Abstract
Transposable elements (TEs) are major components of eukaryotic genomes and have played important roles in genome evolution. Plant introgressive hybridization is widespread in nature and may induce a wide range of genetic and epigenetic variations including reactivation of dormant TEs. Newly elicited TE transpositions may lead to heritable expression alteration in genes adjacent to excision and insertion sites and consequently to phenotypic novelty. Prior studies showed that distant hybridization between sexually incompatible plant species might cause reactivation of TEs. However, these studies used plants with confirmed alien genetic introgression, so it remains unclear whether the cause of TE activation is introgression or the alien pollination process itself. We pollinated rice stigma (Oryza sativa L., cv. Jijing88) with Zizania latifolia pollen. Immediate offspring and subsequent generations were self-fertilized. We conducted whole-genome resequencing on a derivative line that showed heritable phenotypic variations in plant height and grain characteristics. No genetic introgression from the pollen donor was observed, but transpositional reactivation of otherwise quiescent rice endogenous TEs was detected. In total, 33 de novo mobilization events occurred involving 13 TEs, namely a MITE mPing, Pong (the transposase-donor of mPing), and 11 LTR retrotransposons. Transpositions were verified by locus-specific polymerase chain reaction amplification and Southern analysis. Gene expression analysis suggested that at least some of the mobilized TEs caused heritable expression changes in neighboring genes, including genes that mapped to quantitative trait loci (QTLs) associated with grain weight and size. Thus, pollination by Zizania without entailing introgression can induce mobilization of endogenous TEs and cause heritable gene expression alterations. Our results suggest that pollination by related but sexually incompatible species may generate genetic and phenotypic novelty via modifications induced by enhanced mobility of TEs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transposable elements (TEs) are parasitic to their host genome but may also have biologically important functions. TEs can be classified as DNA transposons (class II) and retrotransposons (class I) according to their models of transposition. Transposons move via a “cut-and-paste” model and their copy numbers therefore often remain unaltered. By contrast, retrotransposons transpose via an RNA intermediate using a “copy-and-paste” model (Bennetzen 2000) and thus are a major cause of genome-size differences between related plant species. The rapid accomplishment of whole-genome sequences for increasing numbers of species has revealed that TEs are major genomic components of all organisms. For example, TEs account for 12 % of the genome in Drosophila, 45 % in humans, 80 % in maize, and >90 % in hexaploid wheat (Bennetzen 2000; Flavell 1986; Gao et al. 2004; Kidwell and Lisch 1997; McCarthy et al. 2002). Uncontrolled mobility of TEs would conceivably result in genomic catastrophe and would be maladaptive to their host. To maintain genome stability, the activity of TEs must be suppressed for normal growth and development of their host to be maintained. However, the repressive control of TEs can be compromised under certain stressful conditions, leading to transcriptional enhancement and transpositional reactivation, which would be subsequently followed by re-repression (Guerreiro 2012; Liu et al. 1995; Shapiro 2010; Sabot et al. 2011; Sahu et al. 2013). TE mobilization produces immediate structural genomic changes and disruption of gene function and may also modulate heritable changes in the expression of adjacent genes (Jordan et al. 2003; Kamal et al. 2006; Lowe et al. 2007a; Naito et al. 2009; Wang et al. 2013a; Bourque et al. 2008). TE activity can therefore generate phenotypic variations in myriad ways, and therefore, TEs are considered as important drivers of genome evolution. For example, in Drosophila, TEs are responsible for approximately 80 % of spontaneous phenotypic mutations (Guerreiro 2012).
Accumulated evidence indicates that distant hybridization between genetically diverged individuals of different, or even the same, species may cause transcriptional reactivation and/or mobilization of TEs in the resulting hybrids. For example, rampant mobilization of a retrotransposon occurred in interspecific marsupial hybrids and caused chromosomal expansion and karyotype repatterning, which, in turn, facilitated reproductive isolation (O’Neill et al. 1998). In Drosophila virilis, four different TEs (Ulysses, Penelope, Paris, and Helena) were mobilized through the well-known process of hybrid dysgenesis (Petrov et al. 1995). The formation of three hybrid species in sunflower was accompanied with the amplification of retrotransposons, which caused substantial differences in genome size. Notably in plants, some intraspecific crosses may also elicit mobilization of previously quiescent TEs. For example in rice, crosses between certain cultivars have induced transpositional activity of MITEs Stowaway Os-1 and Mashu (Kantama et al. 2013). These observations are in line with the well-known “genome shock” theory proposed by McClintock nearly three decades ago (McClintock 1983).
Introgressive hybridization is a special kind of distant hybridization in which some portions of one species genome (donor parent) are integrated into the other (recipient parent). This occurs widely between sympatric natural plant populations of the same or related different species (Rieseberg and Wendel 1993; Song et al. 2002). Introgressive hybridization is believed to be an important force in genome evolution of many plant taxa (Rieseberg et al. 2003; Wendel 2000). In plant breeding, introgression of alien DNA segments from wild species into crops is widely used and represents one of the most effective methods for enhancing germplasm. Although the major and immediate effect of introgression is direct transfer of useful alleles from the donor species into the recipient one, several studies suggest that introgression may also impact the recipient genome by generating de novo genetic and epigenetic variations. For example, introgression of uncharacterized foreign DNA segments into cultured animal cells has caused genome-wide perturbation of DNA methylation patterns and lead to changes in gene expression (Remus et al. 1999). Introgression of minute chromatin segments of Zizania latifolia (Manchurian wild rice) into cultivated rice has caused extensive and wide-ranging genetic and epigenetic changes including mobilization of several TEs in the resultant recombinant inbred lines (Dong et al. 2006; Liu et al. 1999a; Liu et al. 1999b; Liu and Wendel 2000; Shan et al. 2005; Wang et al. 2010a; Wang et al. 2010b; Wang et al. 2005; Wang et al. 2013b). However, it remains unclear whether the genetic and epigenetic changes were caused by alien chromatin integration or whether the alien pollination process itself may have caused the changes. This question is pertinent because one previous study showed that alien pollination of rice by pollen from a very remote and virtually unrelated plant species, Oenothera biennis L., generated a mutator-like genotype that continuously produces genetic and phenotypic variations (Wang et al. 2009). However, this dramatic example might be a case of contingency, and it is unknown whether these observations are more widely applicable.
In this study, we chose to examine the effects of alien pollination in experiments between rice and Z. latifolia because extensive data are available from a set of introgression lines between these two species (Dong et al. 2006; Liu et al. 1999a; Liu et al. 1999b; Liu and Wendel 2000; Shan et al. 2005; Wang et al. 2010a; Wang et al. 2010b; Wang et al. 2005; Wang et al. 2013b). We used a widely grown japonica rice cultivar, Jijing88, which had undergone several rounds of additional selfing in our hands, as the recipient parent. We aimed to address whether the process of alien pollination itself might cause mobilization of rice TEs independently of the effects of Zizania introgression.
Materials and Methods
Plant Materials, Measurement of Phenotype and DNA Extraction
We produced a stabilized introgression line, X9, from introgressive hybridization between rice (Oryza sativa ssp. japonica cv. Jijing88) and a sexually incompatible wild species of the tribe Oryzeae, Z. latifolia. A “repeated pollination” method was used as previously described (Liu et al. 1999b; Shan et al. 2005) (Fig. 1a), followed by selfing for 10 generations to produce X9. We measured some agricultural traits of the putative introgression line X9 and its rice parent Jijing88, including the number of effective tillers, panicle length, seed width and length, and 1000 seed weight. Trait data were obtained independently at least three times. Means were calculated and statistical differences between lines assessed using Independent-Samples Student’s t-test. Genomic DNA was isolated from expanded young leaves of X9 and both its donor parents using a modified CTAB method (Allen et al. 2006) and phenol extraction. DNA sample quality was determined by a ND-1000 NanoDrop spectrophotometer (Eppendorf, Germany).
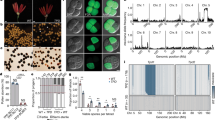
Production and characteristics of the X9 introgression line. a Schematic diagram of X9 production by “repeated pollination”; b whole plant morphology of X9 and the parental line, Jijing88; c phenotypes of randomly selected panicles from Jijing88 and X9; d phenotypes of randomly selected grains from Jijing88 and X9; e statistics comparing X9 to Jijing88 for six morphological traits. Data are shown as mean ± standard deviation and Student’s t test was used to generate the P values in e
Genomic In Situ Hybridization
The in situ hybridization protocol was described by Han et al. (Han et al. 2004), with minor modifications. Genomic DNA of Z. latifolia plants was labeled by nick translation with Texas Red-5-dCTP (Perkin Elmer; cat. no. NEL426). Jijing88 genomic DNA was used as a blocker. Slides were examined with an Olympus fluorescence microscope (BX61) and digitally photographed.
Zizania Pollen Grain Germination and Pollen Tube Growth in Pistil of Rice
Emasculated flowering spikelets of Jijing88 were rapidly pollinated with pollen grains from Z. latifolia. An improved method for observation of pollen grain germination and pollen tube growth in pistil was used, as described by Chen et al. (Chen et al. 2008). Spikelets were isolated 5 min, 2 h, 8 h, and 24 h after pollination and fixed in FAA (37 % formaldehyde, glacial acetic acid, and 70 % ethanol, 1:1:18) for 24 h. Samples were then serially rehydrated in 70, 50, 30, and 10 % alcohol for 2 h each, washed twice in distilled water, and then stained with 0.1 % aniline blue for at least 5 min. The exposed stigma and ovary were then put onto clean glass slides for observation under a Nikon Eclipse80i fluorescence microscope, and images were digitally captured. The same process was employed to examine control samples, specifically emasculated flowering panicles that were pollinated with their own pollen. Control panicles were isolated 5, 10, 20, and 30 min after pollination.
Whole-Genome Resequencing
The rice-Zizania introgression underwent 10 selfing generations; therefore, X9 and its rice parent Jijing88 can be considered as stable pure lines. In this experiment, genomic DNA was extracted from a pool derived from at least 20 individuals of each line. The genomes of X9 and Jijing88 were sequenced using the Illumina HiSequation (2000) platform. Clean data have been deposited at the SRA database http://www.ncbi.nlm.nih.gov/sra/ with accession number SRP041648.
Identification and Characterization of Transposable Element Mobilization
To further investigate TE mobilization in the X9 genome, we applied a paired-end mapping (PEM) detection strategy, as employed previously by Sabot et al. (Sabot et al. 2011). Briefly, we first constructed a TE reference fasta file using known TE sequences from RetrOryza database (http://retroryza.fr/) (Chaparro et al. 2007) and then employed a Perl script to remove all the reference TEs from the Nipponbare genome reference (MSU7.0 http://rice.plantbiology.msu.edu/ index.shtml), creating a new reference genome that lacked TEs. We subsequently mapped X9 and Jijing88 sequence ends to each of the two references (TE reference and TE-free genome) with BWA software (Li and Durbin 2009). Next, we used SAMtools and Perl scripts to select paired-end sequences that had one end highly and uniquely similar to the TE-free genome reference, and the other end highly similar to the TEs reference. This clear pattern of PEM to both the chromosomal region and transposable element was indicative of TE location. Finally, we identified TEs that had mobilized in X9 but not in the rice parent Jijing88 by comparing the precise TE locations in the two genomes. To confirm TE mobilizations in the X9 genome, polymerase chain reaction (PCR) primers were designed as targeting junctions of a set of selected TE insertions using Primer 5 (http://www.premierbiosoft.com), and two types of locus-specific PCR were performed according to TE size. For large TEs (>2 kb), four primers were designed for each transposition locus: two from the TE flanking sequence and two located at the 5′ and 3′ ends of the TE (Fig. 4a). If the TE in question was an LTR retrotransposon, the primers were designed against the LTR region. For small TEs (mPing), we designed two primers flanking the transposition locus (Fig. 4b). All the primer sequences used to verify TE location are listed in Table S1. The PCR reaction conditions were 10 min at 94 °C; 28 cycles of 30 sec at 94 °C, 60 sec at 60 °C, and 2 min at 72 °C; and a final extension for 10 min at 72 °C. PCR products were visualized by ethidium bromide staining after 1 % agarose gel electrophoresis. Amplicons were purified from the gel and sequenced to confirm TE presence. TEs and their insertion sites were obtained from the annotated Nipponbare genome (http://rice.plantbiology.msu.edu/downloads_gad.shtml). Quantitative trait locus (QTL) data related to rice agriculture traits were downloaded from the Gramene QTL Database (http://www.gramene.org/qtl/).
TE Copy Number Estimation by Southern Analysis
DNA samples for Southern analysis were separately digested with XbaI, EcoRI, and HindIII. These enzymes were chosen because their sites were either absent from the studied TEs or occurred infrequently, thus allowing TE copy number to be estimated from blotting patterns. Restriction enzymes were purchased from New England Biolabs Inc. (Beverly, MA, USA). DNA from six X9 individuals was analyzed, along with the donor parents Oryza sativa ssp. japonica cv. Jijing88 and Oryzeae, Z. latifolia. Digested DNA was separated on a 1 % agarose gel then transferred to Hybond N+ nylon membranes (Amersham Pharmacia Biotech) using the alkaline transfer method recommended by the manufacturer. Element-specific PCR primers were designed against selected low-copy TEs (Table S2) and used to amplify probes with Nipponbare genomic DNA as a template. Probes were gel-purified, confirmed by sequencing, and labeled with fluorescein-11-dUTP using the Gene Images Random Prime-Labeling Module (Amersham Pharmacia Biotech). Hybridization signal was detected using the Gene Images CDP-Star Detection Module (Amersham Pharmacia Biotech) after washing at a stringency of 0.2 × SSC, 0.1 % SDS for 2 × 50 min. Membranes were exposed to X-ray film for 1–3 h depending on signal intensity.
RNA Isolation and Real-Time qRT-PCR Analysis of Genes Adjacent to Transposition Loci
Total RNA was isolated from X9 and Jijing88 at the four-leaf seedling stage with Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. RNA was treated with DNase I (Invitrogen) to eliminate possible genomic DNA contamination prior to reverse transcription with Superscript RNase H-Reverse Transcriptase (Invitrogen). Because grain weight and grain length and width were significantly higher in X9 than in Jijing88, eight genes close to TE transposition loci were chosen for analysis, some of which were also associated with QTLs related to grain weight and size. Gene-specific qRT-PCR primers were designed using Primer 5 (Table S3). The qRT-PCR reaction conditions were 1 min at 95 °C and 40 cycles of 5 s at 95 °C, 15 s at 60 °C, and 30 s at 72 °C. Relative expression of the analyzed genes was calculated using the 2-ΔΔC T method (Livak and Schmittgen 2001).
Statistics
Phenotypic statistical significance was determined using the Independent-Samples Student’s t test instance in SPSS 11.5 for Windows (http://www.spss.com/statistics/). The same method was used to test whether expression of eight genes close to TE sites was significantly different between X9 and Jijing88. And all genome mapping graphics were obtained by R version 3.0.1.
Results
Identification and Characterization of a Rice Line Derived from Alien Pollination by Z. latifolia Followed by Self-Fertilization
We produced a phenotypically stable rice line (X9) from alien pollination of the Jijing88 rice cultivar with Z. latifolia pollen (Fig. 1a). A single plant was selected from the immediate offspring that had distinct phenotypes when compared with Jijing88; this was successively selfed for 10 generations to produce the X9 line. Jijing88 is a pure-line japonica rice cultivar that has been the most widely grown variety in Northeast China over the last decade. Purity of the plants used for the alien pollination experiment was ensured by several successive generations of selfing and was validated by extensive genotyping with molecular markers. Therefore, differences in overall plant stature and in at least six morphological traits between X9 and Jijing88 (Fig. 1b–e) suggested occurrence of heritable genetic and/or epigenetic changes due to alien pollination of Jijing88 by Zizania pollen.
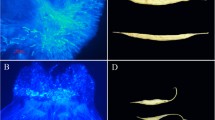
To test whether introgression of Zizania chromatin might have occurred in X9, we first examined X9 by cytological analysis. Results showed that X9 had a somatic chromosome number of 2n = 24, identical to the recipient rice cultivar Jijing88. Furthermore, genomic in situ hybridization (GISH) of X9 using Zizania genomic DNA as a probe and a 100✕ excess of Jijing88 genomic DNA as a blocker showed no evidence of Zizania chromosomal segment introgression into X9 (Fig. 2a). The cytological analysis therefore indicated no evidence for “real hybridization”, i.e., merging of the two species’ genomes. To test this further, we repeated the alien pollination experiment to examine the fate of the Zizania pollen on the Jijing88 rice stigma. Zizania pollen could germinate on the rice stigma but were incapable of entering the rice stigma to form pollen tubes (Fig. 2b). This suggests that, at least for the rice cultivar used here (Jijing88), real hybridization between rice and Zizania through this manipulation is unlikely.
Genome composition of Jijing88 and X9, and pollen grain germination and growth. a Chromosome structure of Jijing88 (a) and X9 (b); b Jijing88 (a–d) and Zizania (e–h) pollen grain germination and pollen tube growth in pistil of Jijing88. Sampling times were 5, 10, 20, and 30 min after pollination for Jijing88 and 5 min, 2 , 8 , and 24 h after pollination for Zizania
Mobilization of Diverse TEs in X9 Revealed by Whole-Genome Resequencing
Previous reports indicated that rampant mobilization of diverse TEs occurred in introgression lines of rice-Zizania that were produced by similar procedures as in this study but which had genetic evidence of introgressed Zizania chromatin (Liu and Wendel 2000; Shan et al. 2005; Wang et al. 2010a; Wang et al. 2010b; Wang et al. 2013b). We therefore wished to determine whether reactivation of TEs might also have occurred in the X9 line although which harbored no observed Zizania introgression. To test this possibility, we conducted whole-genome resequencing on X9 and Jijing88 using Illumina technology (Shendure and Ji 2008). In total, 43.5 million 100 base pair (bp) paired-end reads were obtained for the two genotypes, of which 41.7 million reads were successfully mapped to the Nipponbare reference genome (MSU7.0 http://rice.plantbiology.msu.edu/index.shtml) using BWA software, yielding 11.8× and 9.8× coverage of the Jijing88 and X9 genomes, respectively. We used PEM to determine TE locations and identified 33 novel transpositional events by 13 different TEs in X9 that were different in Jijing88. The reactivated TEs included MITE mPing, its transposase-donor, Pong, and 11 LTR retrotransposons (Table 1). The 33 new transpositional events occurred in 11 of the 12 rice chromosomes, with chromosome 2 as the exception (Fig. 3; Table S4). Notably, most of the transpositions occurred in gene-rich regions, with 15 new insertions and five excisions mapped either within genes or their upstream flanks, <2 kb from the transcription initiation sites (Fig. 3 and Table S4). Moreover, four transpositions mapped within QTLs associated with grain length, grain width, and grain weight in rice (Fig. 3), which coincided with the alteration of these traits in ×9 (Fig. 1c, d).
Verification of TE Mobilization in X9 by Locus-Specific PCR Amplification and Southern Analysis
Of the 33 TE mobilization events in X9 revealed by whole-genome resequencing, eight insertions and four excisions involving eight different TEs were selected for confirmation by independent analysis. The mobilization events were tested by locus-specific PCR amplifications with primers anchored to the insertion or excision junctions (Table S1). We designed four primers for each of the seven large TEs (whole size >2 kb), namely Lullaby, Osr3, Osr42, RN55_37, RN157_332, Tos17, and Pong (Fig. 3). In these cases, if a new, homozygous insertion event occurred in X9, the specific band would be amplifiable from Jijing88 but not from X9 with either or both primer combinations P1/P2 and P3/P4; however, the reverse should be true for primer combination of P1/P4, depending on whether the inserted element remained intact or underwent additional structural changes (e.g., ectopic recombination). We found that the seven analyzed TEs met these criteria, therefore verifying that de novo insertions by these TEs did occur in X9 (Fig. 4a). For the small MITE mPing (430 bp), we designed primers to amplify its whole length. We analyzed five loci for this element, comprising three excisions and two insertions, and in all cases, the mobilization events were verified (Fig. 4b). We also isolated and sequenced the locus-specific PCR products from all eight TEs. The expected TE-flank junctions were verified in all cases.
Conformation of TE locations by locus-specific PCR and Southern analysis. a Schematic diagram indicating primer distributions and agarose gel electrophoresis analysis of amplicons from locus-specific PCR for four randomly selected large TEs; b schematic diagram indicating primer distribution for mPing verification and agarose gel electrophoresis analysis indicating excision and insertion of mPing; c Southern analysis of introgression line X9 and its rice parental line Jijing88. TE probes were hybridized to DNA samples digested with Xba I, EcoR I, or Hind III. With respect to Jijing88, increased copy number in X9 is indicated by a red arrow, and reduced copy number in X9 is indicated with a blue arrow
We next performed Southern blot analysis for seven low-copy TEs amenable to this analysis, which included one transposon, namely Pong, and six retrotransposons, namely Lullaby, Osr3, Osr42, RN55-37, RN157-332, and Tos17. Mobilization events were detected for all these TEs in X9 (as shown by gain or loss of hybridization fragments), while no events were detected in Jijing88 (as shown by monomorphic patterns) (Fig. 4c). All but one (RN55-37) of these analyzed TEs did not contain a homolog in Zizania, indicating their occurrence in rice was after the divergence of Oryza and Zizania.
TE Excision or Insertion Caused Heritable Changes in the Expression of Adjacent Genes in X9
To test if the mobilized TEs in X9 have functional consequences, we used real-time qRT-PCR analysis to assess the expression of eight genes associated with TE excision or insertion sites in X9 but not in Jijing88 (Table 2 and Fig. 3). Gene expression of all eight genes was significantly altered in X9 relative to Jijing88, with four being significantly upregulated and four downregulated (Fig. 5). Notably, three of the four upregulated genes were located in QTLs associated with grain length and 1000 seed weight (Fig. 3). Both of these traits were significantly altered in ×9 relative to Jijing88 (Fig. 1c–e); this is unlikely to be coincidental, but further verification is needed to confirm the causal correlations.
Discussion
Pollination by Zizania as the Underlying Cause for the Mobilization of TEs
A series of previous studies documented that introgressive hybridization between rice and Zizania induced TE mobilization, as well as other types of genetic and epigenetic changes, in the resultant introgression lines (Liu et al. 2004; Shan et al. 2005; Wang et al. 2010b; Wang et al. 2005; Wang et al. 2013b). It is notable that the introgression lines used in all these previous studies were derived from a single hybridization event and harbor introgressed chromatin of Zizania (Shan et al. 2005; Wang et al. 2005). It was therefore assumed that the integration of Zizania DNA was responsible for the observed genetic and epigenetic changes, due to similar mechanisms as reported in animal cell lines (Remus et al. 1999; Muller et al. 2001). However, the finding that alien pollination of rice by pollen of a very remote and virtually unrelated plant species, O. biennis L., generated a mutator-like genotype that produces transgenerational genetic and phenotypic variations, including mobilization of several TEs (Wang et al. 2009), raised the possibility that the alien pollination process per se without foreign DNA integration may also cause inability of the host genome. Our aim in this investigation was to further examine the effect of alien pollination process on host genome stability in terms of TE transpositional activity.
We found that different TEs were mobilized in the introgression line X9 but not in its parent Jijing88, and we speculate four possible causes for this, as follows: (i) intrinsic mobility of some TEs in this specific rice cultivar (Jijing88) and hence segregation of preexisting heterozygous TE loci in X9; (ii) contamination by pollen or seeds from other rice cultivars during production of X9; (iii) introgression of Zizania chromatin; and (iv) pollination by Zizania. We can confidently rule out (i) as a casual factor based on the following evidences: (a) in our previous efforts to induce TE activity by various means (e.g., Lin et al. 2006; Yang et al. 2012), we did not observe activity of any of the TEs studied here in the rice cultivar. In fact, as documented by other studies (Naito et al. 2009; Teramoto et al. 2014), rice cultivars with intrinsic TE mobility under normal growing conditions are extremely rare and so far only one (Gimbozu) was identified. (b) As detailed in the “Materials and Methods” section, Jijing88 was a pure-line rice cultivar and plants used for the pollination experiment had been purposely selfed for several additional generations. In addition, segregation of preexisting heterozygosity can produce loss of a TE copy, but not gain of a copy. We can also rule out (ii) as a causal factor because we strictly bagged all the panicles of Jijing88 after emasculation and grew all the plants in an isolated greenhouse. Moreover, seed or pollen contamination would be a 100 and 50 % contamination, respectively, for the polymorphic loci between Jijing88 and the contaminator; both Simple Sequence Repeats (SSR) and Amplified Fragment Length Polymorphisms (AFLP) molecular marker (Carr et al. 2003) analysis revealed no evidence of this (primers listed in Tables S5, S6). In addition, whole-genome resequencing revealed that none of the 202 retrotransposon loci were missing in X9 when compared to Jijing88. At the onset of this study, we expected to find introgression of Zizania chromatin (iii), but neither cytological analysis nor whole-genome resequencing provided such evidence. Still, we cannot rule out the possibility that introgression had actually occurred at the S0 generation of X9, i.e., immediately following the “repeated pollination” manipulation (Liu et al. 1999a, b) but were segregated out (purged) during the successive selfing. Nevertheless, we consider this as also highly unlikely based on the following two lines of evidence: (i) our previous results have shown that, once occurred, the introgressed Zizania chromatin in the rice genome is stable across many generations (Wang et al. 2005); and (ii) our Zizania pollen germination experiment indicated that the landed Ziania pollens on the stigma of this specific rice genotype (Jijing88), albeit germinated to pollen tubes, could not elongate to enter the rice embryo sac. Taken together, we consider alien pollination by Zizania (iv) as the only conceivable cause for TE mobilization. Although the exact elicitors remain to be identified, there are several possibilities. For example, although pollen tube of Zizania were incapable of extending and hence the fertilization was blocked, some substances (e.g., signaling molecules and small RNAs) can be released and brought into the zygote upon subsequent self-pollination of the rice plants. Indeed, several studies have showed that small RNAs can be released during pollen development and play important roles and affect activity of the target TEs (Castel and Martienssen 2013; Dunoyer et al. 2013). Therefore, it is reasonable to infer that some small RNAs of Zizania pollen may have been responsible to inducing the TEs transpositional activities in X9, sensu the “genome shock” theory (McClintock 1983) .
Functional Consequences of Mobilized TEs
The impact of TEs upon host gene expression was first discovered more than 50 years ago. However, at that time, TEs were widely regarded as purely genetic parasites and were thought to be selfish elements that were rarely co-opted by the genome for a beneficial purpose (Doolittle and Sapienza 1980; Orgel and Crick 1980). Indeed, the presence of TEs can severely impair genome function, for example by direct disruption of functional genes (Kazazian et al. 1988) or by the promotion of ectopic homologous recombination, which can lead to potentially harmful duplications, deletions, and genome rearrangements (Hedges and Deininger 2007). Conversely, accumulated evidence suggests that some TE activity can be functionally beneficial especially in times of stress (Jordan et al. 2003; Kamal et al. 2006; Lowe et al. 2007b; Naito et al. 2009; Wang et al. 2013a; Bourque et al. 2008). TEs can affect the expression of proximal genes via several mechanisms, such as exposing genes to new promoters and enhancers (Bejerano et al. 2006; Bourque et al. 2008; Jordan et al. 2003; Naito et al. 2009; van de Lagemaat et al. 2003), disruption of original promoter sequences, reduction of transcriptional noise through the spread of epigenetic silencing (Ahmed et al. 2011), and read-through antisense transcription (Kashkush et al. 2003). Here, we provide new evidence that the expression level of genes near to TE loci can be modulated: all the eight TE-proximal genes examined exhibited significant, but not dramatic, alterations in expression level. This is consistent with the previous data indicating that rapid amplification of certain types of TEs has only modest impacts on the expression of host genes (Naito et al. 2009).
Implications of Alien Pollination-Induced TE Mobilization for Plant Genome Evolution and Crop Improvement
The role of hybridization in plant genome evolution is still under debate, with opposing opinions considering hybridization to be evolutionary noise (Gaeta et al. 2007) or, conversely, an engine for generating biodiversity (Arnold 1992). Nevertheless, it is generally accepted that introgressive hybridization provides an efficient means for transfer of important genes from donor to recipient genomes (Gaeta et al. 2007; Leitch and Leitch 2008). However, the ability of the introgressive hybridization process itself to generate de novo genetic variation remains poorly studied (Brennan et al. 2009). In this study, we found that pollination of rice by Zizania induced mobilization of TEs in the absence of introgression. These data provide additional supporting evidence for the earlier proposition that alien pollen may produce a kind of “biological stress” that can induce genome variations (Wang et al. 2009). In particular, we noted that several of the activated TEs inserted preferentially into or close to genic regions, leading to heritable changes in the expression of these genes. In addition, we found that some of the new TE insertion sites were located in QTLs connected with several important agronomic traits, suggesting that newly induced TE transpositions have a role in causing phenotypic novelties. It is conceivable that “accidental” pollinations may occur under natural settings, where alien pollen of related species are sympatric. Therefore, our observations suggest that alien pollination may be one cause of natural genetic variation, particularly when TE mobility is involved. This process could be used to generate genetic diversity in crops, as demonstrated by the increased plant height and enlarged seed size in the X9 line. In particular, we conceive that this manipulation might be combined with the phenomenon of intraspecific crossing-induced TE activities in rice (Kantama et al. 2013), whereby the TE activity might be further enhanced. Further research will be needed to explore the mechanistic basis of this novel phenomenon.
References
Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H (2011) Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res 39:6919–6931
Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325
Arnold ML (1992) Natural hybridization as an evolutionary process. Annu Rev Ecol Syst 23:237–261
Bejerano G, Lowe CB, Ahituv N et al (2006) A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441:87–90
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Bourque G, Leong B, Vega VB et al (2008) Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18:1752–1762
Brennan AC, Bridle JR, Wang AL, Hiscock SJ, Abbott RJ (2009) Adaptation and selection in the Senecio (Asteraceae) hybrid zone on Mount Etna, Sicily. New Phytol 183:702–717
Carr J, Xu M, Dudley JW, Korban SS (2003) AFLP analysis of genetic variability in New Guinea impatiens. Theor Appl Genet 106:1509–1516
Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14:100–112
Chaparro C, Guyot R, Zuccolo A, Piégu B, Panaud O (2007) RetrOryza: a database of the rice LTR-retrotransposons. Nucleic Acids Res 35:D66–D70
Chen S-Q, Zhong W, Liu M-X, Xie Z-W, Wang H-H (2008) Pollen grain germination and pollen tube growth in pistil of rice. Rice Sci 15:125–130
Dong Z, Wang Y, Zhang Z et al (2006) Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor Appl Genet 113:196–205
Doolittle WF, Sapienza C (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603
Dunoyer P, Melnyk C, Molnar A, Slotkin RK (2013) Plant Mobile Small RNAs. Cold Spring Harbor Perspectives in Biology 5
Flavell R (1986) Repetitive DNA and chromosome evolution in plants. Philos Trans R Soc B Biol Sci 312:227–242
Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19:3403–3417
Gao L, Mccarthy EM, Ganko EW, Mcdonald JF (2004) Evolutionary history of Oryza sativa LTR retrotransposons: A preliminary survey of the rice genome sequences. BMC Genomics 5
Guerreiro MPG (2012) What makes transposable elements move in the Drosophila genome. Heredity 108:461–468
Han F, Liu B, Fedak G, Liu Z (2004) Genomic constitution and variation in five partial amphiploids of wheat—Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor Appl Genet 109:1070–1076
Hedges D, Deininger P (2007) Inviting instability: transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res Fundam Mol Mech Mutagen 616:46–59
Jordan IK, Rogozin IB, Glazko GV, Koonin EV (2003) Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19:68–72
Kamal M, Xie X, Lander ES (2006) A large family of ancient repeat elements in the human genome is under strong selection. Proc Natl Acad Sci U S A 103:2740–2745
Kantama L, Junbuathong S, Sakulkoo J, De Jong H, Apisitwanich S (2013) Epigenetic changes and transposon reactivation in Thai rice hybrids. Mol Breeding 31:815–827
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332(10):164–166
Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A 94:7704–7711
Leitch AR, Leitch IJ (2008) Genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Lin X, Long L, Shan X, Zhang S, Shen S and Liu B (2006) In planta mobilization of mPing and its putative autonomous element Pong in rice by hydrostatic pressurization. J Exp Bot 57(10):2313–2323
Liu B, Piao H, Zhao F, Zhao J, Liu Z, Huang B (1999a) DNA methylation changes in rice induced by Zizania latifolia (Griseb.) DNA introgression. Hereditas 131:75–78
Liu B, Piao HM, Zhao FS, Zhao JH, Zhao R (1999b) Production and molecular characterization of rice lines with introgressed traits from a wild species Zizania latifolia (Griseb.). J Genet Breed 53:279–284
Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43:874–880
Liu WM, Chu WM, Choudary PV, Schmid CW (1995) Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res 23:1758–1765
Liu Z, Wang Y, Shen Y, Guo W, Hao S, Liu B (2004) Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia. Plant Mol Biol 54:571–582
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 25:402–408
Lowe CB, Bejerano G, Haussler D (2007a) Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci 104:8005–8010
Lowe CB, Bejerano G, Haussler D (2007b) Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A 104:8005–8010
Mccarthy EM, Liu J, Lizhi G, Mcdonald JF (2002) Long terminal repeat retrotransposons of Oryza sativa. Genome biology 3
Mcclintock B (1983) The significance of responses of the genome to challenge. Physiology Or Medicine Literature Peace Economic Sciences, 180
Muller K, Heller H, Doerfler W (2001) Foreign DNA integration. Genomewide perturbations of methylation and transcription in the recipient genomes. J Biol Chem 276(17):14271–14278
Naito K, Zhang F, Tsukiyama T et al (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461:1130–U232
O’neill RJW, O’neill MJ, Graves JM (1998) Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–72
Orgel LE, Crick FH (1980) Selfish DNA: the ultimate parasite. Nature 284:604–607
Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER (1995) Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A 92:8050–8054
Remus R, Kämmer C, Heller H, Schmitz B, Schell G, Doerfler W (1999) Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J Virol 73:1010–1022
Rieseberg LH, Raymond O, Rosenthal DM et al (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216
Rieseberg LH, Wendel JF (1993) Introgression and its consequences in plants. Hybrid zones and the evolutionary process, 70–109
Sabot F, Picault N, El‐Baidouri M et al (2011) Transpositional landscape of the rice genome revealed by paired‐end mapping of high‐throughput re‐sequencing data. The Plant Journal 66:241–246
Sahu PP, Pandey G, Sharma N, Puranik S, Muthamilarasan M, Prasad M (2013) Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep 32:1151–1159
Shan X, Liu Z, Dong Z et al (2005) Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol Biol Evol 22:976–990
Shapiro JA (2010) Mobile DNA and evolution in the 21st century. Mobile DNA 1
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145
Song Z, Lu B, Zhu Y, Chen J (2002) Pollen competition between cultivated and wild rice species (Oryza sativa and O. rufipogon). New Phytol 153:289–296
Teramoto S, Tsukiyama T, Okumoto Y, Tanisaka T (2014) Early embryogenesis-specificeExpression of the rice transposon Ping enhances amplification of the MITE mPing. Plos Genetics 10
Van De Lagemaat LN, Landry J-R, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19:530–536
Wang H, Chai Y, Chu X et al (2009) Molecular characterization of a rice mutator-phenotype derived from an incompatible cross-pollination reveals transgenerational mobilization of multiple transposable elements and extensive epigenetic instability. BMC Plant Biol 9:63
Wang HY, Tian Q, Ma YQ et al (2010a) Transpositional reactivation of two LTR retrotransposons in rice‐Zizania recombinant inbred lines (RILs). Hereditas 147:264–277
Wang N, Wang H, Wang H et al (2010b) Transpositional reactivation of the Dart transposon family in rice lines derived from introgressive hybridization with Zizania latifolia. BMC Plant Biol 10:190
Wang X, Weigel D, Smith LM (2013a) Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet 9:e1003255
Wang Y-M, Dong Z-Y, Zhang Z-J et al (2005) Extensive de novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.). Genetics 170:1945–1956
Wang Z-H, Zhang D, Bai Y et al (2013b) Genomewide variation in an introgression line of rice-zizania revealed by whole-genome re-sequencing. PLoS One 8:e74479
Wendel JF (2000) Genome evolution in polyploids. In. Plant molecular evolution. Springer, 225–49
Yang X, Yu Y, Jiang L, Lin X, Zhang C, Ou X, Kenji Osabe, Liu B (2012) Changes in DNA methylation and transgenerational mobilization of a transposable element (mPing) by the Topoisomerase II inhibitor, Etoposide, in rice. BMC Plant Biol 12:48
Acknowledgments
This work was supported by the State Key Basic Research and Development Plan of China (2011CB100205, 2013CBA01404), National Natural Science Foundation of China (No. 30970232 & 31170208), Morden Seed Industry Development Special Fund and the Programme for Introducing Talents to Universities (B07017), and Graduate Student Innovation Fund (12SSXT130).
Author Contributions
XYL and YZD conceived and designed the experiments. WY, JTT, SS, WJ, WZH, ZD, LN, XCM, and LXY performed the experiments. BL contributed reagents/materials/analysis tools. WY, SY, and YZD wrote the paper.
Conflict of Interests
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ying Wu, Tingting Jiang, and Yue Sun contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Primers used for locus-specific PCR amplification. (DOC 38 kb)
Table S2
Primers used to generate probes for Southern analysis. (DOC 32 kb)
Table S3
Primers used for qRT-PCR analysis. (DOC 35 kb)
Table S4
Positioning of TE insertion and excision. (DOC 72 kb)
Table S5
Primers used to amplify fragments of SSR and the chromosomal information. (DOC 162 kb)
Table S6
AFLP adapters/primers list. (DOC 45 kb)
ESM 1
(GIF 86 kb)
Rights and permissions
About this article
Cite this article
Wu, Y., Jiang, T., Sun, Y. et al. Mobilization of Diverse Transposable Elements in Rice Induced by Alien Pollination Without Entailing Genetic Introgression. Plant Mol Biol Rep 33, 1181–1191 (2015). https://doi.org/10.1007/s11105-014-0819-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0819-9