Abstract
Lablab is a regionally important multipurpose legume crop used for human consumption, animal feed, and soil conservation. Despite these qualities, the potential value of this crop has not been fully utilized, and very little research attention has been given to it. The main objective of the study was molecular genetic diversity analysis of Lablab collections using 15 SSR markers. The molecular genetic diversity study of 91 Lablab collections revealed a total of 225 alleles with an average of 14.80 alleles per locus. All markers across the entire population were found to be highly polymorphic and informative with PIC values ranging from 0.78 to 0.92 with a mean value of 0.85. The average expected heterozygosity and gene diversity were 0.75 and 0.86 respectively, indicating a high level of diversity. Analysis of molecular variance showed that 94% of the total genetic variation was attributed to within populations, while only 6% was attributed to among populations. The fixation index value (0.061) recorded indicates the presence of moderate population differentiation as a result of high gene flow (Nm = 3.820) among populations. Due to high gene flow, Cluster, PCoA, and structure analysis did not exactly categorize the populations into genetic groups corresponding to their geographic origin. The observed relatively higher genetic diversity in Konso and West Wellega populations among the eight populations indicates that these areas could be considered hot-spots for genetic diversity and possible germplasm evaluation. Generally, genetic diversity obtained from this study provides inputs for Lablab conservation and improvement in Ethiopia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lablab (Lablab purpureus (L.) Sweet: Fabaceae) is one of the most ancient legume crops in the world and is widely distributed throughout tropical and subtropical regions of Asia and Africa (Maass et al. 2010; Kimani et al. 2012,). It is a monotypic genus in the family Fabaceae characterized as a semi-erect, bushy, perennial herb, but usually cultivated as an annual. It is a predominantly self-fertilizing, diploid crop with 2n = 2x = 22 chromosomes (Kukade and Tidke 2014; She and Jiang 2015) and an estimated genome size of 423 Mbp (Chang et al. 2018).
Lablab is an indigenous African legume, a remarkable drought and salinity-tolerant crop, and, thus, can be grown in a wide range of environmental conditions and soil types (D’Souza and Devaraj 2010; Maass et al. 2010). It is an important multipurpose legume crop that contributes towards food, feed, nutritional and economic security, and soil conservation, and is also rich in bioactive compounds with pharmacological potential, particularly against SARS-Cov2 (Habib et al. 2017; Weldeyesus 2017; Liu et al. 2020; Minde et al. 2021).
The crop is cultivated as a sole crop and intercropped with cereals, such as maize or sorghum, and different legume crops (Maass et al. 2010; Nord et al. 2020). Its dense green cover serves as a cover crop, preventing desiccation and minimizing erosion caused by wind and rain (Kimani et al. 2012; Naeem et al. 2020; Kongjaimun et al. (2022). As a legume crop, it is also useful for biologically fixing atmospheric nitrogen (Rangaiah and Dsouza 2016).
In Ethiopia, Lablab is mostly grown in the Konso zone of the southern part of the country, and some parts of the Amhara and Benishangul-Gumuz regions (Westphal 1974; Tesfaye 2007). The crop is cultivated as a hedge crop or on farmland for its edible seeds, and it grows at altitudes ranging from 400 to 2350 m.a.s.l. Considerable agro‐morphological variability in plant height, leaf size, maturity, number of seeds per pod, seed color, size, and shape have been reported (Pengelly and Maass 2001).
Lablab is considered a minor and neglected crop in most parts of Africa. Due to this, there is an increasing risk of genetic erosion for the cultivated Lablab varieties as well as the naturally occurring wild species in Africa. There are several reasons for this, including a lack of research attention and a decrease in cultivation areas and demand caused by the replacement of crops with superior economic importance in Africa (Maass et al. 2010).
According to Robotham and Chapman (2017), the greatest genetic diversity was found in Africa, making Ethiopia one of the probable centers of domestication. Its landraces have exhibited distinct genetic diversity from the rest of the world’s Lablab collections (Robotham and Chapman 2017; Maass et al. 2017). Hence, the country could be an abundant source of Lablab landraces. To effectively use landraces in breeding, it is crucial to genetically characterize them to understand the extent and pattern of diversity within and between populations. This helps assess the available genetic diversity for conservation and identifies genes valuable for future breeding progress.
Molecular markers reveal genetic similarities and differences without being influenced by environmental factors. A number of molecular markers such as RAPD (Liu 1996), AFLP (Maass et al. 2005; Kimani et al. 2012), and microsatellite (SSR) (Wang et al. 2007; Robotham and Chapman 2017; Amkul et al. 2021) have been used in different Lablab research and genetic diversity analyses of different germplasm sets in several countries. Among the various types of molecular markers, simple sequence repeat (SSR) markers have the advantage of simplicity, effectiveness, abundance, reproducibility, co-dominant inheritance, extensive genomic coverage, and ease of detection by polymerase chain reaction (PCR). They are consequently considered the most powerful molecular markers for resolving genetic diversity, population structure analysis, and cultivar identification in many crop plants (Powell' et al. 1996).
Although Lablab is grown in Ethiopia and the existence of morphological diversity and wild relatives have been reported (Pengelly and Maass 2001; Tesfaye 2007; Maass 2016; Robotham and Chapman 2017), very limited effort has been made to assess the existing genetic diversity of this crop in the country, which is considered an orphan or underutilized crop. Only few and mostly the same germplasm accessions from Ethiopia, distributed globally by the International Livestock Research Institute (ILRI), have been included in such diversity studies (Maass et al. 2005; Robotham and Chapman 2017; Kamau et al. 2021; Kongjaimun et al. 2022). This is the first time of more comprehensive characterization of Lablab genetic resources from Ethiopia, applying molecular markers. Therefore, this paper’s purpose is to profile genetic diversity of Lablab germplasm collected in Ethiopia. It also aims towards decision-making of plant genetic resources (PGR) management and towards developing breeding strategies.
Materials and Methods
Plant Materials
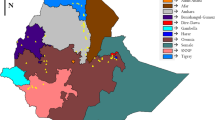
A total of 91 accessions of Lablab were used for this experiment. Ten accessions were exotic materials, which were obtained from Bako Agricultural Research Center, Ethiopia, who had previously received them from ILRI. Twenty-two accessions were acquired from the Ethiopian Biodiversity Institute (EBI), and the remaining 59 accessions were collected from Konso, North Wollo, Gamo Gofa, West Wellega, and West Gojjam zones (Fig. 1). The collected seeds were planted in pots at the National Agricultural Biotechnology Research Center (NABRC) greenhouse, located at 9° 3′ N latitude and 38° 30′ E longitudes with an altitude of 2400 m.a.s.l. Ten seeds from each of the accessions were grown in a greenhouse, and fresh leaves were collected from 2-week-old plants for genomic DNA (gDNA) extraction.
DNA Extraction and Quality Check
Two weeks after planting, an equal amount of bulk leaf samples was collected from five random plants of every accession as suggested by Gilbert et al. (1999). About 100 mg of fresh leaves were placed in 2 ml autoclaved and labeled Eppendorf tubes and freeze-dried for 24 h at − 80 °C. After 24 h, the leaves were further dried in liquid nitrogen and then ground to the powder form for 3 min using a Geno Grinder (MM-200, Retsch). Genomic DNA was extracted using the plant DNA extraction protocol based on the method of Diversity Array Technology (DArT) (DArTs 2019) with some minor modifications. The DNA pellet was air-dried and dissolved in 100 µl of nuclease-free water and kept at room temperature until the DNA pellet was dissolved.
The DNA was quantified using a nano drop spectrophotometer (ND-8000, Thermo Scientific). The level of DNA purity was determined by the 260/280 absorbance ratio. The quality of DNA was further assessed using 1% agarose gel in 1 × TAE buffer using a standard lambda DNA (Biolabs, New England). For gel preparation, 1% agarose powder was dissolved in 1 × TAE buffer. The mixture was boiled in a microwave oven at 100 °C. After agarose was completely dissolved and cooled to 50–60 °C, it was cast on the gel tray with a comb. After solidifying, the gel was placed in a gel tank containing 1 × TAE buffer. Five microliters of DNA from each sample were taken and mixed with 2 µl loading dye, which contained gel red and loaded in the well. The gel was run at constant voltage of 100 V for 40 min. The gel was visualized under UV light and subsequently photographed using a BioDoc-It™ imaging System (Cambridge, UK). Samples with high band intensity, lesser smear, and purity with 1.8 to 2 at 260/280 nm were selected for further PCR analysis. Purified and working concentration of DNA was stored in the refrigerator (− 20 °C) till the next use.
Primer Selection and Optimization
A total of 20 Lablab-specific SSR primers were used for PCR amplification. PCR optimization and testing of SSR primers were done using 12 representative Lablab accessions. The 20 SSR markers used were selected from Zhang et al. (2013), Robotham and Chapman (2017), and Keerthi et al. (2018) based on their high values of polymorphic information content (PIC). Out of the 20 tested SSR primers, 15 were selected for final analysis on the basis of reliability, polymorphism, and their specificity to target region (Table 1).
PCR and Gel Electrophoresis
Lyophilized primers for the target genes were reconstituted using nuclease-free water to obtain 100 µM stock solutions. All primers were stored at − 20 °C and finally diluted to working concentration of 10 µM. PCR reaction was carried out with a thermal cycler (GeneAmp®PCR System 9700) in a total volume of 12.5 µl reaction containing 6.25 µl one Taq 2 × Master Mix (M04821) Biolabs England, with standard buffer (which contain all PCR reaction components, MgCl2, PCR buffer, dNTPs, and Taq DNA polymerase), 0.5 µl forward primer, 0.5 µl reverse primer, 0.25 µl DMSO, 3 µl nuclease-free water, and 2 µl genomic DNA. The PCR was programmed at initial denaturation (preheating) step of 3 min at 94 °C followed by 35 cycles of a denaturation at 94 °C for 1 min, annealing at 50.2–58 °C depending of the primers for 2 min and elongation at 72 °C for 1 min, with a final elongation at 72 °C for 10 min followed by a holding step at 4 °C. PCR amplification of each primer was optimized using “gradient” methodology.
PCR products were loaded on 3% agarose gel (w/v) with gel red containing 6 × loading dye. Electrophoresis was performed in 1 × TAE buffer at 100 constant volts for 3 h and 30 min. The gel was stained with gel red and visualized under UV light using a BioDoc-it™ imaging system (Cambridge, UK). DNA fragment sizes were estimated by comparing the DNA bands with a 100 and 50 base pair DNA ladder as molecular ruler.
Data Scoring and Analysis
The amplified products were scored based on fragment band size using PyElph 1.4 software package (Pavel and Vasile 2012). Clearly resolved and unambiguous bands were scored for every primer and sample. Bands with the same fragment size were treated as identical fragments.
Different statistical software packages were utilized to compute the standard indices of genetic diversity. Locus-based diversity indices, including major allele frequency (MAF), number of alleles (Na), gene diversity (GD), polymorphic information content (PIC), and heterozygosity, were computed using the PowerMarker ver. 3.25 software (Liu and Muse 2005). Population diversity indices such as number of effective alleles per locus (Ne), Shannon information index (I), fixation index (F), gene flow (Nm) and percent polymorphism (% P), observed heterozygosity (Ho), expected heterozygosity (He), fixation index (F), and estimate of the deviation from Hardy–Weinberg equilibrium (HWE) over the entire populations were computed with GenAlEx ver. 6.501 software (Peakall and Smouse 2006). Furthermore, the analysis of molecular variance (AMOVA) was done to partition the total genetic variation within and among the eight geographically defined populations and estimate variance components using the same software (Peakall and Smouse 2006). AMOVA uses the estimated F-statistics such as genetic differentiation (Fst), fixation index or inbreeding coefficient (Fis), and overall fixation index (Fit) to compare the genetic differentiation among and within populations. The magnitude among and within population differentiation was quantified using F-statistics (Fit, Fis, and Fst) also known as fixation indices (Wright 1951).
According to Wright (1951), Fst values ranging from 0 to 0.05 are considered low, 0.05 to 0.15 moderate, and 0.15 to 0.25 large, while greater than 0.25 indicate very large genetic differentiations.
Rarified allelic richness (Ar) and private rarified allelic richness (Arp) were computed using HP-Rare 1.1 software (Kalinowski 2005).
To examine the genetic relationship between the different accessions, Unweighted Pair Group Method with Arithmetic Mean (UPGMA)-based neighbor-joining tree and hierarchical clustering were performed using DARwin var. 6.0 (Perrier and Jacquemoud-Collet 2006). A dendrogram was generated based on the dissimilarity matrix as input data to visualize the pattern of cluster among the accessions. To further examine the pattern of variation among samples and resolve the power of coordination, principal coordinate analysis (PCoA) was carried out using GenAlex ver.6.501 software.
The population structure and admixture patterns of the 91 accessions were determined by the Bayesian model-based clustering method of Pritchard et al. (2000) using Structure ver. 2.3.4 software. To estimate the true number of population clusters (K), a burn-in period of 100,000 was used in every run, and data were collected over 200,000 Markov chain Monte Carlo (MCMC) replications for K = 1 to K = 10 using 20 iterations for each K. The structure output results were zipped into one zip archive, and the zipped file was uploaded into the web-based program STRUCTURE HARVESTER ver. 0.6.92 (Earl and vonHoldt 2012). The most likely K value was determined by applying the ΔK method of Evanno et al. (2005) using the web-based STRUCTURE HARVESTER ver. 0.6.92 (Earl and von Holdt 2012). A bar plot for the best K was determined using Clumpak (beta version) (Kopelman et al. 2015).
Results
Microsatellite Marker Level of Polymorphism
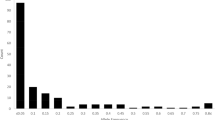
Fifteen microsatellite markers revealed 225 alleles across all the accessions that were dissimilar in fragment sizes, with a mean of 14.8 alleles per locus. The allele frequency distribution reflects that 47.1% of the alleles were rare (0.01 to 0.05), 28% ranged from 0.05 to 0.10, and 24.9% were abundant (higher than 0.10) (Table 2).
Major allele frequency (MAF) per locus ranged from 0.32 for marker c21512_g2_i1 to 0.13 Lpxu-002 with an average of 0.22 (Table 3). The highest gene diversity (0.92), allelic richness (9.25), polymorphic information content (0.92), number of alleles (21), and effective number of alleles (6.87) were recorded for KTD272 (Table 3). The lowest gene diversity (0.80), allelic richness (4.5), polymorphic information content (0.78), number of alleles (9.00), and effective number of alleles (3.53) were obtained for marker Lpxu-009 (Table 3). All the markers were highly informative with the PIC varied from 0.78 (Lpxu-009) to 0.92 (KTD272) (average = 0.85). The observed heterozygosity (Ho) varied from 0.00 to 0.98, with a mean of 0.27, and expected heterozygosity (He) ranged from 0.69 (Lpxu-009 and KTD 241) to 0.84 (Lpxu-013), with a mean of 0.75 (Table 3). All SSR markers showed highly significant (p < 0.0001) deviation from the Hardy–Weinberg equilibrium test.
Genetic Variability Within and Among Populations
The number of observed alleles (Na) was higher for accessions collected from Konso (9.53) followed by north Wollo (7.67) and West Wellega (7.40). Similarly, accessions collected from Konso showed the highest value for the number of effective alleles (6.40), Shannon’s information index (1.96), private allele richness (1.67), and expected heterozygosity (0.83). The inbreeding coefficient (fixation index) value ranged from 0.59 for Metekel accessions to 0.70 for Gamo Gofa and West Wellega accessions (Table 4).
Genetic Relationships Between the Populations
The magnitude of genetic distances between populations coming from different geographical origins showed more differentiation between Metekel, West Gojjam, and the rest of populations (GD range from 0.121 to 0.951). The highest value of GD (0.951) was observed between West Gojjam and Metekel populations followed by West Gojjam and Exotic materials (0.908). The smallest genetic distance (GD = 0.121) was found between populations from West Gojjam and North Wollo (Table 5). All Ethiopian populations were quite different from the exotic materials.
Analysis of Molecular Variance (AMOVA)
AMOVA was conducted to determine the extent of the variation within and among populations. Variation within populations accounted for higher variation (94%) than the variation among population (6%). The differences were highly significant (P < 0.001). A moderate genetic differentiation coefficient among populations (Fst = 0.061) was recorded with a high gene flow (3.82) among them (Table 6).
Diversity Patterns and Population Structure
The cluster analysis categorized the 91 Lablab accessions into three major clusters (C1, C2, and C3) forming different sub-clusters (Fig. 2). Most of the individuals fell within C1 (48%; 44 accessions), followed by C2 (33%; 30 accessions) and C3 (19%; 17 accessions). Eighty percent of Exotic materials were grouped together in C1 but in separate sub-clusters. C1 is composed of accessions from all populations. Eighty percent of accessions in C2 were from geographically nearest populations of North Wollo, West Gojjam, and North Gonder. C3 comprised almost entirely of Konso accessions (88%).
The genetic relatedness of the 91 Lablab accessions was further investigated using principal coordinate analysis (PCoA) (Fig. 3). However, the first three coordinates explained only 22.55% of the genetic variation, with 9.04%, 6.78%, and 6.36% for dimensions 1, 2, and 3, respectively. Patterns of genotype distribution on a two-dimensional plot, therefore, showed a nearly uniform distribution of the accessions from different collection sites, indicating a weak population structure.
Based on the true number of cluster (K) suggested by Evanno et al. (2005), the real structure showed a clear peak of the populations at K = 4 (Fig. 4A). This means that the most likely number of clusters to group the 91 accessions into sub populations was four (Fig. 4B). The Clumpak result (bar plot) showed wide genetic admixtures and failed to show a clear structure based on clustering of the pre-defined populations, and hence, there was no clear geographic origin-based structuring of populations (Fig. 4B).
Inferred population structure of 91 Lablab accessions in eight populations using 15 SSR markers. A The highest delta K = 4 (peak) value following Evanno et al. (2005). B Estimated population structure along with geographical locations. G.Gofa Gamo Gofa, W.Wolega West Wolega, N.Gonder North Gonder, W.Gojam West Gojjam
Discussion
Marker Polymorphism
Molecular characterization of plant genetic diversity and relationships using microsatellite markers is useful because of their co-dominance and ability to reveal a high number of alleles per polymorphic locus. The current study detected 225 alleles across all 91 accessions. On average, there were 14.8 alleles across the 15 loci, where individual SSR markers counted 9 to 21 alleles. This study recorded a higher total number of alleles (225) as compared to 133 alleles by Keerthi et al. (2018). Similarly, the mean number of alleles (14.8) detected was higher than those reported by others (Zhang et al. 2013; Robotham and Chapman 2017; Keerthi et al. 2018), ranging from 2.65 to 7.4. The differences in the mean and total numbers of alleles between the present research and previous studies could be attributed to the genetic materials and the number of genotypes (sample size) used as well as the number and efficacy of the SSR markers. Likewise, the mean number of effective alleles (4.75) was higher than reported in a previous study (Keerthi et al. 2018).
The high value of PIC in the present study indicates the markers used were highly informative. Also, high allelic variation was available in the marker loci within the studied Lablab accessions. Consequently, these markers should be useful for further genetic analysis such as genetic diversity, genetic linkage map construction, and QTL mapping of Lablab genotypes. Although the 15 SSR markers used were selected from Zhang et al. (2013), Robotham and Chapman (2017), and Keerthi et al. (2018) based on their high PIC values, the mean PIC value obtained here was also higher. The range from 0.13 to 0.32 of the major allele frequency (MAF) across accessions (Table 3) indicates again that the SSR markers applied are very informative and can be used in genetic diversity studies and in marker-assisted breeding of Lablab.
Heterozygosity indicates how much genetic variation exists in a population and how the variation is distributed across the alleles of analyzed markers. The observed heterozygosity (Ho) is the proportion of heterozygous individuals in a population sample, while expected heterozygosity (He) is the probability of an individual being heterozygous in any locus (Hirpara and Gajera 2018). The observed heterozygosity revealed low values in relation to expected heterozygosity at the majority of markers (Table 3), indicating a high amount of homozygosity. The observed lower heterozygosity reflects low levels of outcrossing. Robotham and Chapman (2017) also reported very low observed heterozygosity (0.205), suggesting the inbreeding index characteristic of a predominantly self-fertilizing crop with little outcrossing.
All the markers (100%) exhibited significant deviations from Hardy–Weinberg equilibrium (HWE) between Ho and He in which all of them showed excess heterozygosity across all markers. The lower observed heterozygosity level in this study and other studies might be due to the autogamous nature of Lablab, which contributes to low heterozygosity levels. The average genetic diversity (0.86) detected among the 91 Lablab accessions (Table 3) showed high levels of variation among the studied Lablab accessions. This study suggests that the significant increase in genetic diversity among Lablab accessions indicates potential for enhancing Lablab agronomic traits through breeding and underscores the importance of conserving Lablab germplasm.
Population Genetic Diversity
The high genetic variation observed particularly among the tested Konso and West Wellega accessions (Table 4) could be used as a potential source of important traits in future Lablab breeding programs as private alleles provide a unique genetic variability in certain loci (Kalinowski 2005).
The Metekel population had the largest genetic distances with the populations of West Gojjam, North Gonder, North Wollo, and Konso, in descending order. This could come from a low genetic material exchange between these populations and/or particular plant types existing. For instance, all the known cultivated two-seeded accessions described by Maass et al. (2017) and Maass and Chapman (2022) originated from Metekel (pers. comm. BL Maass). On the other hand, low genetic distances were also observed between populations of North Gonder and North Wollo (0.164), as well as Konso and Gamo Gofa (0.189), indicating a high frequency of identical alleles among accessions, thus, and leading to a certain genetic homogeneity. This could be attributed to the proximity of Konso and Gamo Gofa geographical regions; hence, there could be higher chances of exchanging and sharing of seed through agricultural systems/office and farmers. However, it cannot be explained for the other pairs of regions that are far away from each other. Interestingly, the exotic accessions had a relatively high genetic distance to all Ethiopian populations, being West Gojjam (0.91) the highest and North Gonder (0.43) the lowest. Some of this exotic germplasm might have already been mixed with traditional Ethiopian populations; for example, ILRI 147 (i.e., cv. Highworth) is one of the 50 most popular accessions distributed by seven international genebanks, among them ILRI (International Livestock Research Institute) (Galluzzi et al. 2016).
The low genetic differentiation among populations (6%) (Table 6) is possibly due to gene flow as a result of germplasm exchange by farmers that are geographically or culturally relatively close to each other. This leads to an increase in the distribution of certain genes among different populations. Similarly, Kimani et al. (2012) reported 99% variation within and only 1% variation among Lablab populations from Kenya using amplified fragment length polymorphism (AFLP) markers. Kamotho et al. (2016), on the other hand, found 85% of genetic variation within and 15% among more comprehensive Kenyan Lablab populations using SSR markers. The low level of genetic variability among Ethiopian Lablab populations may be a result of gene flow (introduction and migration of alleles or genotypes) from one population to another through seed exchange.
The study also showed only moderate genetic differentiation among Lablab populations (Fst = 0.061). A low level of genetic differentiation among populations could be indicative of high gene flow (Nm = 3.820). According to Slatkin (1987) and Diego and Jolla (1987), Nm values are grouped into three categories: Nm > 1.00 high, 0.25–0.99 intermediate, and 0.000–0.249 low. Therefore, the high Nm value of 3.820 from this study indicates high gene flow among populations, which supports the AMOVA result showing low variation among the eight geographically defined populations.
The cluster analysis categorized the 91 Lablab accessions into three main clusters (C1, C2, and C3) by forming different sub-groups. The clustering model showed that only weak relationships existed between the pattern of genetic diversity and geographical origins of collections. Accessions collected from different geographical populations clustered together, which may be attributed to the existence of gene flow among neighboring populations. In all of the clusters, many accessions were grouped with other geographically distant populations. This indicates accessions in one cluster might have evolved from different lines of ancestry or independent events of evolutionary forces such as genetic drift, mutation, migration, selection, and germplasm exchange might have separated them into related but different gene pools (Keneni et al. 2012). Most of exotic materials were grouped in C1 by forming different sub-groups with Ethiopian populations. Some of this exotic germplasm might have already been mixed with traditional Ethiopian populations.
Similar to dendrogram clustering, principal coordinate analysis (PCoA), a two-dimensional display of clustering, did not reveal distinct population clusters based on their geographic regions of origin. Instead, there was a mixing of genotypes across different coordinates, indicating a weak pattern of population differentiation.
Moreover, this result is clearly reflected in the model-based population genetic structure analysis, which found evidence of potential genetic admixture among the Lablab genotypes. It revealed the existence of weak sub-structuring (K = 4) of the eight populations of Lablab. This finding may indicate germplasm exchange between farmers, leading to gene flow across adjoining populations.
Conclusion
All previous molecular genetic diversity analyses in Lablab were conducted using a very limited number of accessions from Ethiopia. For the first time, a comprehensive set of germplasm accessions from Ethiopia have been characterized by applying SSRs. Successful molecular characterization of this neglected crop Lablab can help breeders to focus on the available genetic resource and to implement improvement programs. The abundance of different alleles observed among the populations provides evidence for novel alleles that can be efficiently exploited through future breeding programs for the trait(s) of interest. The existence of relatively higher genetic diversity in Konso and West Wellega populations also reveals that these areas could be considered hot spots for genetic diversity as well as sources of desirable genes for genetic improvement and conservation. Finally, we suggest conducting additional study using high-density markers that include accessions from additional regions of Ethiopia in order to acquire a more comprehensive understanding of the genetic structure and diversity of Lablab across the entire nation.
Data Availability
No datasets were generated or analysed during the current study.
References
Amkul K, Sookbang JM, Somta P (2021) Genetic diversity and structure of landrace of Lablab (Lablab purpureus (L.) sweet) cultivars in Thailand revealed by SSR markers. Breed Sci 71:176–183. https://doi.org/10.1270/jsbbs.20074
Chang Y, Liu H, Liu M et al (2018) The draft genomes of five agriculturally important African orphan crops. Gigascience 8:1–16. https://doi.org/10.1093/gigascience/giy152
D’Souza MR, Devaraj VR (2010) Biochemical responses of Hyacinth bean (Lablab purpureus) to salinity stress. Acta Physiol Plant 32:341–353. https://doi.org/10.1007/s11738-009-0412-2
Diego S, Jolla L (1987) A Multispecies Approach to the Analysis of 41:385–400
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Galluzzi G, Halewood M, López Noriega I, Vernooy R (2016) Twenty-five years of international exchanges of plant genetic resources facilitated by the CGIAR genebanks: a case study in international interdependence. Biodivers Conserv 25:1421–1446. https://doi.org/10.1007/s10531-016-1109-7
Gilbert JE, Lewis RV, Wilkinson MJ, Caligari PDS (1999) Developing an appropriate strategy to assess genetic variability in plant germplasm collections. Theor Appl Genet 98:1125–1131. https://doi.org/10.1007/s001220051176
Habib HM, Theuri SW, Kheadr EE, Mohamed FE (2017) Functional, bioactive, biochemical, and physicochemical properties of the Dolichos Lablab bean. Food Funct 8:872–880. https://doi.org/10.1039/c6fo01162d
Hirpara DG, Gajera HP (2018) Molecular heterozygosity and genetic exploitations of Trichoderma inter-fusants enhancing tolerance to fungicides and mycoparasitism against Sclerotium rolfsii Sacc. Infect Genet Evol 66:26–36. https://doi.org/10.1016/j.meegid.2018.09.005
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Kamau EM, Kinyua MG, Waturu CN, Kiplagat O, Wanjala BW, Kariba RK, David RK (2021) Diversity and population structure of local and exotic Lablab purpureus accessions in Kenya as revealed by microsatellite markers. Glob J Mol Biol 3(8):1–16
Kamotho GN, Kinyua MG, Muasya RM et al (2016) Assessment of genetic diversity of Kenyan dolichos bean ( Lablab purpureus l. Sweet ) using simple sequence repeat ( SSR ) markers. Int J Agric Environ Bioresearch 1:26–43
Keerthi CM, Ramesh S, Byregowda M, Vaijayanthi PV (2018) Simple sequence repeat (SSR) marker assay-based genetic diversity among dolichos bean (Lablab purpureus L. Sweet) advanced breeding lines differing for productivity per se traits. Int J Curr Microbiol Appl Sci 7:3736–3744. https://doi.org/10.20546/ijcmas.2018.705.433
Keneni G, Bekele E, Imtiaz M et al (2012) Genetic diversity and population structure of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Mol Biol Report 30:654–665. https://doi.org/10.1007/s11105-011-0374-6
Kimani EN, Wachira FG, Kinyua M (2012) Molecular diversity of Kenyan Lablab bean (Lablab purpureus (L.) Sweet) accessions using amplified fragment length polymorphism markers. Am J Plant Sci 03:313–321. https://doi.org/10.4236/ajps.2012.33037
Kongjaimun A, Takahashi Y, Yoshioka Y, Tomooka N, Mongkol R, Somta P (2022) Molecular analysis of genetic diversity and structure of the Lablab (Lablab purpureus (L.) Sweet) gene pool reveals two independent routes of domestication. Plants 12(1):57. https://doi.org/10.3390/plants12010057
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Kukade SA, Tidke JA (2014) Reproductive biology of Dolichos lablab L. (Fabaceae). Indian J Plant Sci 3:22–25
Liu CJ (1996) Genetic diversity and relationships among Lablab purpureus genotypes evaluated using RAPD as markers
Liu K, Muse SV (2005) PowerMaker: an integrated analysis environment for genetic maker analysis. Bioinformatics 21:2128–2129. https://doi.org/10.1093/bioinformatics/bti282
Liu YM, Shahed-Al-Mahmud M, Chen X et al (2020) A carbohydrate-binding protein from the edible Lablab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell Rep 32:108016. https://doi.org/10.1016/j.celrep.2020.108016
Maass BL (2016) Origin, domestication and global dispersal of Lablab purpureus (L.) Sweet (Fabaceae): current understanding. Legum Perspect 5–8
Maass BL, Jamnadass RH, Hanson J, Pengelly BC (2005) Determining sources of diversity in cultivated and wild Lablab purpureus related to provenance of germplasm by using amplified fragment length polymorphism. Genet Resour Crop Evol 52:683–695. https://doi.org/10.1007/s10722-003-6019-3
Maass BL, Knox MR, Venkatesha SC et al (2010) Lablab purpureus-a crop lost for Africa? Trop Plant Biol 3:123–135. https://doi.org/10.1007/s12042-010-9046-1
Maass BL, Robotham O, Chapman MA (2017) Evidence for two domestication events of hyacinth bean (Lablab purpureus (L.) Sweet): a comparative analysis of population genetic data. Genet Resour Crop Evol 64:1221–1230. https://doi.org/10.1007/s10722-016-0431-y
Maass BL, Chapman MA (2022) The Lablab genome: Recent advances and future perspectives. Chapter 13, pp. 229-253 in: Chapman MA (ed) Underutilised crop genomes. Series: Compendium of Plant Genomes. Springer, Cham, Germany. https://doi.org/10.1007/978-3-031-00848-1_13
Minde JJ, Venkataramana PB, Matemu AO (2021) Dolichos Lablab-an underutilized crop with future potentials for food and nutrition security: a review. Crit Rev Food Sci Nutr 61:2249–2261. https://doi.org/10.1080/10408398.2020.1775173
Naeem M, Shabbir A, Ansari AA et al (2020) Hyacinth bean (Lablab purpureus L.) – an underutilised crop with future potential. Sci Hortic (Amsterdam) 272. https://doi.org/10.1016/j.scienta.2020.109551
Nord A, Miller NR, Mariki W et al (2020) Investigating the diverse potential of a multipurpose legume, Lablab purpureus (L.) Sweet, for smallholder production in East Africa. PLoS One 15:. https://doi.org/10.1371/journal.pone.0227739
Pavel AB, Vasile CI (2012) PyElph - a software tool for gel images analysis and phylogenetics. BMC Bioinformatics 13:. https://doi.org/10.1186/1471-2105-13-9
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pengelly BC, Maass BL (2001) Lablab purpureus (L.) Sweet - diversity, potential use and determination of a core collection of this multi-purpose tropical legume. Genet Resour Crop Evol 48:261–272. https://doi.org/10.1023/A:1011286111384
Perrier X, Jacquemoud-Collet JP (2006) DARwin software https://DARwin.cirad.fr
Powell’ W, Morgante2 M, Andre3 C et al (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Kluwrr Academic Publishers
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Rangaiah D V, Dsouza M (2016) Hyacinth bean (Lablab purpureus): an adept adaptor to adverse environments
Robotham O, Chapman M (2017) Population genetic analysis of hyacinth bean (Lablab purpureus (L.) Sweet, Leguminosae) indicates an East African origin and variation in drought tolerance. Genet Resour Crop Evol 64(441):139–148. https://doi.org/10.1007/s10722-015-0339-y, https://doi.org/10.1007/s10722-015-0356-x
She CW, Jiang XH (2015) Karyotype analysis of Lablab purpureus (L.) Sweet using fluorochrome banding and fluorescence in situ hybridisation with rDNA probes. Czech J Genet Plant Breed 51:110–116. https://doi.org/10.17221/32/2015-CJGPB
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236(80):787–792. https://doi.org/10.1126/science.3576198
DARTs (2019) Plant DNA extraction protocol for DArT. 2–3
Tesfaye A (2007) Plant diversity in Western Ethiopia: ecology , ethnobotany and conservation. PhD Thesis
Wang ML, Morris JB, Barkley NA et al (2007) Evaluation of genetic diversity of the USDA Lablab purpureus germplasm collection using simple sequence repeat markers. J Hortic Sci Biotechnol 82:571–578. https://doi.org/10.1080/14620316.2007.11512275
Weldeyesus G (2017) Forage productivity system evaluation through station screening and intercropping of Lablab forage legume with maize under Irrigated lands of smallholder farmers. African J Agric Res 12:1841–1847. https://doi.org/10.5897/ajar2016.11989
Westphal E (1974) Pulses in Ethiopia, their taxonomy and agricultural significance. Kew Bull 30:590. https://doi.org/10.2307/4103104
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354. https://doi.org/10.1111/j.1469-1809.1949.tb02451.x
Zhang G, Xu S, Mao W et al (2013) Development of EST-SSR markers to study genetic diversity in hyacinth bean (Lablab purpureus L.). Plant Omics 6:295–301
Acknowledgements
The authors would like to acknowledge the Ethiopian Institute of Agricultural Research, McKnight Foundation Collaborative Crop Research Program through the Ethiopian Legume Diversity Project implemented by the Department of Plant Biology and Biodiversity Management, and the Addis Ababa University, Institute of Biotechnology of the Addis Ababa University, for their financial and other associated supports for this research. We are also thankful to the Ethiopian Biodiversity Institute (EBI) and Bako Agricultural Research Center for providing Lablab germplasm used in this study. BL Maass is acknowledged for critical comments on previous versions of the manuscript.
Funding
This research was funded by the Ethiopian Institute of Agricultural Research and the McKnight Foundation Collaborative Crop Research Program through its financial support made to the Ethiopian Legume Diversity Project.
Author information
Authors and Affiliations
Contributions
ZA, TF, and STW contributed to the study conception and design. Material preparation, data collection, and analysis were performed by STW and TD. The first draft of the manuscript was written by STW, TF, ZA, and TD. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants and animals, and ethical approval is not applicable.
Competing Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Workneh, S.T., Feyissa, T., Asfaw, Z. et al. Genetic Diversity and Population Structure of Lablab (Lablab purpureus L. Sweet) Accessions from Ethiopia Using SSR Markers. Plant Mol Biol Rep (2024). https://doi.org/10.1007/s11105-024-01447-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11105-024-01447-4