Abstract
Since its first report almost 200 years ago, fire blight, caused by the gram-negative bacterium Erwinia amylovora, has threatened apple and pear production globally. Identifying novel genes and their functional alleles is a prerequisite to developing apple cultivars with enhanced fire blight resistance. Here, we report 13 strain-specific and environment-dependent minor QTLs linked to fire blight resistance from a segregating Malus sieversii × Malus × domestica mapping population. Interval mapping at 95% confidence and Kruskal–Wallis analysis at P value = 0.005 were used to identify QTLs for three strains of E. amylovora differing in virulence and pathogenicity. The QTLs identified explain a small to moderate part of resistance variability, and a majority was not common between years or E. amylovora strains. These QTLs are distributed in eight linkage groups of apples and comparison of their map position to previously identified fire blight resistance QTLs indicates that most are novel loci. Interaction between experimental conditions in the greenhouse and field, and between years, and differences in virulence levels of strains might be responsible for strain- and year-specific QTLs. The QTLs identified on LG10 for strain Ea273 in 2011 and strain LP101 in 2011, and on LG15 for strain LP101 could be the same QTLs identified previously with strain CFBP1430 in cultivar “Florina” and “Co-op16 × Co-op17” mapping population, respectively. We discuss the potential impact of newly identified minor fire blight QTLs and major gene-based resistance on the rate of mutation in pathogen populations to overcome resistance and durability of resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
New apple cultivars are constantly needed for optimal performance under changing biotic and abiotic stresses and to meet market demands. Identification of novel genes and their functional alleles is essential to breed cultivars with enhanced disease resistance. A wide variety of bacterial diseases cause significant economic loss to fruit growers throughout the world (Harshman et al. 2017); of these, fire blight, caused by the gram-negative bacterium Erwinia amylovora, is a particularly devastating disease of pome fruits, mainly apples and pears (Norelli et al. 2003a). In the USA alone, the economic damage of fire blight to the apple industry was estimated at over $100 million annually through blossom, shoot, or rootstock blight. The actual cost varies yearly and is difficult to estimate due to the recurring cost of sprays and pruning of infected parts, and the multi-year impact of tree and orchard loss. Since its first report in the Hudson Valley, New York almost 200 years ago, fire blight has spread from North America to Europe, western Asia, northern Africa, and New Zealand (van der Zwet et al. 2012). Some preventive measures, including pruning of infected plant parts, restricted movement between infected orchards, and use of antibiotics in combination with fire blight prediction models, and resistant rootstock and scion can slow the spread and reduce the impact of the damage (Norelli et al. 2003a).
Varying levels of resistance to fire blight both in cultivated apples and pears and their wild relatives have been reported (Luby et al. 2002; Aldwinckle et al. 1999). Selection based on visual phenotyping of natural infections is possible, but expensive, and has some intrinsic limitations. For example, due to high heterozygosity in pome fruits, it is difficult to discern heterozygous from homozygous effects, and unfavorable weather for fire blight development will result in a lack of symptoms in susceptible individuals even though bacteria are present (Khan et al. 2012a; Kellerhals et al. 2008). Quantitative trait loci (QTLs) have been identified in different Malus genetic backgrounds with small to major effects on fire blight resistance/susceptibility (Khan et al. 2012a; Malnoy et al. 2012). QTL donor accessions for major fire blight loci on LG3, LG10, and two on LG12 are wild apples Malus robusta 5, M. fusca, and M. × arnoldiana, and “Evereste” and M. floribunda 821, respectively; the QTL on LG7 was identified from Malus × domestica cultivar “Fiesta” (Emeriewen et al. 2014, 2017; Fahrentrapp et al. 2013; Gardiner et al. 2012; Parravicini et al. 2011; Khan et al. 2007). Incorporating these QTLs into susceptible but commercially important and consumer-favored backgrounds is a challenge when using traditional plant-breeding techniques, as the poor fruit quality of most of these donors is inherited and can take up to five generations to select out. The aggressiveness of E. amylovora strains also varies, resulting in different levels of fire blight infection (Malnoy et al. 2012; Lee et al. 2010; Norelli et al. 2003b; Norelli et al. 1986). The majority of the major QTLs identified so far are strain-specific and not valid for multiple pathogen strains, as are often present in orchards, and can break down when used extensively (Malnoy et al. 2012; Khan et al. 2012a). For example, highly aggressive E. amylovora strains (E2002a, LA637) can overcome FB_MR5 resistance (Peil et al. 2011). High selection pressure from resistance alleles on the pathogen, along with its capacity to rapidly mutate, can lead to this breakdown of resistance. Exposing mapping populations and germplasm to multiple strains can not only demonstrate the stability and effectiveness of QTLs to a large number of bacterial strains, but will lead to the identification of novel QTLs. Resistance genes with minor effects, both strain-specific and non-specific, can provide a wider spectrum of resistance and may induce mutations more slowly within the pathogen population (Wöhner et al. 2014; Khan et al. 2012a; Norelli et al. 2003b).
Identification of new sources of both major strain-specific and non-specific minor resistance genes is important for ideal broad-spectrum resistance. Durable resistance barriers can be created by combining multiple monogenic and polygenic resistances with complementary action toward different strains of the same pathogen to provide broad-spectrum resistance for several diseases in pre-breeding germplasm stocks. Monogenic and polygenic resistance could be pyramided through marker-assisted pseudo-backcrossing, cis-genically, or via genome-editing (Bortesi and Fischer 2015; Khan and Korban 2012; Khan et al. 2007; Baumgartner et al. 2015; Broggini et al. 2014; Khan et al. 2012a). Regardless of the strategy to improve resistance, characterization of sources of resistance and development of DNA tests to readily identify novel functional genes and their allelic variants is required as current resistances lose effectiveness. We evaluated a mapping population (GMAL 4593) derived by crossing M. × domestica cultivar “Royal Gala” and an accession of M. sieversii (PI 613981), using three strains of E. amylovora, in field and greenhouse to identify QTLs linked to fire blight resistance.
Materials and Methods
Plant Material
A total of 169 progeny plants from GMAL 4593 mapping population, together with their parents, were used for fire blight resistance phenotyping and QTL identification. The GMAL 4593 population was derived from a cross between M. × domestica “Royal Gala” as the female parent and an elite accession of M. sieversii “KAZ 95 18-07” (PI 613981) as male parent, previously described by Norelli et al. (2017). The progeny and parents were bud grafted onto seedling rootstocks with at least three replications of each genotype. Initially, grafted plants were planted in a peat-perlite-vermiculite mixture in 13-cm diameter pots, grown in a greenhouse (ca. 20 °C (14–25 °C)) in Geneva, NY and trained to a single shoot (Aldwinckle and Preczewski 1976). Subsequently, 3–4 replicates of each genotype of the same potted plants were planted in the field in Kearneysville, WV in a randomized block design.
Bacterial Strains and Inoculum
E. amylovora (Ea) strains used in this study are listed in Table 1. Inoculum was prepared by growing Ea strains in liquid Kado 523 (Kado and Heskett 1970) broth at 28 °C, 250 rpm for 14–18 h. Concentration of inoculum was adjusted to approx. 1 × 1010 cfu/ml based on optical absorbance at 620 nm using a standard curve and subsequent dilution with sterile 0.05 M potassium phosphate buffer, pH 6.5. Inoculum was maintained on ice until used for inoculation.
Inoculation and Evaluation of Disease Resistance
Greenhouse plants were inoculated and evaluated for fire blight resistance in 2008. Greenhouse-grown plants were then shipped to Kearneysville, WV, planted in the field autumn 2008, and evaluated for their fire blight resistance in spring 2010 and 2011. Shoots of both greenhouse- and field-grown plants were inoculated by transversally bisecting the 2–4 youngest leaves of actively growing shoot-tip leaf whorls with scissors dipped in a suspension of a single Ea strain (approx. 1 × 109 cfu/ml) (Norelli et al. 2003b). The unit of replication in the greenhouse trial was individual, single-shoot-potted plants, whereas in the field trials, the unit of replication was individual shoots on plants with multiple shoots. On field-grown plants, shoots were selected on separate scaffold branches to limit systemic spread between inoculated shoots, and inoculated shoots were labeled to facilitate multiple observations and/or inoculation with different Ea strains. Evaluation of resistance/susceptibility was done as described in Norelli et al. (2003b) and Harshman et al. (2017). Total shoot length and fire blight lesion length from the point of inoculation to the furthest point of visible necrosis were measured 6–10 weeks after inoculation when fire blight lesions had ceased extension and a distinct margin was visible between necrotic and healthy stem tissue. Percentage of the current season’s shoot necrosis (PSN) was calculated by dividing the lesion length (cm) by the shoot length (cm) and multiplying the quotient by 100.
Statistical Analysis
Datasets for each year and E. amylovora strain were treated separately for analysis. Phenotypic data collected for entire mapping population was checked for outliers, normality, and frequency distribution. Analysis of variance (ANOVA) was performed for percentage of shoot necrosis (PSN) to test genotypic differences and to estimate variance components (Khan et al. 2006). Best linear unbiased predictions (BLUPs) were also estimated to summarize the different replicates per genotypes, using the LME4 package in R software (R Core Team 2014). The model used for BLUP estimates was as follows: Model=lme4 [Trait∼1| Genotype) + (1|Rep)]. The normality was checked using the Shapiro test in R software (R Core Team 2014). The datasets corresponding to the inoculation in 2008 and 2011 with Ea273 present a normal distribution. Mathematical transformations (sqrt, arsin sqrt, log, exp) were used in the datasets corresponding to the inoculation in 2010 and 2011 with AFRS273 strain but none of these transformations resulted in a normal distribution. Frequency distributions of the PSN were plotted using R software (R Core Team 2014). The statistical differences between the years and strains were tested with the Wilcoxon-paired test and the correlations between PSN, shoot length, and datasets from years and strains were assessed with Pearson’s correlation using R software (R Core Team 2014).
Genetic Linkage Map
The apple genetic map previously developed by Norelli et al. (2017) was used in the present study. This genetic map is composed of 578 SSR, SNPlex, HRM, and GBS markers, constructed with JoinMap 4.1 software (van Ooijen 2006).
QTL Analysis and Heritability Estimates
BLUP estimates for PSN for strains and years were used for QTL mapping analysis in MapQTL 5 software (van Ooijen 2009). Kruskal–Wallis test, a single marker analysis, interval, and multiple QTL mapping were used to identify QTLs. Interval mapping was performed for the dataset following a normal distribution (2008 and 2011 with Ea273) and Kruskal–Wallis test was performed for the other dataset (2010 and 2011 with AFRS273 and 2011 with LP101). The logarithm of odds (LOD) genome-wide threshold was calculated by permutation analysis to estimate the position of significant QTLs with 95% confidence. The map location of the QTLs was visualized on MapChart 2.30 (Voorrips 2002). Variance components were used to estimate heritability. Heritability was estimated as the ratio between the genotypic and the phenotypic variances: H2 = σ2g/σ2p, where σ2p = (σ2g + σ2e/n), σ2g is the genotypic variance, σ2e is the environmental variance, and n is the mean number of replicate per genotype. We estimated phenotypic variation explained by the QTLs, markers flanking the QTLs, and the magnitude of the effect of the presence and absence of the marker alleles. Genetic map positions of identified QTLs were also compared with QTLs reported in the literature to confirm if they are the same or novel.
Results
Genotype, Strain, and Year Interaction
The progeny of the GMAL 4593 mapping population showed varying responses to three E. amylovora strains (Fig. 1). Percentage of shoot necrosis (PSN) of fire blight for the population inoculated with strain Ea273 was similar between 2008 and 2011 (Wilcoxon paired test P value = 0.4). However, for inoculation with AFRS273, PSN differed between 2010 and 2011 (Wilcoxon paired test P value < 0.001), with more symptoms in 2011. The environmental conditions in the orchards were different between experiments conducted in 2010 and 2011. No significant difference was found for temperatures in 2010 and 2011 but humidity was significantly higher (Student’s t test P value < 0.05) in 2011 than in 2010 (Online Resource 1). In 2011, simultaneous infection with the three strains under similar environmental conditions showed a higher PSN in the population challenged with AFRS273 than with Ea273, which showed a higher PSN than with LP101 (Wilcoxon paired test P value < 0.001). AFRS273 produced greater PSN than the two other strains when comparing all genotypes common between the three strains in 2011 inoculations. Only 69% of these genotypes showed higher PSN with Ea273 than LP101 (Fig. 2).
Best linear unbiased predictors (BLUPs) for mean shoot length necrosis (%) of fire blight for the GMAL 4593 apple mapping population, depending on Erwinia amylovora strain (Ea273, AFRS273, LP101) in 3 years (2008, 2010, 2011). Letters correspond to the significant (P value < 0.05) difference with Wilcoxon-paired test
Best linear unbiased predictors (BLUPs) of shoot length necrosis (%) of fire blight for the apple mapping population (GMAL 4593) inoculated in the orchard with three E. amylovora strains, LP101 (light gray), Ea273 (gray) and AFRS273 (dark gray) in 2011. Each bar corresponds to a unique genotype. In the first group, genotypes present a higher shoot length necrosis (%) when infected by AFRS273, then LP101 and then Ea273. The second group includes genotypes that show higher shoot length necrosis (%) when infected by AFRS273, followed by Ea273, and then LP101
Percentage of shoot necrosis (PSN) of fire blight for the Ea273 strain from 2008, 2010, and 2011 showed a normal distribution. The progeny plants inoculated with strain AFRS273 in 2010 had the highest variation at 79% of variation between the minimal and the maximal PSN. Genotypes inoculated with strain AFRS273 in 2011 skewed toward high PSN, indicating severe fire blight infection while LP101 infection had more genotypes below 40% necrosis, indicating low to mild severity of fire blight infection compared to other datasets (Fig. 3). There is no significant difference between the mean PSN between 2008 and 2011 for strain Ea273 (Table 2), but there are differences for individual genotypes (Online Resource 4). The genotypes with high PSN differ from year to year. In general, there was no significant correlation between the BLUPs of PSN and the corresponding shoot length. There was moderate positive correlation (0.25–0.5) between strain and year datasets, except for LP101-2011 and Ea273-2008, where it was weak positive correlation. There is positive significant correlation (0.51) between PSN from Ea273 inoculations in 2011 and LP101 inoculations in 2011 (Online Resource 2). The broad-sense heritability of necrosis (%) varied between 0.54 and 0.75, depending on the strain and the year. Broad-sense heritabilities for fire blight resistance from strain Ea273 inoculation were 0.70 and 0.74 in 2008 greenhouse and 2011 field experiments, respectively. However, for strain AFRS273, there were large differences in average fire blight necrosis (67.5% for 2010 and 89.56% for 2011) and heritability estimates in 2010 (0.72) and 2011 (0.53). Only Ea273 had both greenhouse and field experiments, other strains had only field evaluations (Table 2).
Frequency distribution of the best linear unbiased predictors (BLUPs) of percentage of shoot length necrosis (PSN) of fire blight for the apple mapping population GMAL 4593. Three Erwinia amylovora strains (Ea273, AFRS273, LP101) were used to inoculate the population in three years (2008, 2010, 2011). Black arrows indicate PSN for parents of the respective progeny
Identification of QTLs Linked to Fire Blight Resistance
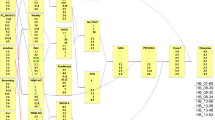
A total of 13 marker-trait associations were significant in eight linkage groups for three E. amylovora strains for 3 years. Three of these QTLs were significant at 95% in interval mapping analysis, while the remaining ten QTLs were significant (P value = 0.005 or above) in Kruskal–Wallis analysis. Four QTLs were detected for the LP101 strain, one on LG5, two on LG10, and one on LG15. For Ea273 strain, two QTLs were identified in 2011 on LG9 and 10, and one QTL was identified in 2008 on LG8. Six QTLs were detected for AFRS273 strain, three were in 2010, and three in 2011 (Table 3 and Fig. 4). Among the QTLs detected with the three strains in each of the 3 years, only the QTLs detected on LG8 with strain AFRS273 in 2011 and with strain Ea273 in 2008 are physically close and could correspond to the same resistance loci. QTL detected on LG5 with LP101 in 2011 and on LG8 with strain Ea273 in 2008 were physically close to QTLs detected for shoot length (Online Resource 3).
Fire blight resistance QTLs identified in GMAL 4593 apple mapping population for 3 years and three Erwinia amylovora strains. Best linear unbiased predictors (BLUPs) of percentage of the shoot necrosis (PSN) of fire blight were used for QTLs for interval mapping (IM) and Kruskal–Wallis (KW) analyses. LOD threshold to declare a significant QTL in IM was set as 4.5 (95%) and KW was **** 0.005. Note: The QTLs detected with the E. amylovora strain Ea273 using IM are represented in green: the lines correspond to the 1-LOD and 2-LOD confidence intervals. The QTLs detected with AFRS273 and LP101 using KW are represented in blue and pink, respectively, the boxes correspond to the markers significant at least at P = 0.005 and the confidence interval lines for the significant QTL are up to P = 0.1
Discussion
Novel Fire Blight QTLs
A total of 13 novel marker-trait associations linked to fire blight resistance were identified from a Malus sieversii × Malus × domestica mapping population through interval mapping at 95% confidence and Kruskal–Wallis analysis at P value = 0.005. These QTLs were distributed in eight linkage groups. Although several QTLs were identified on the same linkage groups as previously identified QTLs, the majority are far enough from previously identified fire blight resistance QTLs to assume that they are new loci representing novel resistance mechanisms. Fire blight QTLs identified on LG10 at 15.9 cM with strain Ea273 in 2011 could be the same as the QTL identified with strain CFBP1430 for lesion length after first and second weeks in cultivar “Florina” (Le Roux et al. 2010). This QTL had approximately 15% phenotypic variation explained (PVE) in both studies. QTL identified by Le Roux et al. (2010) was between common SSR markers CH02b07 and Hi03f06 but closer to CH02b07, as in this study. However, the precise location varies between studies, possibly due to the effect of population size. The QTLs identified for strain LP101 in 2011 using KW test at 73.8 and 87.9 cM seem to be novel fire blight QTLs on LG10. The QTL identified using KW test on LG15 with LP101 is also in a similar region (12.7 cM) as one reported by Khan et al. (2013) in “Co-op16 × Co-op17” mapping population for lesion lengths in the first and second weeks after inoculation, between 4.5 and 25.3 cM peak at MdSNPui07111 and NZ02b14.06 markers, respectively. However, there are no common markers across both studies for accurate comparison of mapping positions. This QTL was identified using Ea strain CBFP1430 and explained 17.4 and 15.4% PVE. Although no QTL was identified consistently over 2 years, two QTLs on LG8 in close proximity, with markers S8_29532144 and GD_SNP01764 close to the QTL peak, identified for strains AFRS273 and EA273 in 2011 and 2008, respectively, could be the same. Although there was a significant positive correlation (0.51) between PSN data from Ea273 and LP101 in 2011, no overlapping QTL was detected. This QTL explained 12.5% phenotypic variation in IM analysis. QTLs on LG10, LG15, and LG8 could be promising for further validation. We cannot rule out the effect of population size and marker density on identification of small-effect QTLs, accurate map position, and consistency over years and strains (Khan et al. 2012b; Beavis 1998). This can be confirmed by adding more recombinant individuals and fine mapping the QTL regions. More than 30 major and minor fire blight resistance loci conferring strain-specific and broad-spectrum resistance have been identified in the literature so far; some may not be distinct loci (Khan et al. 2012a; Malnoy et al. 2012). Overlapping mapping positions could indicate that some loci identified in different studies may be the same. However, R genes tend to cluster, and loci in close proximity may indicate R genes in close proximity, thus distinct loci. The stability of some of the identified loci has to be tested over multiple years, and they may need to be validated in different genetic backgrounds before potential application.
Effect Size of QTLs and G × E × Strain
The efficacy of most of the QTLs found in this study appeared to be affected by the environment and E. amylovora strain. These QTLs explain a small to moderate part of the variability observed for resistance/susceptibility to infection by E. amylovora, and a majority was not common between years and E. amylovora strains. The broad-sense heritability for fire blight resistance estimated here was somewhat lower than reported previously by Le Roux et al. (2010), Khan et al. (2006), and Calenge et al. (2005), potentially due to different experimental conditions. Broad-sense heritability values for strain Ea273 inoculation in 2008 greenhouse and 2011 field experiments were similar. The major difference between mean fire blight necrosis and heritability estimates for strain AFRS273 between 2010 and 2011 in the field experiment might be due to differences in temperature and humidity. The heritability values indicate that the majority of phenotypic variation within this population could be attributed to genetic variability. However, as the total variation in fire blight resistance explained by the QTLs is less than the broad-sense heritability, some additional small-effect QTLs may not have been identified. Although the size of the population used is satisfactory to map strong or moderate-effect QTLs, it may not be sufficient to precisely map the position and estimate the contribution of small-effect QTLs, as population size is one major factor influencing QTL detection (Beavis 1998). Increasing the number of progeny and improvement of the map coverage might lead to the identification of additional, small-effect QTLs in this population (Le Roux et al. 2010). Previous studies have noted that QTLs with minor effects detected in one population can have a major effect in another population or with a different strain. For example, the major QTL detected on LG3 by Peil et al. (2007) was also detected as a minor QTL by Calenge et al. (2005) and the major QTL detected on LG12 (Calenge et al. 2005) was also detected as a minor QTL by Durel et al. (2009). Minor QTLs exhibit strong interaction with the environment and are difficult to deploy in commercial backgrounds through breeding. Interaction between experimental conditions in the greenhouse and field, between years, and differences in virulence levels of strains might be responsible for strain- and year-specific QTLs. Although there was no significant difference between the PSN of fire blight between 2008 and 2011 for strain Ea273, the individual genotypes differ significantly from year to year. These differences in fire blight necrosis could be due to environmental differences. In 2008, the experimentation took place in the greenhouse, and in 2011, the experiment was carried out in orchards, which could have triggered different resistance mechanisms. Additionally, the trees would be a year older in the orchard and probably have more shoots, impacting the vigor and thus the infection rate. Significant (P value < 0.05) difference in humidity between 2011 and 2010 (Online Resource 1) indicates that moisture might be an important factor for differences in the infection rate of E. amylovora and specificity of QTLs to the years. Usually QTLs conferred by major R-genes are robust and have less effect from the environment in the same genetic mapping population. E. amylovora strains have varying levels of pathogenicity and virulence, resulting in different levels of fire blight infection due to different resistance mechanisms (Smits et al. 2014; Malnoy et al. 2012; Norelli et al. 1984). E. amylovora strain-specific QTLs have been reported in previous fire blight resistance QTL mapping studies. For example, fire blight QTLs on linkage group (LG)3, LG7, LG10, and two on LG12 have major effects and showed strain specificity (Emeriewen et al. 2014; Fahrentrapp et al. 2013; Parravicini et al. 2011; Khan et al. 2007). R genes have been identified for the QTLs FB_MR5 from Malus × robusta 5 on LG3 and FB_E from the crab apple “Evereste” on LG12 (Fahrentrapp et al. 2013; Parravicini et al. 2011). A CC-NBS-LRR gene was found responsible for the major QTL FB_MR5. Transformation of the CC-NBS-LRR gene into the fire blight-susceptible cultivar “Gala” showed significantly lower infection upon artificial inoculation with E. amylovora (Broggini et al. 2014; Fahrentrapp et al. 2013; Peil et al. 2007). Some E. amylovora strains can overcome the major resistance of M. × robusta R5 (Wöhner et al. 2014). In the long run, small effects may slow down the rate of mutation to overcome resistance in pathogen populations and thus can be more durable than resistance based on major genes (Laloi et al. 2017). Therefore, we speculate that strain-specific minor effect alleles may provide a wider spectrum of resistance to fire blight and induce slower mutation rates within E. amylovora population.
Conclusions
An effective and sustainable management strategy for fire blight requires an integrated control strategy aimed at reducing primary inoculum, interfering with infection through the application of antibiotics, biological agents, or chemicals, as well as enhancing host resistance. Apple cultivars with improved fire blight resistance, either to specific Erwinia strains, or wide resistance across strains, can reduce losses, increase orchard profitability, and limit the impact and cost of antibiotic use, for positive economic effect on apple production globally. Pyramiding multiple strain-specific and non-specific resistances could lead to the development of ideal broad-spectrum and durable resistance to the strains present in production areas through marker-assisted pseudo-backcrossing, cis-genically, or via genome-editing (Nishitani et al. 2016; Malnoy et al. 2016; Broggini et al. 2014; Kellerhals et al. 2008).
References
Aldwinckle HS, Preczewski JL (1976) Reaction of terminal shoots of apple cultivars to invasion by Erwinia amylovora. Phytopathology 66:1439–1444. https://doi.org/10.1094/Phyto-66-1439
Aldwinckle HS, Gustafson HL, Forsline PL (1999) Evaluation of the core subset of the USDA apple germplasm collection for resistance to fire blight. Acta Hortic (489):269–272. https://doi.org/10.17660/ActaHortic.1999.489.46
Baumgartner IO, Patocchi A, Frey JE, Peil A, Kellerhals M (2015) Breeding elite lines of apple carrying pyramided homozygous resistance genes against apple scab and resistance against powdery mildew and fire blight. Plant Mol Biol Report 33(5):1573–1583
Beavis WD (1998) QTL analyses: power, precision, and accuracy. Molecular dissection of complex traits 1998:145–162
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33(1):41–52. https://doi.org/10.1016/j.biotechadv.2014.12.006
Broggini GA, Wöhner T, Fahrentrapp J, Kost TD, Flachowsky H, Peil A, Hanke MV, Richter K, Patocchi A, Gessier C (2014) Engineering fire blight resistance into the apple cultivar “Gala” using the FB_MR5 CC-NBS-LRR resistance gene of Malus × robusta 5. Plant Biotechnol J 12(6):728–733. https://doi.org/10.1111/pbi.12177
Calenge F, Drouet D, Denancé C, Van de Weg WE, Brisset M-N, Paulin J-P, Durel C-E (2005) Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor Appl Genet 111(1):128–135. https://doi.org/10.1007/s00122-005-2002-z
Durel C-E, Denancé C, Brisset M-N (2009) Two distinct major QTL for resistance to fire blight co-localize on linkage group 12 in apple genotypes ‘Evereste’ and Malus floribunda clone 821. Genome 52(2):139–147. https://doi.org/10.1139/G08-111
Emeriewen OF, Richter K, Kilian A, Zini E, Hanke MV, Malnoy M, Peil A (2014) Identification of a major quantitative trait locus for resistance to fire blight in the wild apple species Malus fusca. Mol Breed 34(2):407–419. https://doi.org/10.1007/s11032-014-0043-1
Emeriewen OF, Peil A, Richter K, Zini E, Hanke MV, Malnoy M (2017) Fire blight resistance of Malus × arnoldiana is controlled by a quantitative trait locus located at the distal end of linkage group 12. Eur J Plant Pathol 148(4):1011–1018. https://doi.org/10.1007/s10658-017-1152-6
Fahrentrapp J, Broggini GA, Kellerhals M, Peil A, Richter K, Zini E, Gessler C (2013) A candidate gene for fire blight resistance in Malus × robusta 5 is coding for a CC–NBS–LRR. Tree Genet Genomes 9(1):237–251. https://doi.org/10.1007/s11295-012-0550-3
Gardiner S, Norelli J, De Silva N, Fazio G, Peil A, Malnoy M, Horner M, Bowatte D, Carlisle C, Wiedow C, Wan Y (2012) Putative resistance gene markers associated with quantitative trait loci for fire blight resistance in Malus ‘Robusta 5’accessions. BMC Genet 13(1):25. https://doi.org/10.1186/1471-2156-13-25
Harshman JM, Evans KM, Allen H, Potts R, Flamenco J, Aldwinckle HS, Wisniewski ME, Norelli JL (2017) Fire blight resistance in wild accessions of Malus sieversii. Plant Dis 101(10):1738–1745. https://doi.org/10.1094/PDIS-01-17-0077-RE
Kado CI, Heskett MG (1970) Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology 60:969–976. https://doi.org/10.1094/Phyto-60-969
Kellerhals M, Patocchi A, Duffy B, Frey J (2008) Modern approaches for breeding high quality apples with durable resistance to scab, powdery mildew and fire blight. Ecofruit-13th International Conference on Cultivation Technique and Phytopathological Problems in Organic Fruit-Growing: Proceedings to the Conference from 18th February to 20th February 2008 at Weinsberg, Germany, pp. 226–231
Khan MA, Korban SS (2012) Association mapping in forest trees and fruit crops. J Exp Bot 63(11):4045–4060. https://doi.org/10.1093/jxb/ers105
Khan MA, Zhao YF, Korban SS (2013) Identification of genetic loci associated with fire blight resistance in Malus through combined use of QTL and association mapping. Physiologia Plantarum 148(3):344-353. https://doi.org/10.1111/ppl.12068
Khan MA, Duffy B, Gessler C, Patocchi A (2006) QTL mapping of fire blight resistance in apple. Mol Breed 17(4):299–306. https://doi.org/10.1007/s11032-006-9000-y
Khan MA, Durel CE, Duffy B, Drouet D, Kellerhals M, Gessler C, Patocchi A (2007) Development of molecular markers linked to the ‘Fiesta’ linkage group 7 major QTL for fire blight resistance and their application for marker-assisted selection. Genome 50(6):568–577. https://doi.org/10.1139/g07-033
Khan MA, Zhao YF, Korban SS (2012a) Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in Rosaceae. Plant Mol Biol Rep 30(2):247–260. https://doi.org/10.1007/s11105-011-0334-1
Khan MA, Han Y, Zhao YF, Troggio M, Korban SS (2012b) A multi-population consensus genetic map reveals inconsistent marker order among maps likely attributed to structural variations in the apple genome. PLoS One 7(11):e47864
Laloi G, Vergne E, Durel CE, Le Cam B, Caffier V (2017) Efficiency of pyramiding of three quantitative resistance loci to apple scab. Plant Pathol 66:412–422
Le Roux P-MF, Khan MA, Broggini GAL, Duffy B, Gessler C, Patocchi A (2010) Mapping of quantitative trait loci for fire blight resistance in the apple cultivars ‘Florina’ and ‘Nova Easygro’. Genome 53(9):710–722. https://doi.org/10.1139/G10-047
Lee AS, Ngugi HK, Halbrendt NO, O’Keefe G, Lehaman B, Travis JW, Sinn JP, McNellis TW (2010) Virulence characteristics accounting for fire blight disease severity in apple trees and seedlings. Phytopathology 100(6):539–550. https://doi.org/10.1094/PHYTO-100-6-0539
Luby JJ, Alspach PA, Bus VG, Oraguzie NC (2002) Field resistance to fire blight in a diverse apple (Malus sp.) germplasm collection. J Am Soc Hortic Sci 127:245–253
Malnoy M, Martens S, Norelli JL, Barny MA, Sundin GW, Smits TH, Duffy B (2012) Fire blight: applied genomic insights of the pathogen and host. Annu Rev Phytopathol 50:475–494. https://doi.org/10.1146/annurev-phyto-081211-172931
Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Kanchiswamy CN (2016) DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904. https://doi.org/10.3389/fpls.2016.01904
Nishitani C, Hirai N, Komori S, Wada M, Okada K, Osakabe K, Yamamoto T, Osakabe Y (2016) Efficient genome editing in apple using a CRISPR/Cas9 system. Sci Rep 6:31481. https://doi.org/10.1038/srep31481
Norelli JL, Aldwinckle HS, Beer SV (1984) Differential host × pathogen interactions among cultivars of apple and strains of Erwinia amylovora. Phytopathology 74:136–139. https://doi.org/10.1094/Phyto-74-136
Norelli JL, Aldwinckle HS, Beer SV (1986) Differential susceptibility of Malus spp. cultivars Robusta 5, Novole, and Ottawa 523 to Erwinia amylovora. Plant Dis 70(11):1017–1019. https://doi.org/10.1094/PD-70-1017
Norelli JL, Jones AL, Aldwinckle HS (2003a) Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis 87:756–765. https://doi.org/10.1094/PDIS.2003.87.7.756
Norelli JL, Holleran HT, Johnson WC, Robinson TL, Aldwinckle HS (2003b) Resistance of Geneva and other rootstocks to Erwinia amylovora. Plant Dis 87:26–32. https://doi.org/10.1094/PDIS.2003.87.1.26
Norelli JL, Wisniewski M, Fazio G, Burchard E, Gutierrez B, Levin E, Droby S (2017) Genotyping-by-sequencing markers facilitate the identification of quantitative trait loci controlling resistance to Penicillium expansum in Malus sieversii. PLoS One 12(3):e0172949. https://doi.org/10.1371/journal.pone.0172949
Parravicini G, Gessler C, Denance C, Lasserre-Zuber P, Vergne E, BRISSET MN, Paocchi A, Durel CE, Broggini GA (2011) Identification of serine/threonine kinase and nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes in the fire blight resistance quantitative trait locus of apple cultivar ‘Evereste’. Mol Plant Pathol 12(5):493–505. https://doi.org/10.1111/j.1364-3703.2010.00690.x
Peil A, Garcia-Libreros T, Richter K, Trognitz FC, Trognitz B, Hanke MV, Flachowsky H (2007) Strong evidence for a fire blight resistance gene of Malus robusta located on linkage group 3. Plant Breed 126:470–475. https://doi.org/10.1111/j.1439-0523.2007.01408.x
Peil A, Flachowsky H, Hanke MV, Richter K, Rode J (2011) Inoculation of Malus × robusta 5 progeny with a strain breaking resistance to fire blight reveals a minor QTL on LG5. Acta Hortic (896):357–362. https://doi.org/10.17660/ActaHortic.2011.896.49
R Core Team (2014) R: a language and environment for statistical computing. Vienna Austria R Found. Stat, Comput
Smits TH, Guerrero-Prieto VM, Hernández-Escarcega G, Blom J, Goesmann A, Rezzonico F, Duffy B, Stockwell (2014) Whole-genome sequencing of Erwinia amylovora strains from Mexico detects single nucleotide polymorphisms in rpsL conferring streptomycin resistance and in the avrRpt2 effector altering host interactions. Genome Announc 2(1):e01229-13. https://doi.org/10.1128/genomeA.01229-13
van der Zwet T, Orolaza-Halbrendt N, Zeller W (2012) Fire blight—history, biology and management. APS Press, St. Paul. https://doi.org/10.1094/9780890544839.fm
Van Ooijen JW (2006) JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V. Wageningen
Van Ooijen JW (2009) MapQTL 6; software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B.V. Wageningen
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wöhner TW, Flachowsky H, Richter K, Garcia-Libreros T, Trognitz F, Hanke MV, Peil A (2014) QTL mapping of fire blight resistance in Malus × robusta 5 after inoculation with different strains of Erwinia amylovora. Mol Breed 34(1):217–230. https://doi.org/10.1007/s11032-014-0031-5
Acknowledgements
We thank Philip Forsline, formerly with the USDA-ARS Plant Genetics Resources Unit, Geneva, NY, for providing GMAL4593 grafted trees. We also thank Roger Lewis, Wilbur Hershburger, Daniel Silverman, and Trevor Parton of the USDA-ARS Appalachian Fruit Research Station, Kearneysville, WV and Herbert Gustafson of the Section of Plant Pathology and Plant-Microbe Biology, School of Integrative Plant Sciences, Cornell University, Geneva, NY for their expert technical assistance.
Funding
This work was funded in part by the Washington Tree Fruit Research Commission, Wenatchee, WA (www.treefruitresearch.com) project AP-08-804 “Mapping Malus sieversii: A valuable genetic resource for apple breeding,” project CP-10-101 “Fire blight resistance in Malus sieversii for scion breeding,” and the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch project accession number 1011771.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Desnoues, E., Norelli, J.L., Aldwinckle, H.S. et al. Identification of Novel Strain-Specific and Environment-Dependent Minor QTLs Linked to Fire Blight Resistance in Apples. Plant Mol Biol Rep 36, 247–256 (2018). https://doi.org/10.1007/s11105-018-1076-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-018-1076-0