Abstract
Transcriptional networks play important roles in the regulation of biological processes through coordinated activation or repression of downstream target genes. Arabidopsis bZIP transcription factors, GBF1, HY5, and HYH, interact and heterodimerize with each other to form the regulatory network in photomorphogenesis. The genome-wide direct target genes of GBF1 and the roles of HY5 and HYH in controlling GBF1’s genome-wide DNA binding ability have been shown earlier. However, the GBF1 regulated genes at global scale, and how HY5 and HYH modulate GBF1-mediated genome-wide gene expression remain unknown. Here, we report the genome-wide gene expression profile in gbf1, gbf1 hy5, and gbf1 hyh mutants. Our results suggest that HY5 and HYH antagonistically regulate GBF1-mediated global gene expression. We validated the microarray analysis with independent qPCR analyses. Functional analysis of GBF1-regulated genes validates previously known roles of GBF1 in important biological processes. Furthermore, the data also highlight possible novel role of GBF1 in several other biological processes. The previous ChIP-chip results and this transcriptome data together demonstrate the complex transcriptional regulatory mechanism of these transcription factors, GBF1, HY5, and HYH, in photomorphogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Studying genetic interactions between genes involved in same biological processes helps to dissect signaling pathways. To understand the function of transcription factors (TF), it is important to look at the alteration in gene expression of downstream target genes. Analyzing genetic interactions between TFs at genomic scale helps to understand global gene regulatory network. Whereas GBF1 works as dual regulator of photomorphogenesis, HY5 and HYH work as positive regulators for photomorphogenesis (Oyama et al. 1997; Holm et al. 2002; Mallappa et al. 2006). These three TFs have bZIP DNA binding domain, and earlier molecular-genetic studies have been shown that these TFs work interdependently to regulate light-regulated gene expression and photomorphogenesis (Holm et al. 2002; Singh et al. 2012; Ram and Chattopadhyay 2013). HY5 and HYH together largely work as synergistic partners for various light-regulated developmental processes as well as for the regulation of global gene expression (Holm et al. 2002; Sibout et al. 2006; Sellaro et al. 2009). However, interrelation of GBF1 with HY5 and HYH has been more complicated (Singh et al. 2012; Ram and Chattopadhyay 2013).
GBF1 shows light-intensity-dependent epistatic relationship with HY5 for light-mediated inhibition of hypocotyl elongation (Singh et al. 2012). For regulation of expression of Rubisco small subunit gene, RBCS1-A, GBF1, and HY5 work antagonistically. However, GBF1 shows antagonistic relationship with HYH for light-mediated inhibition of hypocotyl elongation and for the regulation of RBCS1-A expression. On the other hand, GBF1 works in synergistic/additive manner with HY5/HYH for regulation of other light-regulated gene, CAB1, encoding chlorophyll a-b binding protein 1 (Singh et al. 2012). Protein-protein interaction studies suggest direct interaction between GBF1 and HY5/HYH proteins (Singh et al. 2012). DNA-protein interaction studies show that GBF1 and HY5 form DNA-binding heterodimer at RBCS-1A promoter; however, GBF1-HYH heterodimers do not bind to RBCS-1A promoter.
A recent study has comprehensively analyzed genome-wide direct targets of GBF1 and also highlighted the importance of HY5/HYH for its genome-wide DNA binding by performing ChIP-chip experiments in wild type as well as hy5/hyh mutant backgrounds. The results show that HY5 at greater extent and HYH at lesser extent help GBF1 to bind to its target promoters more efficiently and more specifically at whole genome level (Ram et al. 2014). However, GBF1-mediated global gene expression and the role of HY5/HYH in this process still remain to be investigated. Here, in this study, we have performed genome-wide expression profiling in gbf1, gbf1 hy5, and gbf1 hyh mutant backgrounds and have analyzed the GBF1-mediated gene expression and interrelationship between GBF1 and HY5/HYH at genomic scale.
Results
Analysis of Transcriptome Regulation by GBF1 with HY5 and HYH
Four genetic backgrounds such as gbf1, gbf1 hy5, gbf1 hyh, and wild type were used for genome-wide expression profiling. Earlier studies have shown that HY5 and HYH proteins accumulate to maximum level at 3–4 days in constant white light, and HY5 protein level starts to decrease after 4 days in white light (WL) (Hardtke et al. 2000; Holm et al. 2002). Therefore, we used 4-day-old constant WL-grown Arabidopsis seedlings for RNA extraction. Furthermore, ChIP-chip analysis for GBF1 in presence and absence of HY5/HYH was also performed under same conditions (Ram et al. 2014). For our analysis, we have taken account of the genes, which show more than 1.5-fold expression change with significant score of p < 0.05. A total number of 402 genes showed altered expression in gbf1 mutant as compared to wild type. Among these genes, whereas 190 genes were downregulated, and 212 genes were found to be upregulated (Fig. 1a). In gbf1 hy5 and gbf1 hyh double mutant backgrounds, 720 and 572 genes showed altered expression compared with wild type, respectively (Fig. 1a). A large fraction of genes displaying altered expression in gbf1 also showed altered expression in gbf1 hy5 and gbf1 hyh double mutants (Fig. 1b). This result is consistent with interrelated role of GBF1 with HY5 and HYH in light-mediated seedling development (Singh et al. 2012). In a three-way comparison, the greatest overlap was found between gbf1 hy5 and gbf1 hyh backgrounds (Fig. 1b). Of these common 302 genes, 59 genes were also displayed altered expression in gbf1, suggesting that the rest 243 common genes were because of overlapping function of HY5 and HYH, which is consistent with earlier studies (Holm et al. 2002; Sibout et al. 2006). These findings altogether indicate that the transcriptome analysis has been successful in identifying true candidate genes.
Number of differentially expressed genes and their overlap among genotypes. a Number of significantly upregulated and downregulated genes relative to wild type (WT) shown in genetic backgrounds. A double criteria of p value less than 0.05 and fold change higher than 1.5 were used to consider them as significant. b Venn diagrammatic representation of differentially expressed genes (DEGs) in gbf1, gbf1 hy5, and gbf1 hyh backgrounds as compared to wild type. The figure shows possible overlap/uniqueness sectors of DEGs among three backgrounds
HY5 and HYH Antagonistically Regulate GBF1-Mediated Genome-Wide Gene Expression
To know the effect of HY5 and HYH on GBF1-mediated genome-wide expression, we compared the genes induced or suppressed in gbf1 hy5 or gbf1 hyh double mutants with that of gbf1 single mutant (Fig. 2). While the genes showing induction in gbf1 hy5 (371) were compared with all the genes with altered expression in gbf1, 80 genes were common between these two sets, and of these 80 genes, almost equal numbers of genes were induced or suppressed in gbf1 (Fig. 2a). A similar type of trend was observed when the genes showing suppression in gbf1 hy5 were compared with all the genes having altered expression in gbf1 (Fig. 2a). These results indicate that additional mutation of hy5 in gbf1 mutant background causes an opposite effect on regulation of about half of the common targets.
HY5 and HYH antagonistically regulate GBF1-mediated genome-wide gene expression. a–e The larger circles show total number of genes induced and suppressed in shown backgrounds. The inlaid circles represent the subset of those genes, which are also affected in the gbf1 mutant background. b Numbers of genes that have shown induction or suppression in hy5 mutant in the previous study by Lee et al. 2007 are used for comparison
In order to determine the effect of hy5 and hyh single mutation on gene expression, we normalized gbf1 hy5 and gbf1 hyh double mutants against gbf1 single mutant. The qPCR analysis of ten randomly selected differentially expressed genes (DEGs) in hy5 and hyh single mutant seedlings correlated with expression values of these genes in our microarray analysis of gbf1 hy5 and gbf1 hyh double mutants normalized against gbf1 single mutant (Fig. 5), suggesting that this strategy provided the real gene expression alterations in hy5 and hyh mutant backgrounds. Transcriptome analysis in hy5 mutant background has been carried out in different conditions by different groups (Holm et al. 2002; Lee et al. 2007; Zhang et al. 2010). We compared our microarray analysis with the microarray analysis in hy5 mutant background performed by Lee et al. 2007, since in this case, there was a minimum difference in array platform and experimental conditions compared with our experiment conditions. When the altered expression genes in gbf1 were compared with those in hy5 (Lee et al. 2007), although only a small number of genes were found common, however, of these common genes, more number of genes showed opposite expression pattern between them (Fig. 2b). From our data, we found that total 529 genes showed induction whereas 489 genes showed suppression because of single mutation of hy5 (Fig. 2c), which is comparable with the number of altered expression genes in hy5 mutant background published by Lee et al. 2007. Further, when these altered expression genes were compared with altered expression genes in gbf1 mutant, most of the common altered expression genes showed opposite regulation in both the backgrounds (Fig. 2c), which is consistent with Fig. 2b. Altogether, these analyses clearly suggest that HY5 mainly plays an antagonistic role to GBF1 for genome-wide expression.
Similarly, we normalized gbf1 hyh double mutant against the gbf1 single mutant and found that total 322 genes showed induction, whereas 484 genes showed suppression (Fig. 2d). Further, while we compared these altered expression genes with that of gbf1 mutant, we found that most of the common genes showed opposite regulation in both these backgrounds (Fig. 2d), suggesting an antagonistic function of GBF1 and HYH for these genes. However, while the altered expression genes in gbf1 hyh versus wild type were compared with altered expression genes in gbf1 versus wild type, we observed that among the common genes, more number of genes showed similar regulation and less number of genes showed opposite regulation (Fig. 2e). These observations suggest that additional mutation of hyh in gbf1 mutant background results in no effect or additive effect for more number of genes and antagonistic effect for lesser number of genes.
Altogether, these results suggest that both HY5 and HYH negatively regulate GBF1-mediated gene expression, and HY5 has greater impact on GBF1-mediated genome-wide gene expression compared to HYH. This conclusion is obvious in hierarchical clustering display of transcriptome data (Fig. 3), where it is found that the expression of the representative genes of overrepresented gene categories is opposite in gbf1 mutant and hy5 (gbf1 hy5 versus gbf1) or hyh (gbf1 hy5 versus gbf1) mutant backgrounds. Additionally, in gbf1 hy5 and gbf1 hyh double mutant backgrounds, the expression level of those genes was intermediate to their expression in gbf1 and hy5 (gbf1 hy5 vs. gbf1) and gbf1 and hyh (gbf1 hyh vs. gbf1), respectively (Fig. 3), which further supports the antagonistic interaction of HY5 and HYH with GBF1. To validate these array analyses, we performed real-time PCR gene expression analysis for randomly selected ten genes, which showed either induction or suppression in at least one analysis. For almost all the genes, comparable expression was observed in both the microarray and real-time PCR analyses (Fig. 4). Thereby, it validates the transcriptome analysis.
Graphical view showing GO terms associated with biological process enriched in differentially expressed genes. a gbf1, b gbf1 hy5, and c gbf1 hyh backgrounds relative to wild type (WT). The GO enrichment was performed using BiNGO. Darkest colors represent most significant, whereas lightest colors represent least significant GO term
The Functional Classification of GBF1-Regulated Genes
To look into function of the genes regulated by GBF1, enrichment analysis of Gene Ontology (GO) terms associated with biological processes was done for all the DEGs in all three genetic backgrounds (Fig. 4). Many GO terms related with biological processes such as response to light stimulus, response to stress, cellular metabolic processes, transport, and response to hormones were significantly enriched in DEGs in all the three genotypes. Additionally, some GO terms related to defense response, post-embryonic root development, cell wall organization, and leaf senescence were only enriched in gbf1 mutant background, however not in gbf1 hy5 and gbf1 hyh backgrounds. These results suggest that for some biological processes, GBF1 works with HY5 and HYH, and for other processes, it works independently to HY5 and HYH. Furthermore, some of the GO terms were significantly enriched in gbf1 hy5 and gbf1 hyh mutant, however not in gbf1 mutants, for example, GO terms associated with photosynthesis and stomatal complex morphogenesis (Fig. 4). Also, GO terms related to chloroplast organization and GO terms associated with pigment biosynthesis were significantly enriched only in gbf1 hy5 background. These results correlate well with previously published studies. Our earlier ChIP-chip study found that GBF1 significantly binds to regulatory regions of genes involved in response to light, cell wall biogenesis, developmental processes, response to other organism, ion transport, and response to stress (Ram et al. 2014). The role of the GBF1 in many of these biological processes is well documented such as response to light stimulus and developmental processes such as hypocotyl elongation, flowering and lateral root formation, and leaf senescence (Mallappa et al. 2006, 2008; Smykowski et al. 2010). Table 1 shows DEGs involved in light response. The role of GBF1 has also been shown in cell elongation and expansion (Mallappa et al. 2006), and cell wall biogenesis is directly associated with these growth parameters. Thus, these previous studies further validate our transcriptome analysis. To further look at the function of GBF1 and its interrelationship with HY5 and HYH in these biological processes, we looked at expression level of some of the representative genes involved in these biological processes (Fig. 3). It is found that for many of these representative genes, which were related to these biological processes, GBF1 acts antagonistically with HY5 and HYH, e.g., cell wall, plant defense, drought, and oxidative stress. While we looked at the enrichment of these biological processes in GO term enrichment analysis, these were either not or less significantly enriched in gbf1 hy5 and gbf1 hyh double mutants compared to gbf1 single mutant.
Validation of Differential Expressed Genes
To validate the results of differential gene expression obtained via microarray analysis, quantitative reverse transcription PCR (qRT-PCR) analysis was performed for randomly selected ten genes, which show either induction or suppression in at least one genotype (Table 2). We used hy5 and hyh single mutant lines in qPCR analysis to compare their expression in gbf1 hy5 vs. gbf1 and gbf1 hyh vs. gbf1, microarray analysis, respectively. As shown in Fig. 5, except for two genes, AT2G46830 and AT1G22770, all other analyzed DEGs have similar expression in microarray and qRT-PCR analysis. For the genes AT2G46830 and AT1G22770, there was a correlated expression between microarray and qRT-PCR analysis for gbf1, gbf1 hy5, and gbf1 hyh mutants. Overall, these results show that our microarray analysis has been successful in identifying true expression patterns in various mutants.
Validation of microarray analysis. Indicated genes showing differential expression in any of the analysis in microarray were analyzed for expression change through real-time PCR. To get the idea of expression in hy5 and hyh mutants, expression in gbf1 hy5 and gbf1 hyh was normalized against gbf1 in microarray analysis, and then, it was validated with real-Time PCR analysis in hy5 and hyh mutants. Actin2 was used as an internal control in real-time PCR. Error bars indicate SD, where n = 2
Physiological Validation of Microarray Results
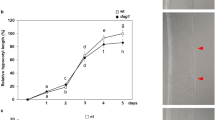
Our GO analysis reveals the role of GBF1 in regulating many genes involved in diverse biological processes, and it further explores interrelationship of GBF1 with HY5 and HYH in these processes. We tried to substantiate some of these observations. Our GO analysis revealed that many GO terms related with post-embryonic root development were enriched in gbf1 single mutant but not in gbf1 hy5 and gbf1 hyh double mutants. To confirm this observation, we analyzed gbf1 hy5 and gbf1 hyh double mutants, along with their parental single mutants for lateral root formation. As shown in Fig. 6a–d, the double mutant of gbf1 hy5 and gbf1 hyh displayed similar number of lateral roots to wild type, which explains why the genes related with post-embryonic root development were not enriched in gbf1 hy5 and gbf1 hyh double mutants compared to wild type. Further, these results show that gbf1 hy5 and gbf1 hyh displayed intermediate number of lateral roots compared to their parental single mutants, suggesting that function of GBF1 is antagonistic to HY5 and HYH for lateral root formation.
Analysis of lateral root formation in gbf1 hy5 and gbf1 hyh double mutants. a, c The root growth of 12-day-old wild type and various mutant plants grown in constant WL (100 μmol/m2/s). b Quantification of number of lateral roots formed in wild type (WT) and various mutants at different days after germination. d Quantification of number of lateral roots formed in wild type (WT) and various mutants
Comparison Between Microarray and ChIP-Chip Data
ChIP-chip analysis for GBF1 identified 2264 genes as its direct targets (Ram et al. 2014). We used previous ChIP-chip data to compare with the DEGs in gbf1, gbf1 hy5, and gbf1 hyh mutants. As shown in Fig. 7, high-level overlap was not found between two datasets. Only 12–14 % of DEGs have GBF1 binding sites, the rest of the genes seem to be indirectly regulated by GBF1. Further, of the 48 genes that have altered expression in gbf1 mutant and are direct targets of GBF1, 19 of them are positively regulated and 29 of them are negatively regulated. These results suggest that at genome-wide scale, GBF1 acts as a transcriptional activator as well as transcriptional repressor.
Venn diagram showing the overlap between the GBF1 direct target genes identified in GBF1OE line in previous ChIP-chip study (Ram et al. 2014) and DEGs in the three backgrounds. Genes in the overlap region between ChIP-chip targets and DEGs in gbf1 background (48 genes) are defined as directly regulated genes. Of these 48 genes, 19 are positively regulated and 29 are negatively regulated by GBF1
Discussion
Previous molecular and genetic studies have established functional interrelationship between GBF1 and HY5 and HYH for light-mediated seedling development (Singh et al. 2012; Ram and Chattopadhyay 2013). Genome-wide DNA binding analysis of GBF1 in presence or absence of its heterodimer partners HY5 and HYH has shown that HY5 and HYH play crucial roles in GBF1’s genome-wide DNA binding (Ram et al. 2014). However, it was not clear how GBF1 regulates genome-wide gene expression and how do HY5 and HYH affect this. To understand functional relationship between GBF1 and HY5 and HYH for genome-wide gene expression regulation, we performed gene expression microarray analysis in gbf1, gbf1 hy5, and gbf1 hyh mutants. Although hy5 and hyh single mutants were not used for microarray analysis, we were able to get the effect of hy5 and hyh single mutations on genome-wide gene expression changes in our study by normalizing gbf1 hy5 and gbf1 hyh double mutants against gbf1 single mutant. The q-PCR analyses in hy5 and hyh single mutants for randomly selected genes validate the strategy since for the most of the tested genes, we found a good correlation between qPCR and microarray analysis (Fig. 5). Comparative analysis of upregulated and downregulated genes in gbf1 with hy5 and hyh mutants shows that among the common regulated genes, most of the genes are oppositely regulated by GBF1 and HY5/HYH (Fig. 2c, d). These findings suggest that GBF1 has an antagonistic function to HY5 and HYH for most of the common target genes. This conclusion is further supported when we looked at the expression level of individual representative genes from overrepresented gene categories (Fig. 3). Comparative analysis between gbf1 and double mutants (gbf1 hy5 and gbf1 hyh) shows that among the common genes, around half of the genes have the same regulation of expression in both the backgrounds and rest half have opposite regulation of expression in both the backgrounds (Fig. 2a, e). In a double mutant, one would expect that expression level of a gene should be the sum of the expression levels in both the parental single mutants. However, in our case, we do not see this. The possible reason could be that expression level of common target genes might be different in both the single mutants. For example, in gbf1, gene X is upregulated by 2-fold and hy5 mutant gene X is downregulated 5-fold, so in double mutant of gbf1 hy5, gene X would be downregulated 3-fold.
Various lines of evidences, including qPCR of randomly selected genes, validate our analysis (Fig. 5). Functional analysis of GBF1-regulated genes in gbf1 mutant confirms its role in many biological processes in which the role of GBF1 has already been known such as response to light stimulus, lateral root development, chlorophyll biosynthesis process, leaf senescence, response to oxidative stress, reactive oxygen species metabolic process, and cell redox homeostasis (Mallappa et al. 2006, 2008; Smykowski et al. 2010). GO analysis suggests the role of GBF1 in defense response, and in our recent independent study, we have found the role of GBF1 in hypersensitive response (HR) and basal defense against bacterial pathogens (Giri et al., unpublished results). Our data further suggest a novel role of GBF1 in many other biological processes such as response to different kinds of stresses; response to various hormones; transport of water, iron, and nitrate ions; and some metabolic processes. These results should pave the way for future functional studies regarding GBF1 function.
Functional classification of HY5 direct targets in a ChIP-chip study showed that the photosynthesis-related genes were the most highly enriched group (Lee et al. 2007). Consistent with this, in gbf1 hy5 double mutant, but not in gbf1 single mutant, we found that many photosynthesis-related GO terms were significantly enriched. Also, many photosynthesis-related GO terms were significantly enriched in gbf1 hyh double mutant, suggesting a similar role of HYH in photosynthesis. In addition to that, many GO terms related with pigment biosynthesis were significantly enriched in gbf1 hy5 double mutant, and consistently, the role of both GBF1 and HY5 has been known in this biological process (Ang and Deng 1994; Mallappa et al. 2006). We have found that GO terms related with lateral root formation are enriched only in gbf1 mutant but not in gbf1 hy5 and gbf1 hyh double mutants. To confirm this observation, we analyzed lateral root formation in single mutants and double mutants and found that, indeed, there is no difference between wild type versus gbf1 hy5 or gbf1 hyh double mutants in regard to number of lateral roots formed, thereby validating our whole microarray analysis at physiological level.
Finally, comparison between ChIP-chip data (Ram et al. 2014) and microarray data reveals that 12–14 % of altered expression genes from the microarray analysis are direct targets of GBF1 (Fig. 7). These results are not striking, as many previous studies reported that less than 10 % of the directly bound gene promoters were among genes with significant transcriptional changes (Lee et al. 2007; Zheng et al. 2009; Morohashi and Grotewold 2009). These results suggest that GBF1 regulates many genes through indirect mechanism, and it may require other TFs or cofactors to change the expression of its target genes. This observation is consistent with recent genomic studies, which highlight that transcriptional regulation mediated by TFs is not a simple process as it was thought to be earlier.
Methods
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana used in this study is the segregated wild type obtained from the genetic crosses of various mutants in Col-0 or Wassilewskija (WS) background. The gbf1-1 mutant (Mallappa et al. 2006) is in Col-0 accession, whereas hy5-ks50 (Oyama et al. 1997) and hyh (Holm et al. 2002) are in WS background. The gbf1 hy5 and gbf1 hyh are described in Singh et al. 2012. Unless stated otherwise, seeds were surface sterilized and plated on Murashige and Skoog medium supplemented with 0.8 % Bacto Agar (Difco) and 1 % sucrose. The plates were then cold treated at 4 °C for 4 days and transferred to light chambers maintained at 22 °C with the desired light intensities.
Genome-Wide Expression Analyses Through Microarray
For microarray analysis, gbf1, gbf1 hy5, gbf1 hyh, and wild-type seedlings were grown for 4 days under constant WL (20 μmol/m2/s) conditions. Two independent biological replicates for each genotype were used for RNA preparation and hybridization. Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen, CA). The integrity and concentration of the RNA were verified by capillary electrophoresis using a Bioanalyzer 2100 (Agilent). Sample preparation for hybridization and detection was done according to Affymetrix protocols. Raw data (CEL files) were obtained from the hybridization of Arabidopsis Affymetrix ATH1 Arrays with the cRNA samples. Microarray data were analyzed using GeneSpring software (Affymetrix) by normalizing gbf1 single mutant against the WT and gbf1 hy5 and gbf1 hyh double mutants against the WT and gbf1 single mutant.
GO Analysis
GO analysis was performed using BiNGO. For this, hypergeometric distribution was performed to find out the statically significant GO terms. Further, Bonferroni family-wise error rate (FWER) was used to control the fast positive rate. The GO terms showing P value <0.01 for a given gene set were considered to be significant.
Quantitative Real-Time PCR
The details of primers for qPCR are described in Supplemental Table 1. Quantitative PCR was performed in Thermal Cycler Applied Biosystem StepOne™. For qPCR, tissues were grown in conditions similar to microarray analysis. The fold expression was calculated by the ratio of expression value of target gene to expression value of Actin2 in various backgrounds relative to the WT.
References
Ang LH, Deng XW (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6:613–628
Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19:4997–5006
Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16:1247–1259
Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19:731–749
Mallappa C, Yadav V, Negi P, Chattopadhyay S (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J Biol Chem 281:22190–22199
Mallappa C, Singh A, Ram H, Chattopadhyay S (2008) GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J Biol Chem 283:35772–35782
Morohashi K, Grotewold E (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5:e1000396
Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11:2983–2995
Ram H, Chattopadhyay S (2013) Molecular interaction of bZIP domains of GBF1, HY5 and HYH in Arabidopsis seedling development. Plant Signal Behav 8:1–3
Ram H, Priya P, Jain M, Chattopadhyay S (2014) Genome-wide DNA binding of GBF1 is modulated by its heterodimerizing protein partners, HY5 and HYH. Mol Plant 7:448–51
Sellaro R, Hoecker U, Yanovsky M, Chory J, Casal JJ (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr Biol 19:1216–1220
Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signalling. PLoS Genet 2:e202
Singh A, Ram H, Abbas N, Chattopadhyay S (2012) Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J Biol Chem 287:25995–26009
Smykowski A, Zimmermann P, Zentgraf U (2010) G-box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis thaliana. Plant Physiol 153:1321–1331
Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2010) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65:346–358
Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21:2563–2577
Acknowledgments
This work was supported by the core grant of NIPGR to SC, and MJ, HR, and AS are recipients of fellowships from Council of Scientific and Industrial Research, Government of India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Accession Number
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE36964 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36964)
Link for reviewers http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lzujlmmmausmaro&acc=GSE36964
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 272 kb)
Rights and permissions
About this article
Cite this article
Ram, H., Jain, M., Singh, A. et al. Functional Relationship of GBF1 with HY5 and HYH in Genome-Wide Gene Expression in Arabidopsis . Plant Mol Biol Rep 34, 211–220 (2016). https://doi.org/10.1007/s11105-015-0910-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0910-x