Abstract
Aims
Soil phosphorus (P) cycling in karst regions is mainly regulated by microbial activities. Natural restoration has been widely adopted in the degraded karst regions of southwest China. However, the response of functional genes and microbial communities involved in soil P cycling to revegetation has not been well characterized.
Methods
We used metagenomic sequencing to investigate the genes and microorganisms related to soil P cycling derived from natural restoration stages (shrubbery, TG; secondary forest, SG; old-growth forest, OG) in the southeast of Guizhou Province, China.
Results
Natural restoration affected the composition of soil P cycling genes. When TG restored into OG, the relative abundance of organic P (OP) mineralization genes increased from 45.78% to 48.38%, while the genes related to inorganic P (IP) solubilization decreased from 27.19% to 25.03%. The effect of soil nutrients on the relative abundance of OP and IP genes was greater than that of aboveground plant diversity. Structural equation modeling further indicated that soil nutrients directly drove the increase in the relative abundance of OP genes and indirectly impacted the relative abundance of IP genes. The results show that Proteobacteria (38.97%–52.72%) and Actinobacteria (13.44%–29.34%) are the main contributors to soil OP and IP cycling genes, but their contributions vary in different restoration stages.
Conclusions
Natural restoration of the degraded karst vegetation changes the acquisition strategy of soil microbial P by enhancing OP but decreasing IP cycling potentials. This study investigate the regulation of P cycling in the ecological restoration of degraded karst regions from microbial perspective.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
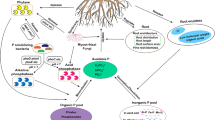
Phosphorus (P) is an essential element for all biotas and participates in many metabolic processes, such as energy transfer, respiration, signal transduction, and macromolecular biosynthesis (Huang et al. 2005). According to data from 41 publications for 258 different soils, P reserves in soils are sufficient to sustain maximum agricultural production worldwide for approximately 350 years (Menezes-Blackburn et al. 2018). Although P is abundant in soils, it exists mainly in unavailable inorganic and organic forms and cannot be used directly by plants and microorganisms. Therefore, the deficiency of bioavailable P is common in terrestrial ecosystems (Elser et al. 2007). Microbes play an integral role in soil P cycling, mediating the bioavailable soil P (Alori et al. 2017; Richardson and Simpson 2011; Rodríguez et al. 2006). Most soil P is sequestered in recalcitrant minerals and organic compounds, and only a small fraction of soil P is utilized by microorganisms and plants in the form of inorganic orthophosphate (H2PO42– or H2PO4–) (Schneider et al. 2019). Therefore, soil microorganisms meet their growth requirements through effective strategies, such as producing organic acids to solubilize inorganic P (IP) and synthesizing hydrolases to mineralize organic P (OP).
The utilization of soil unavailable P by microbes are mainly mediated by several functional gene groups and include OP mineralization, IP solubilization, P uptake and transport, and P-starvation response regulation (Dai et al. 2020). When bioavailable P is deficient, microorganisms upregulate the expression of the genes coding for organic anion production to solubilize IP or enzymes to mineralize OP. Organic anions play their roles via acidification, chelation, and exchange reactions to desorb sparingly available IP forms (Jones 1998; Oburger et al. 2011; Wang et al. 2016), including gluconic acid, acetic acid, fumaric acid, formic acid, and malic acid (Pradhan et al. 2017; Rawat et al. 2021). The gcd genes encoding quinoprotein glucose dehydrogenase (PQQ-GDH) that govern gluconic acid production have been widely studied and reported (Santos-Torres et al. 2021). In addition, the microbial conversion of soil OP compounds into bioaccessible forms is achieved through the action of organophosphorus hydrolases, such as alkaline phosphatases (encoded by genes phoD, phoA, and phoX), acid phosphatases (encoded by gene olpA), phosphodiesterases (encoded by gene ugp), phosphonatases (encoded by gene ptxD), phytases (encoded by gene appA), and C-P lyases (encoded by gene phn) (Pradhan et al. 2017). Among them, alkaline phosphatases (EC: 3.1.3.1) are deemed to be principally derived from soil microbes and are non-specific enzymes that catalyze the hydrolysis of ester-phosphate bonds of many orthophosphate monoesters (Spohn and Kuzyakov 2013; Wasaki et al. 2018; Wei et al. 2019), thereby contributing to P bioavailability in soils. Furthermore, changes in the availability conditions of soil P trigger the expression of microbial functional genes related to the uptake and transport of P and the system of P-starvation response regulation. The genes encoding the system of P uptake and transport allow microbes to assimilate IP under P-low and P-rich conditions through the high-affinity (pst) and low-affinity (pit) IP transporters, respectively (Dai et al. 2020; Rawat et al. 2021). The P-starvation response regulation involves genes phoU, phoR, phoB, etc., that enable soil microorganisms to effectively utilize external P sources during P starvation (Bergkemper et al. 2016; Liang et al. 2020). Recent studies have shown that these genes are associated with P uptake and transport and alkaline phosphatase production, especially under P-low conditions (Dai et al. 2020).
Karst ecosystems account for around 6% of China’s land surface and are widely distributed in the southwest (Jiang et al. 2014). However, they are fragile due to the particularities of soluble carbonate rock, calcium abundance, soil scarcity, water leakage, etc. (Yuan 2001). In the last century, under the pressure of population growth, large areas of forests in karst regions were destroyed by anthropogenic activities such as irrational farming and extensive logging. Consequently, ecological disasters were formed, including soil erosion, vegetation loss, and rocky desertification (Jiang et al. 2014). Since then, the vegetation restoration project named “Grain for Green Project” has been widely implemented in karst regions to overcome ecological degradation (Qi et al. 2013). Many farmlands and plantations have been abandoned and allowed to recover naturally. Previous studies have identified that shrubbery stage, shrubbery-tree stage, secondary forest stage, and the old-growth forest stage are representatives during the natural restoration of the degraded karst vegetation (Yu et al. 2000; Zeng et al. 2007). The restoration process is slow, usually taking more than 50 years to restore shrubbery into an old-growth forest (Yu et al. 2000). During restoration, the changes in the aboveground plant community affect soil properties and microbial communities directly (Mendes et al. 2015; Wang et al. 2019). In turn, soil microbes can also affect plant communities by changing soil nutrient contents (Prober et al. 2015; Zhong et al. 2020). Previous studies have demonstrated the interactions among aboveground plant communities, soil characteristics, and belowground microbial communities during the natural restoration in karst areas. For example, Wang et al. (2022) found that the diversity and composition of soil bacteria and fungi in karst areas changed significantly during secondary succession. Under such conditions, microbial diversity is determined by soil properties, and microbial composition is driven by plant and soil properties. Zhao et al. (2014) revealed declines in soil biota abundances and food web complexity accompanied by the progressive succession of karst vegetation. The declines of the soil microbial and nematode communities may be explained by changes in soil properties (i.e., pH and organic carbon). Numerous studies have revealed that soil microorganisms mediate P cycling through various functions, such as the production of organic anion and enzyme, and the change in microbial communities may affect the transformation and availability of soil P (Huang et al. 2017; Li et al. 2013; Wang et al. 2018). The microbial community composition in different karst restoration stages was previously investigated (Hui et al. 2019; Xue et al. 2017). However, the changes in soil microbial functional genes and microorganisms involved in the transformation and cycling of soil P during the natural restoration of degraded karst vegetation remain unclear.
To investigate the changes in and drivers of soil microbial functional genes involved in P cycling during the natural restoration of degraded karst vegetation, the aboveground plant diversity, soil nutrients, the composition, and the relative abundance of four categories of soil microbial P functional genes in three different restoration stages were measured. It is hypothesized that (i) aboveground plant diversity, soil nutrients, and the relative abundance of the functional genes involved in soil P cycling increase with the restoration process; (ii) the dominant microbial phyla contributing to soil P cycling differ among three restoration stages; (iii) four categories of P cycling genes respond differently to aboveground plant diversity and soil nutrients.
Materials and methods
Site description
The study area is located in Maolan National Nature Reserve in the southeast of Guizhou Province, China (25°09′–25°20′ N, 107°52′–108°05′ E, 550–850 m above sea level). The Reserve has a humid subtropical monsoon climate with an average annual temperature of 15.3℃, an annual relative humidity of 83%, and an annual precipitation of 1,752 mm. The precipitation between April and October accounts for 80% of the annual precipitation. The soil in the studied karst region is dominated by black lime soil, and the vegetation is dominated by subtropical evergreen deciduous broad-leaved mixed forest. The shrubbery (TG), secondary forest (SG), and old-growth forest (OG) stages were selected as the representatives of three typical natural restoration stages in the studied karst region. In the TG stage, the last selective cutting was implemented 11 years ago in 2010, and the propagules were left behind to allow the natural restoration of shrubbery into the old-growth forest. The human interference in the SG stage was the same as the TG stage (selective cutting), and the most recent one happened 37 years ago, in 1984. In the OG stage, the natural recovery lasted more than 100 years, according to historical evidence and inquiries from local elders. Furthermore, they share the same regional species pool during all restoration stages and without human interference during natural restoration. Then, 10 TG plots (10 m × 10 m), 10 SG plots (30 m × 30 m), and 10 OG plots (30 m × 30 m) were randomly selected and constructed according to the standard raised by Condit (1998) in June–August, 2021. The distance between every two adjacent plots is greater than 50 m to eliminate mutual interference.
Plant survey
Each plot was then surveyed for plants following the guidelines proposed by Condit (1998). Plants with a diameter at a breast height greater than 1 cm were investigated and recorded. The plant importance value index (IVI) was calculated based on the following formulas:
Where \({R}_{f}=\frac{{F}_{s}}{{T}_{f}}\), Rf is the relative frequency, Fs is the frequency of the species, Tf is the total frequency of all species; \({D}_{s}=\frac{{N}_{s}}{{A}_{s}}\), Ds is the density of a species, Ns is the number of individuals of that species, As is the area sampled; \({R}_{d}=\frac{{D}_{s}}{{T}_{s}}\), Rd is the relative density, Ts is the total density of all species; \({R}_{do}=\frac{{D}_{os}}{{T}_{d}}\), Rdo is the relative dominance, Dos is the dominance of the species, and Td is the total dominance of all species. The top 10 important plant species in TG, SG, and OG stages are shown in Table S1.
Plant diversity indexes were represented by the total number of species in each plot. Then, the plant Shannon–Wiener index (Shannon), Pielou Evenness index (Pielou), and Rarefied Species Richness index (Rarefied SR) for each plot were calculated based on the abundance information on Plant Species:
Where \({q}_{i}=\frac{(\frac{N-{x}_{i}}{n})}{(\frac{N}{n})}\), S represents the total number of species, pi represents the proportion of the i-th species to the total, xi is the count of species i, \((\frac{N}{n})\) is the binomial coefficient or the number of ways we can choose n from N, qi gives the probability that species i does not occur in a sample of size n. In this study, 20 species of plants were randomly selected from each plot (n = 20), and then Rarefied SR was calculated.
Soil sampling and analyses
Five soil samples (0–20 cm) were taken from each plot and mixed homogenously in early October 2021. Ten soil samples were obtained from each restoration stage, and 30 soil samples were used for chemical and microbial analyses. Each sample was divided into two parts. One was air-dried to determine soil environmental factors, and the other was frozen in liquid N2 immediately for microbial analysis. To determine soil nutrients, soil total carbon (TC), total nitrogen (TN), and total phosphorus (TP) were measured by the dichromate oxidation method (Pribyl 2010), the Kjeldahl method (Bremner and Mulvaney 1982), and the molybdate colorimetry method (Pansu and Gautheyrou 2007), respectively. Soil total calcium (TCa) was measured using an atomic absorption spectrophotometer (ICE 3500, Thermo Scientific, MA, USA) after HNO3-HClO4 digestion (Bao 2007).
DNA extraction, library construction, and metagenomics sequencing
According to the manufacturer’s instruction, the total genomic DNA was extracted from 0.5 g of each soil sample using the FastDNA® Spin Kit for Soil (MP Biomedicals, CA, USA). The concentration and purity of the extracted DNA were determined with a TBS-380 micro-fluorometer (TurnerBioSystems, Sunnyvale, CA, USA) and a NanoDrop 2000 ultra micro-spectrophotometer (Thermo Scientific, Waltham, MA, USA), respectively. Subsequently, the quality of the extracted DNA was checked on 1% agarose gel. The extracted DNA was fragmented to an average size of about 400 bp using Covaris M220 (Gene Company Limited, China) to construct the paired-end library. The paired-end library was constructed using NEXTflex™ Rapid DNA-Seq (Bioo Scientific, Austin, TX, USA). The adapters containing the full complement of sequencing primer hybridization sites were ligated to the blunt end of fragments. Paired-end sequencing was performed on Illumina NovaSeq (Illumina Inc., San Diego, CA, USA) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) using NovaSeq Reagent Kits according to the manufacturer’s instruction (www.illumina.com).
Sequence quality control and genome assembly
A total of 1,813,610,194 raw reads were obtained. The raw reads from metagenome sequencing were used to generate clean reads by removing adaptor sequences and trimming and removing low-quality reads (reads with N bases, with a minimum length threshold of 50 bp and a minimum quality threshold of 20) using the FASTP (Chen et al. 2018) (https://github.com/OpenGene/fastp, version 0.20.0) on the free online Majorbio Cloud Platform (cloud.majorbio.com). After quality control, 1,776,367,626 clean reads were obtained. Then, these high-quality reads were assembled to contigs using MEGAHIT (Li et al. 2015) (parameters: kmer_min = 47, kmer_max = 97, and step = 10) (https://github.com/voutcn/megahit, version 1.1.2), which makes use of succinct de Bruijn graphs. Contigs with a length being or over 300 bp were selected as the final assembling result. These high-quality reads were also submitted to the NCBI database (https://submit.ncbi.nlm.nih.gov/) under the accession number PRJNA951346.
Gene prediction, taxonomy, and functional annotation
MetaGene (Noguchi et al. 2006) (http://metagene.cb.k.u-tokyo.ac.jp/) was used to identify open reading frames (ORFs) in contigs. The predicted ORFs with a length being or over 100 bp were retrieved and translated into amino acid sequences using the NCBI translation table (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG1). We constructed a non-redundant gene catalog using CD-HIT (Fu et al. 2012) (http://www.bioinformatics.org/cd-hit/, version 4.6.1) with 90% sequence identity and 90% coverage. The reads after quality control were mapped to the non-redundant gene catalog with 95% identity using SOAPaligner (Li et al. 2009) (http://soap.genomics.org.cn/, version 2.21), and the gene abundance in each sample was evaluated. The representative sequences of the non-redundant gene catalog were annotated based on the NCBI NR database using BLASTP as implemented in DIAMOND v0.9.19 with an e-value cutoff of 1e–5 using Diamond (Buchfink et al. 2015) (http://www.diamondsearch.org/index.php, version 0.8.35) for taxonomic annotations. Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was conducted using Diamond (http://www.diamondsearch.org/index.php, version 0.8.35) against the KEGG database (http://www.genome.jp/keeg/, version 94.2) with an e-value cutoff of 1e–5. Genome functional analysis emphasized the protein-coding genes involved in soil P cycling. According to previous publications, 119 functional genes with their corresponding KO numbers were selected by searching in the KEGG database and classified into four categories, including i) OP mineralization, ii) IP solubilization, iii) P uptake and transport system, and iv) P starvation response regulation (Table S2). These functional genes were selected from the metagenomes to form a new gene set related to soil P cycling. The function and taxonomy annotation (from kingdom to species) of the selected P cycling genes were also performed according to the assignments against the KEGG and NCBI NR databases using Diamond. The relative contribution of a specific microbial taxon to the P cycling categories, for example, OP mineralization, was calculated by comparing the abundance of this taxon to the total abundance of all taxa involved in soil OP mineralization.
Statistical analysis
Data organizing was performed by Excel 2016. Based on the gene set related to soil P cycling, the similarities and differences in the gene composition for P cycling were explored through the analysis of similarities (ANOSIM) (Clarke 1993) using the “vegan” R package (Oksanen et al. 2018) and presented by nonmetric multidimensional scaling (NMDS) plots using the Bray–Curtis dissimilarity matrix. In addition, in order to compare the composition of the microorganisms involved in soil P cycling between different restoration stages, the taxonomy annotation results (species level) of the soil P cycling gene set were used and analyzed and also presented by NMDS plots as mentioned above. Then, the one-way analysis of variation (ANOVA) and least significant difference (LSD) multiple comparisons (p < 0.05) were conducted to investigate the significant differences in the soil environmental factors, plant diversity indexes, and the genes related to P cycling between the three restoration stages using the SPSS 21.0 statistical software package (IBM, Armonk, NY, USA). The relationships between the relative abundance of genes, including genes involved in soil OP cycling and IP cycling, and soil environmental factors (e.g., soil TC and TN) or plant diversity (e.g., plant Shannon and Rarefied SR) were tested using Spearman’s rank correlations. Structural equation modeling (SEM) was constructed using SPSS Amos version 26.0 (IBM, Armonk, NY, USA) to investigate the direct and indirect effects of soil nutrients and plant diversity on the relative abundance of the shifted P cycling genes. Soil nutrients were composed of soil TC, TN, TP, and TCa, and plant diversity was composed of plant Shannon, Rarefied SR, Pielou, and Species. Suitable fitting of a constructed structural equation model was achieved by maximum likelihood evaluation with a non-significant Chi-square test (Chi-square/df < 3 and p-value > 0.05 for the model) and a comparative fit index (CFI > 0.95).
Results
The composition of genes involved in soil P cycling
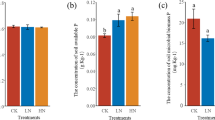
ANOSIM showed significant differences (p < 0.05) in the composition of genes related to soil P cycling between TG, SG, and OG stages. The results indicate that natural restoration can change the composition of microbial genes involved in soil P cycling (NMDS stress, 0.080) (Fig. 1). When the genes were assigned to four P cycling categories, the genes involved in soil OP mineralization had the highest relative abundance in all restoration soils and followed the sequence of OG (48.38%) ≥ SG (47.81%) > TG (45.78%) (there was no significant difference between OG and SG soils) (Fig. 2). The relative abundance of genes responsible for soil IP solubilization was higher in the TG (27.19%) than in the SG (25.47%) and OG (25.03%) soils. The ranges of the relative abundance of genes participating in soil P uptake and transport and P-starvation response regulation were 20.21%–20.81% and 6.22%–6.38%, respectively, and no significant difference was observed between the three restoration soils (Fig. 2).
Changes in the relative abundance of the genes related to soil OP and IP cycling
The changes in the relative abundance of the genes involved in the organic acid formation were consistent with the pattern of the total IP solubilization genes (TG > SG ≥ OG), which contributed the most to the IP solubilization (19.88%–21.84%) (Fig. 3a). Specifically, the relative abundance of acetic acid and fumaric acid genes were higher in the TG soils than in the SG and OG soils (Fig. 3c). However, there was no significant difference in the relative abundance of inorganic pyrophosphatases (1.19%–1.24%) and PQQ-GDH (3.90%–4.17%) between the three restoration soils (Fig. 3a). Six sub-categories were included in OP mineralization (Fig. 3a). The relative abundance of the genes responsible for the synthesis of phosphomonoesterase (18.11%–20.49%), phosphodiesterase (17.86%–18.80%), and organic pyrophosphatase (4.75%–5.33%) contributed top three to OP mineralization, and all of them increased with the restoration process (Fig. 3a). The relative abundance of the genes involved in C-P lyase and phosphonatase were higher in the TG soils than in the SG and OG soils (Fig. 3a). In terms of the genes involved in the production of alkaline phosphatase, the relative abundance of phoD and phoX genes in OG and SG soils was higher than that in TG soil while the relative abundance of phoA genes in OG and SG soils was lower than that in the TG soils (Fig. 3b).
Changes in the relative abundance of the genes involved in the three functional groups of IP cycling and six functional groups of OP cycling (a), formation of alkaline phosphatases (b), and formation of organic acid (c). Data are presented as means ± SD, and different small letters indicate a significant difference (p < 0.05) between three restoration stages based on a one-way ANOVA followed by an LSD test
Taxonomic composition and contributions of genes involved in soil OP and IP cycling
In this study, 82 microbial phyla were assigned to be involved in soil P cycling. For microbes related to soil OP mineralization and IP solubilization, the top 10 identified predominant microbial phyla were the same (Fig. 4). From the TG stage to the OG stage, the relative abundance of Actinobacteria (13.44%–29.34%), Unclassified_d_Bacteria (1.78%–3.22%), Chloroflexi (1.54%–2.07%), Nitrospirae (0.51%–1.79%), and Thaumarchaeota (0.40%–2.05%) increased, whereas those of Proteobacteria (38.97%–52.72%) and Verrucomicrobia (2.62%–5.98%) decreased (Fig. 4). In addition, different microbes contributed differently to the genes involved in soil OP mineralization and IP solubilization. For example, Actinobacteria, Candidatus_Rokubacteria, and Nitrospirae contributed more to the genes involved in soil OP mineralization (1.05%–29.34%) than IP solubilization (0.51%–25.07%), while Proteobacteria, Acidobacteria, and Verrucomicrobia contributed more to the genes involved in soil IP solubilization (3.41%–52.72%) than OP mineralization (2.62%–49.67%).
ANOSIM indicated significant differences (p < 0.05) in the species composition of microbial communities involved in soil P cycling between TG, SG, and OG soils (Fig. S1). Specifically, among the top 20 predominant microbial species involved in soil OP mineralization and IP solubilization, their relative abundances varied significantly, ranging from 0.57%–10.46% (Table S3, Table S4). Among these predominant microbial species, 10–11 (OP mineralization) and 9–12 (IP solubilization) were classified as Proteobacteria. Their relative abundances decreased from 25.44% to 19.51% (OP mineralization) and from 27.07% to 19.73% (IP solubilization), respectively, when the shrubbery is restored into the old-growth forest. In addition, the relative abundance of Actinobacteria among the top 20 predominant microbial species was significantly increased from 8.31% to 13.46% (OP mineralization) and from 5.51% to 12.92% (IP solubilization), respectively. Therefore, the relative abundance of Proteobacteria and Actinobacteria among the top 20 predominant microbial species varied in agreement with the results in Fig. 4.
The relationships between soil nutrients, plant diversity indexes, and the relative abundance of OP and IP genes
Soil nutrients (including soil TC, TN, TP, and TCa) and plant diversity indexes (including Shannon, Rarefied SR, Pielou, and Species indexes) in the three restoration stages are presented in Table 1. The results indicate that plant restoration in the studied karst regions significantly enhanced underground soil nutrients and aboveground plant diversity.
During natural restoration, all four soil nutrient indexes were significantly and positively correlated with the relative abundance of the functional genes responsible for soil OP mineralization (TC, R = 0.543, p < 0.01; TN, R = 0.627, p < 0.01; TP, R = 0.622, p < 0.01; TCa, R = 0.409, p < 0.05) (Fig. 5). Instead, they were all negatively correlated with the relative abundance of IP genes (TC, R = -0.498, p < 0.01; TN, R = -0.570, p < 0.01; TP, R = -0.537, p < 0.01; TCa, R = -0.416, p < 0.05) (Fig. 6). Among the four plant diversity indexes, only Plant Species was significantly correlated with the relative abundance of OP (R = 0.502, p < 0.01) and IP genes (R = -0.508, p < 0.01) (Figs. 5 and 6). Shannon, Rarefied SR, and Pielou indexes were not associated with OP and IP genes.
Relationships between soil TC (a), soil TN (b), soil TP (c), soil TCa (d), plant Shannon (e), plant Rarefied SR (f), plant Pielou (g), plant Species (h) and the relative abundance of OP genes. Black-filled circles are the values of the tested plots. Red solid and dashed lines represent the significant and non-significant results of the correlation analysis, respectively
Relationships between soil TC (a), soil TN (b), soil TP (c), soil TCa (d), plant Shannon (e), plant Rarefied SR (f), plant Pielou (g), plant Species (h) and the relative abundance of IP genes. Black-filled circles are the values of the tested plots. Red solid and dashed lines represent the significant and non-significant results of the correlation analysis, respectively
The driving modes of soil nutrients and plant diversity to the relative abundance of OP and IP genes
The structural equation model (Fig. 7) was constructed based on the results in the TG, SG, and OG stages and with high goodness of fit. The structural equation model showed that the natural restoration of degraded karst vegetation significantly and positively influenced soil nutrients and plant diversity. For the genes responsible for OP mineralization, soil nutrients exhibited a positive and significant effect on their abundance, while plant diversity showed a weak effect. Only OP genes negatively affected the abundance of the genes responsible for IP solubilization, and other variables had no significant effect on them. In addition, a significant tandem pathway was formed by the natural restoration of degraded karst vegetation-soil nutrients-the abundance of OP genes-the abundance of IP genes.
Direct and indirect effects of soil nutrients and plant diversity on the relative abundance of the genes involved in soil OP mineralization and IP solubilization based on the structural equation model. Rectangles represent observed variables, ellipses indicate latent variables, arrows indicate the directions of the effects, and the numbers adjacent to the arrows indicate the effect sizes of the relationships. The widths of the arrows are proportional to the strength of the path coefficients. Continuous and dashed arrows indicate significant (p < 0.05 or p < 0.01) and non-significant effects, respectively. The value of Chi-square/df < 3, CFI > 0.95, and p-value > 0.05 are considered as good goodness of fit
Discussion
The structures of soil microbial communities can be shaped by edaphic properties, such as nutrient content (Bakker et al. 2013; Chong et al. 2010; Chu et al. 2011; Lauber et al. 2009; Schlatter et al. 2015; Yergeau et al. 2007; Yu et al. 2012), and aboveground plant factors, such as vegetation types (Oh et al. 2012; Shi et al. 2015). In this study, plant diversity and soil nutrients increased with restoration time, indicating the positive effect of natural restoration on the restoration of plants and soils in degraded karst regions. The composition of the functional genes of soil P cycling varied across the chronosequence (Fig. 1). Furthermore, no significant difference existed in plant diversity indexes between the SG and OG stages (Table S1). The results indicate that the natural restoration of degraded karst vegetation leads to the saturation of plant diversity at an early stage when the shrubbery (TG) is restored into the secondary forest (SG). This finding was also confirmed by Liu et al. (2021), who found that the restoration of plant diversity was saturated first during the restoration process of aerial seeding in the degraded Mu Us Desert. If plant species characteristics are similar, the increase in species richness means increased diversity of producers with a higher photosynthetic rate per unit area, increased net primary productivity, and more nutrient sources for the soils, such as plant litter and root exudates (Cardinale et al. 2011; Maestre et al. 2012). Therefore, soils can gradually recover, and soil nutrients exhibit a positive linear relationship with plant diversity during the ecological restoration of degraded terrestrial ecosystems (Yan et al. 2020; Zhong et al. 2019). In this study, aboveground plants in three restoration stages were dominated by woody species (Table S1). Thus, increased soil TC, TN, TP, and TCa with natural restoration may be partially attributed to increased plant diversity. This study reveals that natural restoration can restore degraded aboveground plant communities, improve soil nutrients in karst regions, and alter the composition of genes involved in soil P cycling.
Among these four P cycling categories presented in Fig. 2, genes involved in soil OP mineralization had the highest relative abundance in all restoration soils, followed by the genes related to IP solubilization. Similar results were also obtained in grassland, cropland, and tropical forests (Dai et al. 2020; Siles et al. 2022), where the genes responsible for OP mineralization dominate soils. Previous studies have demonstrated that OP mineralization is the main driver of soil P turnover in P-depleted soils (Bergkemper et al. 2016; Dai et al. 2020). In addition, microorganisms require extra energy to obtain P by enhancing the release of phosphatases to mineralize soil OP under P-low conditions (Dai et al. 2020). Soil functional genes related to the system of P uptake and transport and P-starvation response regulation are mainly controlled by soil P supply (Rawat et al. 2021; Santos-Torres et al. 2021). In this study, the relative abundance of the genes related to the two P cycling categories did not differ significantly in the TG, SG, and OG soils (Fig. 2). The reason may be that the three restoration soils in this study all suffered the deficiency of NaHCO3-extractable P (8.91–16.93 mg/kg, unpublished data), which has been confirmed in numerous previous studies (Hui et al. 2019; Ma et al. 2019; Shen et al. 2020).
The deficiency of bioavailable soil P has greatly negatively impacted agricultural production and ecological restoration in the karst regions of southwest China (Zhang et al. 2021; Qian et al. 2022). Especially, the available plant P mainly comes from the microbial transformation in natural revegetation without artificial fertilizer application. In this study, the relative abundance of the genes involved in soil OP mineralization increased from the TG stage to the OG stage (Fig. 2), indicating that the natural restoration of degraded karst vegetation promoted the microbial potential for the transformation of soil OP compounds. Further correlation analysis revealed that the changes in soil OP cycling genes were more affected by soil nutrients than plant diversity (Fig. 5). The structural equation model also indicated that the driving path of soil nutrients to the abundance of OP genes was significant, but that of plant diversity to the abundance of OP genes was not (Fig. 7). These results could be explained by the increased soil organic matter (Table 1), which made the OP the dominant P source in soils during natural restoration. In such cases, soil microorganisms have to produce more phosphatases (i.e., phosphomonoesterase and phosphodiesterase) (Fig. 3a) at the expense of IP solubilizing agents (Ragot et al. 2015, 2017; Rodríguez et al. 2006). Therefore, soil nutrients can directly affect the microbial potential in the mineralization of OP during natural restoration from shrubbery to old-growth forests in karst regions. In alkaline soils, alkaline phosphatases are monomeric enzymes contributing greatly to soil OP mineralization. Soil alkaline phosphatases are mainly derived from the microorganism and are encoded by phoD, phoX, and phoA genes (Lu et al. 2022). In this study, among the three phosphatase-encoding genes, phoD had the highest relative abundance in all studied soils (Fig. 3b). Previous studies also have reported that phoD is the most widespread gene in soils and has become the reference marker in studies on soil P cycling (Ragot et al. 2017). In this study, the relative abundance of the genes coding for alkaline phosphatases and other OP-mineralizing enzymes increases with the restoration process, indicating that the natural restoration of degraded karst vegetation promotes microbial functions in soil OP mineralization.
According to our above-mentioned discussion, the increased soil organic matter can result in OP becoming the dominant soil P source. Therefore, soil microorganisms have to produce more phosphatases at the expense of IP cycling genes (Fig. 2). This postulate is reasonable because both soil nutrients and the relative abundance of IP genes were negatively correlated (Fig. 6). The relative abundance of IP genes was directly negatively affected by OP gene abundance instead of soil nutrients or plant diversity (Fig. 7). Therefore, in karst areas, changes in soil OP and IP cycling genes were mediated by soil nutrients, especially soil organic matter content. Thus, the natural restoration of degraded karst vegetation changed the acquisition strategy of soil microbial P by enhancing OP mineralization but decreasing IP solubilization potentials.
In this study, 82 microbial phyla are involved in soil P cycling, which may imply the widespread existence of microbes participating in soil P cycling. The dominant microbial phyla (i.e., Proteobacteria, Actinobacteria, and Acidobacteria) related to soil OP and IP cycling did not change in all studied soils (Figs. 4, S3, and S4). The results indicate that microorganisms from these microbial phyla have a strong competitive ability and can adapt to various environmental conditions. In addition, another reasonable explanation for the lack of difference in phylum level is that phylum level is so broad that most variation happens at finer taxonomic levels below phylum may not be recognized. These results are similar to previous findings (Zhong et al. 2020; Zhou et al. 2020). However, the community composition of the dominant microbial phyla is greatly affected by the natural restoration of degraded karst vegetation. For example, Proteobacteria was the most abundant bacterial phyla in all studied soils, and its relative abundance decreased from 49.67%–52.72% in TG soils to 38.97%–40.95% in OG soils. Previous studies have revealed that most IP-solubilizing or OP-mineralizing bacteria belong to Gammaproteobacteria or Alphaproteobacteria (Bhattacharyya and Jha 2012; Prosser 2007), and their abundances are reduced with the increasing soil N (Dai et al. 2020). Therefore, the decrease in the relative abundance of Proteobacteria related to P cycling in the SG and OG soils can be explained by the increased soil TN. However, Actinobacteria, which belong to copiotrophic microorganisms (Fierer et al. 2007; Dai et al. 2018), mainly adopt the R selection strategy to drive the rapid response to resource availability and are easy to multiply in soils with high organic C and N (Fierer et al. 2007; Yao et al. 2017). In this study, the relative abundance of Actinobacteria involved in soil P cycling increased with the improvement of soil nutrient conditions. Actinobacteria, Candidatus_Rokubacteria, and Nitrospirae contributed more to OP mineralization genes than to IP solubilization genes, while the performance of Proteobacteria, Acidobacteria, and Verrucomicrobia is opposite (Fig. 4). These results indicated that different microbes have different contributions to the genes involved in soil OP and IP cyclings, implying that diverse microbes may have different functional tendencies in soil P cycling (Li et al. 2018). For example, Actinobacteria play more important roles in OP mineralization than IP solubilization in degraded karst soils, but Proteobacteria prefer to participate in IP solubilization rather than OP mineralization.
Conclusions
The natural restoration of degraded karst vegetation significantly affects the composition of the functional genes and microbial communities involved in soil P cycling. The plant diversity, soil nutrients, and the relative abundance of the genes related to soil OP mineralization increase, whereas the relative abundance of the genes related to soil IP solubilization decreases when the shrubbery is restored into the old-growth forest. The increased soil nutrients rather than plant diversity can mediate the increase of OP gene abundance and the decrease of IP gene abundance. Proteobacteria and Actinobacteria are the main contributors to the genes related to soil OP and IP transformations in all studied soils, and their relative contributions varied with vegetation restoration. In conclusion, the natural restoration of degraded karst vegetation can increase soil nutrients and further change the acquisition strategy of soil microbial P by enhancing OP mineralization and decreasing IP solubilization potentials. These findings provide new insights into the genetic level of soil P cycling and improve understanding of microbial mechanisms underlying soil P cycling in degraded karst regions. Future studies should focus on comparing the composition, diversity, and microbial contribution of the P functional genes in degraded karst soils and normal karst soils to promote the restoration of the degraded karst ecosystem.
Data Availability
The data that support the findings of this study are available from the corresponding author, DM Chen, upon reasonable request.
References
Alori ET, Glick BR, Babalola OO (2017) Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Icrobiol 8:971
Bakker MG, Otto-Hanson L, Lange AJ, Bradeen JM, Kinkel LL (2013) Plant monocultures produce more antagonistic soil Streptomyces communities than high-diversity plant communities. Soil Biol Biochem 65:304–312
Bao S (2007) In: Soil and Agro-Chemistrical Analysis, 3rd edn. Agriculture Press of China, Beijing
Bergkemper F, Schöler A, Engel M, Lang F, Krüger J, Schloter M, Schulz S (2016) Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ Microbiol 18:1988–2000
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microb Biot 28:1327–1350
Bremner J, Mulvaney C (1982) Nitrogen-total. In: A.L., Miller, R.H., Keeney, D.R. (Eds.), Methods of Soil Analysis. American Society of Agronomy and Soil Science Society of America, Madison, pp. 595−641
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L, Balvanera P, O’Connor MI, Gonzalez A (2011) The functional role of producer diversity in ecosystems. Am J Bot 98:572–592
Chen SF, Zhou YQ, Chen YR, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890
Chong CW, Pearce DA, Convey P, Tan GA, Wong RC, Tan IK (2010) High levels of spatial heterogeneity in the biodiversity of soil prokaryotes on Signy Island, Antarctica. Soil Biol Biochem 42:601–610
Chu HY, Neufeld JD, Walker VK, Grogan P (2011) The influence of vegetation type on the dominant soil bacteria, archaea, and fungi in a low Arctic tundra landscape. Soil Sci Soc Am J 75:1756–1765
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Condit R (1998) Tropical forest census plots: methods and results from Barro Colorado Island, Panama and a comparison with other plots. Springer Science & Business Media, New York
Dai ZM, Su WQ, Chen HH, Barberan A, Zhao HC, Yu MJ, Yu L, Brookes PC, Schadt CW, Chang SX, Xu JM (2018) Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agroecosystems across the globe. Glob Chang Biol 24:3452–3461
Dai ZM, Liu GF, Chen HH, Chen CR, Wang JK, Ai SY, Wei D, Li DM, Ma B, Tang CX, Brookes PC, Xu JM (2020) Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J 14:757–770
Elser JJ, Bracken ME, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE (2007) Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152
Huang JL, Su ZC, Xu Y (2005) The evolution of microbial phosphonate degradative pathways. J Mol Evol 61:682–690
Huang LM, Jia XX, Zhang GL, Shao MA (2017) Soil organic phosphorus transformation during ecosystem development: A review. Plant Soil 417:17–42
Hui N, Sun NX, Du HM, Umair M, Kang HZ, Liu XX, Romantschuk M, Liu CJ (2019) Karst rocky desertification does not erode ectomycorrhizal fungal diversity, but alters microbial community structure. Plant Soil 445:383–396
Jiang ZC, Lian YQ, Qin XQ (2014) Rocky desertification in Southwest China: Impacts, causes, and restoration. Earth-Sci Rev 132:1–12
Jones DL (1998) Organic acids in the rhizosphere-a critical review. Plant Soil 205:25–44
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microb 75:5111–5120
Li RQ, Yu C, Li YR, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li M, Zhang J, Wang G, Yang H, Whelan MJ, White SM (2013) Organic phosphorus fractionation in wetland soil profiles by chemical extraction and phosphorus-31 nuclear magnetic resonance spectroscopy. Appl Geochem 33:213–221
Li DH, Liu CM, Luo RB, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676
Li M, Cozzolino V, Mazzei P, Drosos M, Monda H, Hu ZY, Piccolo A (2018) Effects of microbial bioeffectors and P amendements on P forms in a maize cropped soil as evaluated by P-31-NMR spectroscopy. Plant Soil 427:87–104
Liang JL, Liu J, Jia P, Yang TT, Zeng QW, Zhang SC, Liao B, Shu WS, Li JT (2020) Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. ISME J 14:1600–1613
Liu QF, Zhang Q, Jarvie S, Yan YZ, Han P, Liu T, Guo K, Ren LJ, Yue K, Wu HM, Du JJ, Niu JM, Svenning JC (2021) Ecosystem restoration through aerial seeding: Interacting plant-soil microbiome effects on soil multifunctionality. Land Degrad Dev 2021:1–14
Lu JL, Jia P, Feng SW, Wang YT, Zheng J, Ou SN, Wu ZH, Liao B, Shu WS, Liang JL, Li JT (2022) Remarkable effects of microbial factors on soil phosphorus bioavailability: A country-scale study. Global Change Biol 28:4459–4471
Ma B, Zhou XL, Zhang Q, Qin MS, Hu LG, Yang KN, Xie Z, Ma WB, Chen BB, Feng HY, Liu YJ, Du GZ, Ma XJ, Le Roux X (2019) How do soil microorganisms respond to N, P and NP additions? Application of the ecological framework of (co-) limitation by multiple resources. J Ecol 107:2329–2345
Maestre FT, Quero JL, Gotelli NJ, Escudero A, Ochoa V, DelgadoBaquerizo M, Garcia-Gómez M, Bowker MA, Saliveres S, Escolar C, Garcia-Palacios P, Berdugo M, Valencia E, Gozalo B, Gallardio A, Aguilera L, Arredondo T, Blones J, Boeken B, Bran D, Conceicão AA, Cabrera O, Chaieb M, Derak MH, Eldridge DJ, Espinosa CL, Florentino A, Gaitán J, Gatica MG, Ghiloufi W, Gómez-González S, Gutiérrez JR, Hernández RM, Huang X, Huber-Sannwald E, Jankju M, Miriti M, Monerris J, Mau RL, Morici E, Naseri K, Ospina A, Polo V, Prina A, Pucheta E, Ramirez-Collantes DA, Romão R, Tighe M, Tores-Diaz C, Val J, Veiga JP, Wang D, Zaady E (2012) Plant species richness and ecosystem multifunctionaity in global drylands. Science 35:214–218
Mendes LW, Brossi MJD, Kuramae EE, Tsai SM (2015) Land-use system shapes soil bacterial communities in southeastern Amazon region. Appl Soil Ecol 95:151–160
Menezes-Blackburn D, Giles C, Darch T, George TS, Blackwell M, Stutter M, Shand C, Lumsdon D, Cooper P, Wendler R, Brown L, Almeida D, Wearing C, Zhang H, Haygarth PM (2018) Opportunities for mobilizing recalcitrant phosphorus from agricultural soils: a review. Plant Soil 427:5–16
Noguchi H, Park J, Takagi T (2006) MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucl Acids Res 34:5623–5630
Oburger E, Jones DL, Wenzel WW (2011) Phosphorus saturation and pH differentially regulate the efficiency of organic acid anion-mediated P solubilization mechanisms in soil. Plant Soil 341:363–382
Oh YM, Kim M, Lee-Cruz L, Lai-Hoe A, Go R, Ainuddin N, Rahim RA, Shukor N, Adams JM (2012) Distinctive Bacterial Communities in the Rhizoplane of Four Tropical Tree Species. Microb Ecol 64:1018–1027
Oksanen J, Blanchet FG, Kindt R, Legendre P, Mc Glinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2018) Vegan v2.6: community ecology package. 2019–09–01. https://CRAN.R-project.org/package=vegan. Accessed 8 Dec 2022
Pansu M, Gautheyrou J (2007) Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods. Springer Science & Business Media, Heidelberg, Germany
Pradhan A, Pahari A, Mohapatra S, Mishra BB (2017) Phosphate-solubilizing microorganisms in sustainable agriculture: genetic mechanism and application. Advances in soil microbiology: recent trends and future prospects. Springer, Singapore, pp 81–97
Pribyl DW (2010) A critical review of the conventional SOC to SOM conversion factor. Geoderma 156:75–83
Prober SM, Leff JW, Bates ST, Borer ET, Firn J, Harpole WS, Lind EM, Seabloom EW, Adler PB, Bakker JD (2015) Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol Lett 18:85–95
Prosser JI (2007) Microorganisms cycling soil nutrients and their diversity. In: van Elsas JD, Jansson JD, Trevors JT (eds) Modern Soil Microbiology, 2nd edn. CRC Press, New York
Qi XK, Wang KL, Zhang CH (2013) Effectiveness of ecological restoration projects in a karst region of southwest China assessed using vegetation succession mapping. Ecol Eng 54:245–253
Qian Q, Zhang RY, Liu KP, Liang YM, Zhang W, Pan FJ (2022) Soil phosphorus characteristics of different land use types and ecological restoration modes in karst region. Chin J Ecol 41:2128–2136
Ragot SA, Kertesz MA, Bünemann EK (2015) PhoD alkaline phosphatase gene diversity in soil. Appl Environ Microbiol 81:7281–7289
Ragot SA, Kertesz MA, Mészáros É, Frossard E, Bünemann EK (2017) Soil phoD and phoX alkaline phosphatase gene diversity responds to multiple environmental factors. FEMS Microb Ecol 93:fiw212
Rawat P, Das S, Shankhdhar D, Shankhdhar SC (2021) Phosphate-Solubilizing Microorganisms: Mechanism and Their Role in Phosphate Solubilization and Uptake. J Soil Sci Plant Nut 21:49–68
Richardson AE, Simpson RJ (2011) Soil microorganisms mediating phosphorus availability. Plant Physiol 156:989–996
Rodríguez H, Fraga R, Gonzalez T, Bashan Y (2006) Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287:15–21
Santos-Torres M, Romero-Perdomo F, Mendoza-Labrador J, Gutiérrez AY, Vargas C, Castro-Rincon E, Caro-Quintero A, Uribe-Velez D, Estrada-Bonilla GA (2021) Genomic and phenotypic analysis of rock phosphate-solubilizing rhizobacteria. Rhizosphere 17:100290
Schlatter DC, Bakker MG, Bradeen JM, Kinkel LL (2015) Plant community richness and microbial interactions structure bacterial communities in soil. Ecology 96:134–142
Schneider KD, Thiessen Martens JR, Zvomuya F, Keith Reid D, Fraser TD, Lynch KD, O’Halloran LP, Wilson HF (2019) Options for improved phosphorus cycling and use in agriculture at the field and regional scales. J Environ Qua 48:1247–1264
Shen YX, Yu Y, Lucas-Borja ME, Chen FJ, Chen QQ, Tang YY (2020) Change of soil K, N and P following forest restoration in rock outcrop rich karst area. Catena 186:104395
Shi Y, Xiang X, Shen C, Chu H, Neufeld JD, Walker VK, Grogan P (2015) Vegetation-associated impacts on arctic tundra bacterial and microeukaryotic communities. Appl Environ Microb 81:492–501
Siles JA, Starke R, Martinovic T, Parente Fernandes ML, Orgiazzi A, Bastida F (2022) Distribution of phosphorus cycling genes across land uses and microbial taxonomic groups based on metagenome and genome mining. Soil Biol Biochem 174:108826
Spohn M, Kuzyakov Y (2013) Distribution of microbial- and root-derived phosphatase activities in the rhizosphere depending on P availability and C allocation-Coupling soil zymography with 14C imaging. Soil Biol Biochem 67:106–113
Wang YZ, Whalen JK, Chen X, Cao YH, Huang B, Lu CY, Shi Y (2016) Mechanisms for altering phosphorus sorption characteristics induced by low-molecular-weight organic acids. Can J Soil Sci 96:289–298
Wang H, Teng CY, Li HY, Sun XZ, Jiang CL, Lou LP, Yue CL, Zhang ZJ (2018) Microbial community shifts trigger loss of orthophosphate in wetland soils subjected to experimental warming. Plant Soil 424:351–365
Wang KB, Zhang YW, Tang ZS, Shangguan ZP, Chang F, Jia FA, Chen YP, He XH, Shi WY, Deng L (2019) Effects of grassland afforestation on structure and function of soil bacterial and fungal communities. Sci Total Environ 676:396–406
Wang GZ, Liu YG, Cui M, Zhou ZY, Zhang Q, Li YJ, Ha WX, Pang DB, Luo JF, Zhou JX (2022) Effects of secondary succession on soil fungal and bacterial compositions and diversities in a karst area. Plant Soil 475:91–102
Wasaki J, Sakaguchi J, Yamamura T, Ito S, Shinano T, Osaki M, Kandeler E (2018) P and N deficiency change the relative abundance and function of rhizosphere microorganisms during cluster root development of white lupin (Lupinus albus L.). Soil Sci Plant Nutr 64:686–696
Wei XM, Hu YJ, Razavi BS, Zhou J, Shen JL, Nannipieri P, Wu JS, Ge TD (2019) Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol Biochem 131:62–70
Xue L, Ren HD, Li S, Leng XH, Yao XH (2017) Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front Microbiol 8:2377
Yan DF, Bissett A, Gellie N, Mills JG, Lowe AJ, Breed MF (2020) Soil bacterial community differences along a coastal restoration chronosequence. Plant Ecol 221:795–811
Yao F, Yang S, Wang ZR, Wang X, Ye J, Wang XG, DeBruyn JM, Feng X, Jiang Y, Li H (2017) Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Front Microbiol 8:2071
Yergeau E, Bokhorst S, Huiskes AH, Boschker HT, Aerts R, Kowalchuk GA (2007) Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microb Ecol 59:436–451
Yu LF, Zhu SQ, Ye JZ (2000) A study on evaluation of natural restoration for degraded karst forest. Scientia Silvae Sinicae 36:12–19
Yu Y, Wang H, Liu J, Wang Q, Shen TL, Guo WH, Wang RQ (2012) Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River Estuary. Eur J Soil Biol 49:12–21
Yuan D (2001) On the karst ecosystem. Acta Geologica Sinica-English Edition 75:336–338
Zeng FP, Peng WX, Song TQ, Wang KL, Wu HY, Song XJ, Xeng ZX (2007) Changes in vegetation after 22 years’ natural restoration in the karst disturbed area in Northwest Guangxi. Acta Ecol Sin 27:5110–5118
Zhang J, Zheng MH, Zhang YJ, Wang J, Shen H, Lin YB, Tang XL, Hui DF, Lambers H, Sardans Jm Peñuelas J (2021) Soil phosphorus availability affects diazotroph communities during vegetation succession in lowland subtropical forests. Appl Soil Ecol 166:104009
Zhao J, Li SP, He XY, Liu L, Wang KL (2014) The soil biota composition along a progressive Succession of secondary vegetation in a karst area. PLoS ONE 9:e112436
Zhong ZK, Wang X, Zhang XY, Zhang W, Xu YD, Ren CJ, Yang GH (2019) Edaphic factors but not plant characteristics mainly alter soil microbial properties along a restoration chronosequence of Pinus tabulaeformis stands on Mt. Ziwuling, China. Forest Ecol Manag 453:117625
Zhong ZK, Zhang XY, Wang X, Fu SY, Wu SJ, Lu XQ, Ren CJ, Han XH, Yang GH (2020) Soil bacteria and fungi respond differently to plant diversity and plant family composition during the secondary succession of abandoned farmland on the loess plateau, China. Plant Soil 448:183–200
Zhou ZY, Yu MH, Ding GD, Gao GL, He YY, Wang GZ (2020) Effects of Hedysarum leguminous plants on soil bacterial communities in the Mu Us Desert, Northwest China. Ecol Evol 10:11423–11439
Funding
This research was supported by the Basic Research Program in Guizhou Province under grant number (2022)036, the Gui Da Te Gang He Zi Program under grant number (2021)06, the Cultivation Project of Guizhou University under grant number (2020)04, and the Joint Fund of the Natural Science Foundation of China and the Karst Science Research Center of Guizhou Province under grant number U1812401.
Author information
Authors and Affiliations
Contributions
Yu Dai: Conceptualization, Methodology, Writing-Original Draft, Visualization. Danmei Chen: Writing -Review & Editing, Funding acquisition, Supervision, Project administration, Methodology. Lipeng Zang: Project administration, Supervision. Guangqi Zhang: Project administration, Supervision. Qingfu Liu: Project administration, Supervision. Yuejun He: Supervision. Fangjun Ding: Resources. Shasha Wang: Investigation. Chunjie Zhou: Investigation. Yousu Yang: Investigation. Yujuan Li: Investigation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that we have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Responsible Editor: Katharina Maria Keiblinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dai, Y., Chen, D., Zang, L. et al. Natural restoration of degraded karst vegetation shifts soil microbial phosphorus acquisition strategies. Plant Soil 490, 201–215 (2023). https://doi.org/10.1007/s11104-023-06067-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06067-7