR was carried out using the SYBR Premix ExTaq Kit (Takara) following the manufacturer’s instructions. The expression levels were normalized to those of UBIQUITIN1 (UBQ1). The primers are listed in Supplemental Table 1.
Measurement of greening rate
Dark-grown seedlings were transferred to different intensities of continuous white light for 2 days at 22 °C as indicated in the text. The phenotypes of the cotyledons were recorded using a digital camera. Greening rate was scored as the percentage of dark-green cotyledons from 50 to 80 seedlings of each genotype. At least three independent biological repeats were performed for each genotype examined.
Determination of pchlide
Fifty etiolated seedlings were collected and homogenized in 500 µL of ice-cold 80 % acetone and incubated in darkness for 4 h. The samples were centrifuged at 5000 g for 5 min and 150 µL of the supernatant was mixed with 350 µL of glycol. Room temperature fluorescence was excited at 440 nm and scanned from 600 to 720 nm using a fluorescence spectrophotometer (Hitachi).
Trypan blue staining, H2DCFDA fluorescence determination, and analysis of electrolyte leakage
Trypan blue staining, H2DCFDA fluorescence and electrolyte leakage analyses were carried out as previously indicated (Chen et al. 2013). Dark-grown seedlings were transferred to white light for 2 days before staining. Seedlings were mounted on slides, and the cotyledons were photographed using the camera coupled to a dissecting microscope (Olympus).
Results
RVE1 directly binds to the cognate promoter region of PORA
To identify novel upstream regulator(s) of PORA, we generated a pPORA:HIS reporter construct in which the HIS reporter gene was driven by a 1.2-kb PORA promoter sequence, and screened an Arabidopsis GAL4 activation domain fusion library using a yeast one-hybrid approach. We identified five positive colonies that contained sequences from the same gene, RVE1 (At5g17300). We then fused the full-length open reading frame of RVE1 to the activation domain of B42 (AD-RVE1) and co-transformed yeast with the resulting construct and pPORA: LacZ (containing the LacZ reporter gene driven by the PORA promoter). AD-RVE1, but not AD alone, was able to bind pPORA: LacZ and activate LacZ reporter expression (Fig. 1a), confirming the interaction between RVE1 and the PORA promoter in yeast cells.
RVE1 directly binds to the promoter of PORA. a Yeast one-hybrid assay. The LacZ reporter gene driven by the PORA promoter was co-expressed with AD-RVE1 or AD alone in the yeast strain EGY48. b EMSA in which His-RVE1 recombinant proteins were incubated with 32P-labled PORA oligonucleotides in the absence or presence of series of excess amounts of unlabeled wild-type or mutant competitors. PORApEE, PORA promoter fragment containing EE-box; PORApmEE, PORA promoter fragment containing mutated EE-box. Arrow indicates shifted bands of protein-DNA complexes.c ChIP assay. Chromatin samples were isolated from 4-day-old dark-grown Col wild type and 35S: Myc-RVE1 seedlings, and immunoprecipitated with Myc antibody. ChIP DNA was quantified by real-time PCR with primers targeting PORA fragments of the promoter (PORAp) or coding (PORAc) regions. Mean ± SD of three technical replicates. Asterisks denote statistically significant differences (P < 0.01, Student’s t test). The experiments were repeated twice with similar results. The upper panel shows a diagram of PORA and the regions amplified by PCR. The triangle denotes the position of the putative evening element (AAAATATCT) and digitals indicate distance (bp) upstream or downstream of the translation start codon
RVE1 is a Myb-like clock-regulated transcription factor that is homologous to the central clock proteins, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) (Rawat et al. 2009). These proteins can bind a conserved motif, termed evening element (EE, AAAATATCT), in the promoter of target genes (Alabadi et al. 2001; Rawat et al. 2009). The PORA promoter contains a putative EE. To further determine the direct interaction between the PORA promoter and RVE1 in vitro, we conducted an EMSA. When His-RVE1 recombinant protein was incubated with 32P-labeled PORA fragment, an upshifted band was produced. This specific band was abolished by excess un-labeled wild-type oligonucleotides, but not by excess unlabeled oligonucleotides containing mutations within the EE motif (AAACTGCAG) (Fig. 1b). We then generated transgenic plants that overexpressed the Myc-RVE1 fusion under the control of the 35S promoter (35S: Myc-RVE1), and used these and wild-type plants in a chromatin immunoprecipitation (ChIP) assay with MYC antibody. The precipitated DNA was amplified with primer pairs annealing to either the promoter or coding regions of PORA. The promoter fragment containing the EE-box, but not the coding region, was markedly enriched when DNA extracted from the 35S: Myc-RVE1 seedlings was precipitated with Myc antibody compared with the Col wild-type control (Fig. 1c). These results together indicate that PORA is a direct target of RVE1.
RVE1 positively regulates PORA expression
To test whether RVE1 regulates the expression of PORA and its homologous genes, we analyzed the rve1-1 knock-out mutant and RVE1 overexpression transgenic plants (RVE1-OX) (Rawat et al. 2009; Fig. 2a). Seedlings were grown in darkness for 6 days, and the transcript levels of PORA, PORB, and PORC were analyzed by reverse transcription followed by quantitative polymerase chain reaction (qRT-PCR). We found that the expression levels of PORA were drastically decreased in rve1-1, but increased in the RVE1-OX transgenic plants compared with the Col wild-type control (Fig. 2b). Surprisingly, PORB transcript levels were reduced in both the rve1-1 mutant and RVE1-OX plants. The expression of PORC was not dramatically affected (Fig. 2b). These data indicate that RVE1 activates PORA expression in Arabidopsis seedlings.
RVE1 modulates PORA expression. a RVE1 expression levels in Col, rve1, and RVE1-OX seedlings. b Col, rve1, and RVE1-OX seedlings were grown in darkness for 6 days and the expression of PORA, PORB, and PORC was analyzed by qRT-PCR. Relative expression levels were normalized to that of UBQ1. Data are Mean ± SD of three technical replicates. Asterisks denote statistically significant differences compared with the wild-type control (P < 0.01, Student’s t test). The experiments were repeated twice with similar results
Overexpression of RVE1 decreases pchlide accumulation
Pchlide is the substrate of POR in the chlorophyll biosynthesis pathway (Tanaka and Tanaka 2007). We examined whether the level of Pchlide was affected by mutation or overexpression of RVE1. We extracted Pchlide from 6-day-old etiolated seedlings exposed or not to white light for 5 min and determined the fluorescence emission of the isolated compounds using a fluorescence spectrophotometer. As shown in Fig. 3a, Pchlide levels (peak at 635 nm) were similar in rve1-1 and the Col wild type. However, we found that Pchlide levels were much lower in the RVE1-OX plants than in the wild-type seedlings when grown in darkness (Fig. 3a). After 5 min of light illumination, Pchlide and chlorophyllide (peak at 672 nm) levels were much lower in RVE1-OX than in the wild-type control, whereas those of rve1 were slightly increased (Fig. 3b). These data suggest that overexpression of RVE1 decreases Pchlide accumulation.
Pchlide levels in the mutant and transgenic plants of RVE1. a, b Pchlide and chlorophyllide levels in 6-day-old dark-grown seedlings a or in 6-day-old dark-grown seedling transferred to light for 5 min b. Room temperature fluorescence was excited at 440 nm and detected from 600 to 720 nm by a fluorescence spectrophotometer. c Expression of chlorophyll biosynthetic genes in 6-day-old dark-grown Col and RVE1-OX seedlings. Relative expression levels were normalized to that of UBQ1. Data are Mean ± SD of three technical replicates. Asterisks denote statistically significant differences compared with the wild-type control (P < 0.01, Student’s t test). The experiments were repeated twice with similar results
To determine the cause of the low accumulation of Pchlide in RVE1-OX plants, we randomly selected four genes encoding enzymes in the chlorophyll biosynthetic pathway prior to Pchlide formation and examined their transcript levels. These genes includes GSA2 (encoding glutamate 1-semialdehyde aminotransferase), HEMA1 (encoding glutamate-tRNA reductase), CHLH (encoding a subunit of magnesium chelatase), and CRD1 (encoding Mg-proto IX monomethyl cyclase). The mRNA levels of GSA2, CHLH and CRD1 were decreased in RVE1-OX plants compared with the wild type, whereas HEMA1 expression was not affected (Fig. 3c), indicating that RVE1 regulates the transcription of other chlorophyll biosynthetic genes in addition to PORA.
RVE1 promotes seedling greening
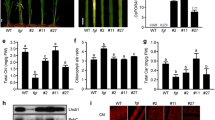
Next, we investigated whether RVE1 is a regulator of the seedling greening process. Etiolated seedlings grown in darkness for a period of time were transferred to white light (150 µmol m−2 s−1), and the percentage of green cotyledons (greening rate) was calculated after 2 days of illumination. When the seedlings were grown in darkness for 4 days and then illuminated for 2 days, all of the rve1 and RVE1-OX seedlings turned green, as did wild-type seedlings (Fig. 4a). However, when 6-day-old etiolated seedlings were transferred to light, the greening rate of the rve1-1 mutant was much lower than that of the wild type, whereas that of RVE1-OX was drastically higher. After 8 days of growth in darkness, the rve1 mutants and wild-type seedlings were unable to turn green upon transfer to light conditions, whereas more than 4 % of RVE1-OX seedlings became green after 2 days of illumination (Fig. 4a). We also tested the effect of light intensity on greening ability using 6-day-old etiolated seedlings. The greening rate of rve1-1 was much lower than that of the wild type, whereas the rate of RVE1-OX was higher than that of the control when subjected to a light intensity of 300 µmol m−2 s−1. Under high light conditions (500 µmol m−2 s−1), more than 20 % of the RVE1-OX seedlings turned green, while the rve1 mutants could not survive (Fig. 4b). These results indicate that RVE1 promotes greening during seedling de-etiolation.
RVE1 regulates seedling greening. a The percentage of green cotyledons of seedlings grown in darkness for the indicated periods of time followed by illumination with 150 µmol m−2 s−1 white light for 2 days. b Percentage of plants with green cotyledons when 6-day-old etiolated seedlings were exposed to the indicated intensities of white light for 2 days. For a and b, data denote Mean ± SD of four replicates. Asterisks denote statistically significant differences compared with the wild-type control (**for P < 0.01, *for P < 0.05, Student’s t test)
RVE1 regulates ROS accumulation and cell death
To test whether the altered greening rate of the rve1 mutant and RVE1-OX seedlings is the result of photobleaching, we investigated total ROS production by detecting the 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) fluorescence of the cotyledons (Chen et al. 2013). When 6-day-old dark-grown seedlings were illuminated for 24 h, H2DCFDA fluorescence (Fig. 5a, shown in green) was similar between rve1 and wild type but barely detectable in RVE1-OX, whereas chlorophyll auto-fluorescence (shown in red) exhibited the reverse pattern (Fig. 5a). Accumulated ROS within the cell may lead to cell death. After 2 days of light exposure, the seedlings were stained with trypan blue to monitor the extent of cell death. We found that the cotyledons of rve1 mutant and wild type exhibited strong blue staining, whereas the RVE1-OX seedlings were barely stained (Fig. 5b). Consistently, the cell death-induced electrolyte leakage was much reduced in RVE1-OX compared with wild type and rve1 mutant (Fig. 5c). Therefore, overexpression of RVE1 inhibits photobleaching and cell death during seedling greening.
RVE1 regulates ROS production and cell death. a Cellular ROS levels in the cotyledons of wild-type (Col), rve1, and RVE1-OE seedlings. H2DCFDA fluorescence (green) indicates ROS and chlorophyll auto-fluorescence is shown in red. b Trypan blue staining of cotyledons of wild-type (Col), rve1, and RVE1-OE seedlings. For a and b, 6-day-old etiolated seedlings were exposed to white light (150 µmol m−2 s−1) for 2 d. Bars denote 200 µm. c Six-day-old etiolated seedlings were exposed to light (150 μmol m−2 s−1) for 12 h and immersed in water, and electrolyte leakage was measured periodically. Data are Mean ± SD, n = 3
RVE1 regulates ROS-responsive gene expression
We previously identified a number of ROS-regulated genes that were directly bound by light signaling transcription factors (Chen et al. 2013). ZAT10 (ZAT ZINC FINGER PROTEIN10) plays a prominent role in ROS signaling and NDB2 (encoding NAD(P)H dehydrogenase) is an oxidative stress-induced gene, while SIB1 (SIGMA FACTOR BINDING PROTEIN1) and ERF4 (ETHYLENE-RESPONSIVE TRANSCRIPTION FACTOR4) respond to singlet oxygen generation (Miller et al. 2008; Laloi et al. 2007; Ho et al. 2008). To examine whether RVE1 modulates the expression of these ROS-regulated genes, 6-day-old etiolated seedlings were illuminated with white light for 1 or 3 h. As shown in Fig. 6, the transcript levels of ZAT10, SIB1, ERF4, and NBD2 were gradually increased after light exposure. After 3 h of illumination, the expression levels of these genes were much lower in RVE1-OE seedlings than in those of the wild type. However, after 1 or 3 h of light treatment, the expression of ZAT10, ERF4, and NBD2 was drastically increased in the rve1 mutant compared with the wild type (Fig. 6). These data indicate that RVE1 represses ROS-responsive gene expression during de-etiolation.
RVE1 regulates ROS-responsive gene expression. qRT-PCR showing the relative expression of various ROS-responsive genes, including ZAT10, SIB1, ERF4, and NDB2. Six-day-old etiolated seedlings were kept in darkness or transferred to white light (150 µmol m−2 s−1) for 1 and 3 h. Relative expression levels were normalized to that of UBQ1. Data are Mean ± SD of three technical replicates. The experiments were repeated twice with similar results
Discussion
The central clock proteins, CCA1 and LHY, have been extensively studied and ensure accurate timekeeping of the circadian clock during all seasons (Pruneda-Paz and Kay 2010; Zhang and Kay 2010). RVE1 controls daily rhythms of auxin production by integrating the circadian and auxin signaling pathways (Rawat et al. 2009). Two RVE1 homologous, RVE2/CIRCADIAN 1 (CIR1) and RVE7/EARLY-PHYTOCHROME-RESPONSIVE 1 (EPR1), seem to act primarily as clock outputs via unknown mechanisms (Zhang et al. 2007; Kuno et al. 2003). In addition to regulating clock rhythms, members of the RVE family are involved in other plant responses. For instance, constitutively expressed CIR1 displayed delayed flowering, longer hypocotyls, and reduced seed germination (Zhang et al. 2007). Loss of RVE8 also caused early flowering and elongated hypocotyls (Rawat et al. 2011).
Our study provides evidence that RVE1 regulates chlorophyll biosynthesis and seedling greening through, at least in part, modulating PORA expression. First, yeast one-hybrid, EMSA, and ChIP assays showed that RVE1 physically binds to the promoter of PORA through the EE-box cis-element (Fig. 1). Second, using qRT-PCR analysis, we showed that RVE1 promotes PORA expression (Fig. 2). Third, overexpression of RVE1 resulted in higher greening rate, less ROS accumulation, and cell death, and reduced ROS-responsive gene expression than the wild type (Figs. 4, 5, 6). In addition, we observed that the greening rate and the expression of some ROS-regulated genes were altered by RVE1 mutation. There might be functional redundancy of RVE1 with other homologous proteins. Future studies using higher order mutants involving other RVE genes are required to address this possibility.
CCA1 and PSEUDO-RESPONSE REGULATORs (PPRs) have been reported to mediate the seedling greening response via unidentified mechanisms (Kato et al. 2007). Overexpression of CCA1 promotes the greening of dark-grown seedlings, whereas constitutive expression of PPR1 has the opposite effect (Kato et al. 2007). CCA1 might act partly by modulating the level of PORA, since CCA1 is also able to bind directly to the PORA promoter and regulate its expression (Ni et al. 2008).
POR enzymes play critical roles in chlorophyll biosynthesis and affect plant greening during the dark-to-light transition (Runge et al. 1996; Sperling et al. 1997). These enzymes can be regulated at the post-translational level. A chloroplast membrane-localized protein CHAPERONE-LIKE PROTEIN OF POR1 interacts with POR and stabilizes its isoforms (Lee et al. 2013). Previous studies showed that the transcript level of PORA is directly modulated by EIN3 and DELLA proteins (Zhong et al. 2009; Cheminant et al. 2011). Our study identified RVE1 as a novel direct regulator of PORA expression. RVE1, EIN3, and DELLAs may act independently or coordinately to maintain proper PORA levels in response to exogenous signals (such as light) and endogenous cues (such as ethylene and gibberellic acid). It has been shown that PORA and PORB were similarly regulated by light and rhythm (Matsumoto et al. 2004). However, the expression of PORB was reduced by both RVE1 mutation and overexpression. Other factors are likely involved in regulating PORB transcript level.
Besides directly regulating PORA expression, RVE1 also negatively modulates the expression of several chlorophyll biosynthesis genes through an unknown mechanism that reduces Pchlide accumulation in the cotyledons of dark-grown seedlings. Upon illumination, excess Pchlide may generate ROS or free radicals, resulting in photo-oxidization in cells (Reinbothe et al. 1996, Buhr et al. 2008). Therefore, the increased greening rate for RVE1-OX seedlings is caused by both the high expression of PORA and the reduced levels of Pchlide in darkness, which reduce the amount of photobleaching that occurs after illumination. Consistent with this notion, the cotyledons of RVE1-OX seedlings barely accumulated ROS or exhibited cell death and had low levels of ROS-responsive genes expression after light exposure (Fig. 5). Thus, RVE1 likely acts as a crucial mediator that promotes chlorophyll biosynthesis and chloroplast development and maximizes the ability of plants to transition from heterotrophic to autotrophic growth.
References
Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293:880–883
Buhr F, Bakkouri M, Valdez O, Pollmann S, Lebedev N, Reinbothe S, Reinbothe C (2008) Photoprotective role of NADPH: protochlorophyllide oxidoreductase A. Proc Natl Acad Sci USA 105:12629–12634
Cheminant S, Wild M, Bouvier F, Pelletier S, Renou JP, Erhardt M, Hayes S, Terry MJ, Genschik P, Achard P (2011) DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23:1849–1860
Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic bHLH/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25:1657–1673
Clough SJ, Bent AF (1998) Floral dip: a simple method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Frick G, Su Q, Apel K, Armstrong GA (2003) An Arabidopsis porB porC double mutant lacking light-dependent NADPH: protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J 35:141–153
Heyes DJ, Hunter CN (2005) Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem Sci 30:642–649
Ho LHM, Giraud E, Uggalla V, Lister R, Clifton R, Glen A, Thirkettle-Watts D, Van Aken O, Whelan J (2008) Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol 147:1858–1873
Ikegami A (2007) The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J Biol Chem 282:19282–19291
Kato T, Murakami M, Nakamura Y, Ito S, Nakamichi N, Yamashino T, Mizuno T (2007) Mutants of circadian-associated PRR genes display a novel and visible phenotype as to light responses during de-etiolation of Arabidopsis thaliana seedlings. Biosci Biotechnol Biochem 71:834–839
Kuno N, Moller SG, Shinomura T, Xu XM, Chua NH (2003) The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15:2476–2488
Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K (2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA 104:672–677
Lee JY, Lee HS, Song JY, Jung YJ, Reinbothe S, Park YI, Lee SY, Pai HS (2013) Cell growth defect factor1/CHAPERONE-LIKE PROTEIN OF POR1 plays a role in stabilization of light-dependent protochlorophyllide oxidoreductase in Nicotiana benthamiana and Arabidopsis. Plant Cell 25:3944–3960
Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318:1302–1305
Masuda T et al (2003) Functional analysis of isoforms of NADPH: protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol 44:963–974
Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T (2004) Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol 135:2379–2391
Miller G, Shulaev V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133:481–489
Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105:9433–9438
Ni Z, Kim D, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ (2008) Altered circadian rhythms regulate growth vigor in hybrids and allopolyploids. Nature 457:327–331
op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg É, Göbel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332
Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155:1545–1551
Pruneda-Paz JL, Kay SA (2010) An expanding universe of circadian networks in higher plants. Trends Plant Sci 15:259–265
Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, Zhao Y, Ljung K, Harmer SL (2009) REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA 106:16883–16888
Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-RESPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7:e1001350
Reinbothe S, Reinbothe C, Apel K, Lebedev N (1996) Evolution of chlorophyll biosynthesis—the challenge to survive photooxidation. Cell 86:703–705
Runge S, Sperling U, Frick G, Aple K, Armstrong GA (1996) Distinct roles for light-dependent DADPH: protochlorophyllide oxidoreductases (POR) A and B during greening in higher plants. Plant J 9:513–523
Sperling U, van Cleve B, Frick G, Aple K, Armstrong G (1997) Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J 12:649–658
Su Q, Frick G, Armstrong G, Aple K (2001) PORC of Arabidopsis thaliana: a third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol Biol 47:805–813
Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58:321–346
Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. Arab Book. doi:10.1199/tab.0145
Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR 1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24:1984–2000
Zhang EE, Kay SA (2010) Clocks not winding down: unraveling circadian networks. Nat Rev Mol Cell Biol 11:764–776
Zhang X, Chen Y, Wang ZY, Chen Z, Gu H, Qu LJ (2007) Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J 51:512–525
Zhong SW, Zhao MT, Shi TY, Shi H, An FY, Zhao Q, Guo HW (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106:21431–21436
Acknowledgments
We thank Dr. Stacey Harmer (University of California, Davis) for providing the rve1-1 and RVE1-OX seeds. This work was supported by the grants from the National Natural Science Foundation of China (31170221, 31325002), and the Ministry of Agriculture of China (2014ZX08009-003) to R. L.
Author information
Authors and Affiliations
Corresponding author
Additional information
Gang Xu and Haiyan Guo have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, G., Guo, H., Zhang, D. et al. REVEILLE1 promotes NADPH: protochlorophyllide oxidoreductase A expression and seedling greening in Arabidopsis . Photosynth Res 126, 331–340 (2015). https://doi.org/10.1007/s11120-015-0146-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-015-0146-5