Abstract
Aims
In the production of the natural medicinal plant American ginseng, replantation typically fails due to continuous cropping obstacles. However, the cause is still not clear and needs more research.
Methods
Soil samples were collected from (a) maize fields where American ginseng had never been planted, (b) fields where American ginseng had just been harvested, and (c) fields where maize had been planted for 2, 4 and 6 years respectively after American ginseng. We investigated the physicochemical properties, the enzymatic activities, and the soil microbial community structure and composition of the samples.
Results
We found that the content of soil salt, NH4+-N, and NO3−-N increased significantly in samples associated with the production of American ginseng, whereas the soil pH, carbon-to-nitrogen ratio, alkaline phosphatase, and cellulase activity all significantly decreased and gradually recovered to the pre-planting level. Moreover, the bacterial diversity decreased, while fungal diversity and richness increased; fungal richness continued to increase in farmlands replanted maize. The relative abundance of some microbial communities was changed significantly and was gradually restored with a longer time to replant maize. Pearson’s correlation analysis shown that significantly changed microbial communities were significantly associated with changes in soil pH, soil salt and nitrogen content, alkaline phosphatase, and cellulase activity.
Conclusions
Changes in soil pH, soil salt and nitrogen content caused changes in microbial community structure and composition, as well as cellulase and alkaline phosphatase activity. These changes may cause the continuous cropping obstacles of American ginseng and may be improved by planting maize.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
American ginseng (Panax quinquefolius L.) is a remarkable natural herbal medicine. It is believed to possess eutherapeutic effects that enhance central nervous system function, protect the cardiovascular system, and improve immunity, due to its anti-inflammatory and antioxidant properties (Kitts et al. 2000; Wang et al. 2015a, b; Wu et al. 2016; Yu et al. 2014). The plant is also employed in the treatment of diabetes (Sen et al. 2012). As an important herbal medicine, the global American ginseng market reached US$ 85 million in 2018, and it is expected that the market will reach 7,033 tons in 2019~2024 (Zhang 2019). China is the third largest production country for American ginseng and produced 3,600 tons of American ginseng in 2016 (Zhang et al. 2020). However, an important issue in its production is continuous cropping obstacle (He et al. 2009). This usually causes the seedling survival rate to drop below 30 % in continuous cropping (Jian et al. 2008). Therefore, its planting mode is typically cultivated continuously in 4-year cycles and improved the soil condition by planting maize after harvesting American ginseng in China.
Continuous cropping obstacle is generally believed to be caused by numerous biotic and abiotic factors, such as the deterioration of soil physicochemical properties (Kaur and Singh 2014; Reeves 1997; Zydlik and Zydlik 2013), autotoxicity of plants (Van Wyk et al. 2017; Zhang et al. 2007; Zhang and Lin 2009), and changes in the diversity and composition of microbial communities (Buyer et al. 2010; Dong et al. 2017; Ma et al. 2010). Soil microbial communities typically play a significant role in agricultural production, as they maintain soil health and quality (Jangid et al. 2008; Van Wyk et al. 2017). Considerable differences can exist in the abiotic characteristics, microbial abundances, rates of microbial activity, and microbial community composition within even extremely close soil environments that share the same geographies (Fierer 2017). Some studies have reported that the continuous cropping obstacle of multiple crops is critically influenced by changes in soil microbial community structure and composition (Gao et al. 2019; Tan et al. 2017). Furthermore, multiple studies have reported that the modification of resident soil microbial flora is related to changes in soil physicochemical properties, soil enzymatic activities (Becker et al. 2017; Schnecker et al. 2015; Zheng et al. 2019), planted species, and the type of soil environment (Berg and Smalla 2009; Dong et al. 2017).
The physicochemical properties of soil are essential factors that influence the composition of soil microbial communities. For example, soil pH has a greater influence on the composition of bacterial and archaeal communities than fungi (Blagodatskaya and Anderson 1998; Lauber et al. 2009; Zhalnina et al. 2015). Some studies have reported that changes in soil pH greatly impact the soil microbial community structure and contribute to continuous cropping obstacles (Gao et al. 2019; Tan et al. 2017). In addition, many other factors can directly or indirectly influence the spatial structure of soil microbial communities, such as soil moisture (Brockett et al. 2012; Drenovsky et al. 2004), nitrogen and phosphorus content (Bowles et al. 2014; Cederlund et al. 2014; Fierer 2017). Nitrogen is an important component of soil quality and is a necessary nutrient for plant growth and development (Jia et al. 2008; Zhang et al. 2015). Nitrogen content is also a critical factor that affects the composition and diversity of microbial communities (Koyama et al. 2014; Pan et al. 2014; Zheng et al. 2019). It is essential for the growth and metabolism of microbes (Wang et al. 2017) and serves as an indispensable energy and nutrient source for microorganisms. Nitrogen is taken up by most plant species in the form of nitrate (NO3−) or ammonium (NH4+) (Cramer and Lewis 1993; Horchani et al. 2010). As the oxidation of NH4+ to NO3− progresses and releases H+, an increase in soil nitrate levels is typically accompanied by lower soil pH (Fageria and Baligar 2005). In most cases, changes in these soil physicochemical properties can also directly or indirectly influence the growth and development of plants and overall crop production. Furthermore, the salt content of soil is an essential factor that impacts the health of soil and plants, as well as the composition of microbial communities (Daliakopoulos et al. 2016; Shrivastava and Kumar 2015). A number of soil physicochemical processes and enzymatic activities might also be affected by the soil salt content (Pathak and Rao 1998; Wichern et al. 2006).
Numerous studies have investigated how soil physicochemical properties and microbial community composition influence enzyme activities pertaining to the regulation of extracellular enzyme levels. Fungi are thought to have a greater influence than bacteria on enzyme activities associated with the decomposition of organic matter, as well as in the generation of products that can provide labile resources for bacteria (Bowles et al. 2014; Schnecker et al. 2015). Extracellular enzymatic activity is a good indicator of carbon decomposition in soil (Sinsabaugh et al. 2005, 2014) and is thought to be closely related to C and N processes (Banerjee et al. 2016; Burns et al. 2013; Henry 2013). For example, soil microbes generate cellulases, a group of hydrolytic enzymes that decompose polysaccharides (Deng and Tabatabai 1994). Enzymes associated with microbial N-acquisition include b-1,4-N-acetyl-glucosaminidase, leucine amino peptidase, and urease (Eivazi and Tabatabai 1977; Hui et al. 2013; Tabatabai and Bremner 1972) and associated with P-acquisition include acidic phosphatase, neutral phosphatase, and alkaline phosphatase (Hui et al. 2013). Previous studies have reported that the activity of alkaline phosphatase was related to continuous cropping obstacles (Meng et al. 2012; Wang et al. 2012).
Previous studies of continuous cropping obstacles of American Ginseng revealed that changes in the soil microbial communities during cultivation resulted in imbalanced microbial communities and a reduction in metabolic functionality (Qi et al. 2010; Ying et al. 2012). There is an obvious phenomenon of decreased bacterial diversity and increased fungal diversity in the soils of American ginseng cultivated in continuous crop versus traditional crop systems (Dong et al. 2017). However, the reasons for the changes in the structure and composition of the microbial community during continuous cropping of American Ginseng are not clear, and the theoretical basis for planting maize after American Ginseng can effectively improve this problem remains unclear. For this study, the physicochemical properties and enzymatic activities were compared among soils from (a) maize fields where American ginseng had never been planted, (b) fields where American ginseng had just been harvested, and (c) fields where maize had been planted for 2, 4 and 6 years respectively after American ginseng. The diversity and composition of bacterial and fungal communities were compared using high-throughput sequencing. The microecological causes of continuous cropping obstacles of American Ginseng were analyzed to provide scientific recommendations to improve the sustainable development of the medicinal plant industry. We hypothesized that (1) the planting of American ginseng changes some key soil physicochemical properties, which makes farmland unsuitable for the growth of American ginseng; (2) these changes influence related soil enzyme activities and microbial community structure and composition and leads to continuous cropping obstacles of American Ginseng; and (3) these changes are gradually restored to baseline conditions as the replanting maize time length increases after harvesting American Ginseng.
Materials and methods
Study sites and soil sample collection
Soil samples were collected in October 2015 in Liuba County, Shaanxi Province (106°54'32.50"E-106°54'41.32"E, 33°36'59.28"N-33°37'4.44"N), which is one of the main areas in China for American ginseng cultivation. Soil samples were randomly collected from (a) maize fields where American ginseng had never been planted (group A), (b) fields where American ginseng had just been harvested (group B), and (c) fields where maize had been planted for 2-years (group C), 4-years (group D), and 6-years (group E) respectively after American ginseng. In the process of experiment, organic fertilizer (2.0 kg·m− 2) was applied as base fertilizer in October, and added 1.0 kg·m− 2 in May of each year. In each field, samples were collected via a five-point sampling method to obtain representative samples. For each group, five soil samples were collected and combined to form a single sample, which was repeated three times, resulting in a total of 15 samples. The samples were aseptically extracted from the soil surface growth layer (0~20 cm depth), homogenized by sieving to 2 mm into a 50 ml centrifuge tube, and then immediately stored at 4 °C for further analysis, as well as at -80 °C, for sequencing.
Determination of soil physicochemical properties
An analysis of the physicochemical properties of the soil sample was performed at the Key Laboratory of Ministry of Education for Medicinal Resources and Natural Pharmaceutical Chemistry (NEL-ECCD) in Xi’an, China. The soil moisture (SM) was determined using a Sartorius MA 100 moisture test apparatus. Soils were air-dried to analyze the soil electric conductivity (SE), the soil salt content (SS), total soil organic carbon (SOC), total nitrogen (TN), total phosphorus, and available phosphorus (AP). Fresh soil samples were used to determine the soil pH and the content of soil ammonium nitrogen (NH4+-N, AN) and nitrate nitrogen (NO3−-N, NN).
Following the homogenization of soil samples by sifting through a 1 mm sieve, the pH was measured via potentiometry in deionized water (soil: water = 1:5 (w:v)) using a pH meter. The SE was determined using a conductivity meter. According to the standard curve between the conductivity and concentration of the potassium chloride solution at room temperature, the following formula, C (g/L) = (S + 41.2653)/2120.76, was used to calculate the SS based on soil electric conductivity. The SOC was determined by the K2Cr2O7 titrimetric method (Yeomans and Bremner 1988). The TN of the soil sample was determined by the K2Cr2O7-H2SO4 digestion method (Flowers and Bremner 1991). The soil C/N ratio (CN) was calculated based on the SOC and TN. The soil NN was determined by the phenol disulfonic acid method (Sahrawat and Prasad 1975). The AN was determined by a KCl extraction-indophenol blue colorimetry method (Wu et al. 2013). The TP was determined via the H2SO4-HClO4 digestion method (Wei 2009). The AP was determined using the NaHCO3 leaching molybdenum antimony colorimetric technique (Pu et al. 2014).
Determination of soil enzymatic activities
Urease activity (EC3.5.1.5) was assayed by colorimetric analysis of sodium phenate-sodium hypochlorite (Kandeler and Gerber 1988; Van Wyk et al. 2017). The activities of sucrase (EC3.2.1.26) and cellulase (EC3.2.1) were colorimetrically determined by DNS based on the decreasing sugar content (Li et al. 2016). Total phosphatase activity was colorimetrically estimated using disodium phenyl phosphate, which is a method based on the determination of the organic group content that is generated by the enzymatic reaction of disodium phenyl phosphate as the matrix. To facilitate determination, acid phosphatase (EC 3.1.3.2) with an acetate buffer (pH = 5.0 ~ 5.4), neutral phosphatase with a citrate buffer (pH = 7.0), and alkaline phosphatase (EC3.1.3.1) with a borate buffer (pH = 9 ~ 10) were used to ensure that the reaction occurred at an optimal pH (Tabatabai 1994). The dehydrogenase activity of the soil was detected through a TTC (triphenyl tetrazolium chloride) reduction method (Friedel et al. 1994). Catalase (EC1.11.1.6) activity was assayed by potassium permanganate titration (Sinha 1972).
Genomic DNA extraction and PCR amplification
The soil samples stored at -80 °C were used to extract DNA and to conduct PCR amplification and Illumina MiSeq sequencing; three replicates were used. The genomic DNA of the soil was extracted using a Soil DNA™ Kit (Sigma-Aldrich, Germany) according to the manufacturer’s recommendations. The purity and concentration of the extracted DNA were determined using a spectrophotometer (Thermo Fischer Scientific, CA, USA), and integrity was determined using a 0.8 % agarose gel. Aliquots (40 µL) of high-quality DNA extracts were obtained for further analysis.
Subsequently, the bacterial V4 hypervariable regions of the 16S rRNA gene and fungal ITS1 region of the 18S rRNA gene were amplified via PCR from the microbial genome DNA using 520F (5’-AYTGGGYDTAAAGN G-3’) and 802R (5’-TACNVGGGTATCTAATCC-3’) primer pairs and ITS1F (5’- CTTGGTCATTTA GAGGAAGTAA-3’) and ITS2 (5’- GCTGCGTTCTTCATCGATGC-3’) primer pairs, respectively.
PCR of the ITS1 region of the fungal 18 S rRNA was performed in triplicate in the same reaction mixtures as the V4 region; the A3 and D3 samples used 3 µL (20 ng/µL) of template, while the other samples used 2 µL (20 ng/µL) of template. The annealing temperature was 56 ℃ and was amplified using 28 cycles. The PCR product was excised from a 2.0 % agarose gel and purified using the QIAquick Gel Extraction Kit.
Illumina MiSeq sequencing and processing
Sequencing was performed using the Illumina MiSeq platform. Multiplexed DNA libraries were established by means of a DNA end-trimmed “A” base introduced at the 3’ end, tagged adapter attachment and DNA fragment enrichment. Barcoded V4 and ITS1 amplicons were sequenced using the paired-end method by Illumina MiSeq with a six cycles index read. Following the statistical computations and analysis of the raw data, effective and high-quality sequences were screened for further analysis.
Next, the bacterial and fungal OTUs were identified using the same 97 % identity threshold, clustered, checked for chimeras, and assembled into incidence tables using UCHIME in mothur to remove the chimeric sequences and using UCLUST method in QIIME to cluster the high-quality sequences (Edgar 2010). A representative sequence was selected from each OTU using default parameters. Finally, the taxonomic assignment was made using the Greengenes Database and Unite database, respectively (Abarenkov et al. 2010; DeSantis et al. 2006). An OTU table was generated to record the abundance and taxonomy of each OTU in each sample. OTUs containing less than 0.001 % of total sequences across all samples were discarded. To minimize the difference in sequencing depth across samples, an averaged, rounded rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under 90 % of the minimum sequencing depth for further analysis.
Bioinformatics and statistical analysis
The rarefaction and rank abundance curves were established to evaluate the reliability and uniformity coefficients of sequencing. Sequence data analyses were primarily performed using QIIME and R packages (v3.2.0). OTU-level alpha diversity indices, such as the Chao1 richness estimator, ACE metric (abundance-based coverage estimator), Shannon diversity index, and Simpson index, were calculated using the OTU table in QIIME. OTU-level ranked abundance curves were generated to compare the richness and evenness of OTUs between samples. Beta diversity analysis was performed to investigate the structural variation of microbial communities across samples using UniFrac distance metrics (Lozupone and Knight 2005; Lozupone et al. 2007) and visualized via principal coordinate analysis (PCoA), nonmetric multidimensional scaling (NMDS), and unweighted pair-group method with arithmetic means (UPGMA) hierarchical clustering (Ramette 2007).
Differences in the UniFrac distances for pairwise comparisons between groups were determined using Student’s t-test and the Monte Carlo permutation test with 1000 permutations and visualized through box-and-whisker plots. Principal component analysis (PCA) was also conducted based on genus-level compositional profiles (Ramette 2007). The taxonomy compositions and abundances were visualized using MEGAN (Huson et al. 2011) and GraPhlAn (Asnicar et al. 2015). Taxa abundances at the phylum, class, order, family, genus and species levels were statistically compared between samples or groups using Metastats (White et al. 2009), and visualized as violin plots.
Results
Soil physicochemical properties
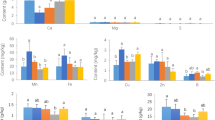
The soil physicochemical properties are shown in Fig. 1. One-way ANOVA revealed that there were differences between different groups. Compared with group A, the SM and SOC of group B showed no significant change (Fig. 1a, e), while the SE, SS, TN, and NN were remarkably higher (p < 0.01). Furthermore, the SE, SS and NN came close to the levels of group A with a longer time to replant maize. The TN decreased remarkably (p < 0.01) in group C and then remarkably increased to the level of group B (p < 0.01) with the increase in replanting maize time; the TN of group E was significantly higher than that of group B (p < 0.05) (Fig. 1c, d, f, h). Simultaneously, the AN and AP exhibited a similar increasing trend between groups A and B, which recorded no significant difference, followed by a gradual decline observed in AN with a longer time to replant maize (Fig. 1g, j). Conversely, the soil pH was remarkably lower (p < 0.01) for group B than group A; soil pH was gradually regained in group C and remarkably higher (p < 0.01) in group D. Soil pH steadily increased with a longer time to replant maize, although group E never reached the pH level of group A (Fig. 1b). Similarly, the CN and TP were significantly lower (p < 0.05) across groups in group B compared to group A, continued to decline in group C, were gradually restored in group D, and returned to the level of group A in group E (Fig. 1k, i).
Changes in soil physicochemical properties. Soil moisture (SM) (a), soil pH (b), soil electric conductivity (SE) (c), soil salt content (SS) (d), total soil organic carbon (SOC) (e), total nitrogen content (TN) (f), carbon-to-nitrogen ratio (CN) (g), ammonium nitrate content (NH4+-N, AN) (h), nitrate nitrogen content (NO3−-N, NN) (i), total phosphorus content (j) and available phosphate (AP) (k) of both never planted American ginseng farmland (Group A), recently harvested American ginseng farmland (Group B), replanted maize for 2 years (Group C), 4 years (Group D) and 6 years (Group E) after planting American ginseng. Data are means ± standard deviation (n = 3). Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a remarkable difference at p ≤ 0.01
Soils enzymatic activity
The enzymatic activities of the soils of the five groups were compared and assayed, as shown in Fig. 2. When comparing groups A and B, we observed a remarkable difference in that the neutral phosphatase activity was significantly higher (p < 0.01) (Fig. 2a), whereas the alkaline phosphatase activity (p < 0.01) (Fig. 2b) and cellulase activity were considerably lower (p < 0.01) (Fig. 2d). No significant difference was found between groups regarding the total phosphatase activity (Fig. 2c), dehydrogenase activity (Fig. 2e), urease activity (Fig. 2f), acid phosphatase activity (Fig. 2g), sucrase activity (Fig. 2h), or catalase activity (Fig. 2i).
Changes in soil enzymatic Activities. Neutral phosphatase (a), alkaline phosphatase (b), total phosphatase (c), cellulase (d), dehydrogenase (e), urease (f), acid phosphatase (g), sucrase (h) and catalase (i) of both never planted American ginseng farmland (Group A), recently harvested American ginseng farmland(Group B), replanted maize for 2 years (Group C), 4 years (Group D) and 6 years (Group E) after planting American ginseng. Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a significant difference at p ≤ 0.01
The alkaline phosphatase activity of group C was remarkably lower than that of group B (p < 0.01) and was considerably higher (p < 0.01) for group D. Although it tended to increase with replanting maize, the alkaline phosphatase activity of group E did not attain the level measured in soils that were never planted with American ginseng (Fig. 2b). In contrast, the cellulase activity of group C was significantly higher (p < 0.05) than that of group B. Higher cellulase activity was found to be directly related to replant maize time length; the cellulase activity of group E approached the levels of group A (Fig. 2d). In addition, the dehydrogenase activity exhibited a gradual increasing trend and was remarkably higher (p < 0.01) in group D and significantly higher (p < 0.05) in group E (Fig. 2e).
Sequencing and microbial community alpha diversity
A total of 634,124 and 1,718,305 valid chimera sequences from bacteria and fungi, respectively, were obtained by high-throughput sequencing analyses of the V4 region of the 16 S rDNA gene and the ITS region of the 18 S rDNA, including 602,263 (94.98 %) and 1,504,849 (87.58 %) high-quality sequences, respectively, in the fifteen soil samples. The number of reads ranged from 22,704 to 47,955 and 80,576 to 123,250, respectively, which resulted in 9,544 and 1,160 OTUs (97 % cutoff) (Table S1). The lengths of the greatest number of reads were distributed across the ranges 224 ~ 226 and 223 ~ 281. The 31.24 % bacterial community OTUs and 52.16 % fungal community OTUs were shared in all five groups of soil samples, which accounted for the majority of the reads.
The OTU rarefaction and rank abundance curves indicated that a sufficient amount and quantity of sequencing data was obtained (Fig. S1), particularly for bacteria. Through the analyses of alpha diversity (Chao, Ace, Shannon, Simpson) (Table S2), we found that the levels of bacterial diversity in the soil tended to be lower after planting American ginseng and higher in the farmlands of replanting maize after American ginseng; however, the richness tended to be higher after planting American ginseng and lower in the farmlands of replanting maize (Fig. S2a, b, c, d). Nevertheless, levels of soil fungal diversity and richness both tended to be higher following the planting of American ginseng and were persistently higher for the farmlands of replanting maize (Fig. S2e, f, g, h), although no significant difference was found.
The composition of bacterial and fungal communities
A total of 47 distinct bacterial phyla were detected across all fifteen samples. The most abundant sequences were affiliated with the phyla Bacteroidetes (14.2 %-28.2 % of total relative abundance) and Proteobacteria (12.7 %-23.7 %), followed by Planctomycetes (11.5 %-19.6 %), Acidobacteria (10.6 %-13.6 %), Actinobacteria (8.1 %-14.1 %), Gemmatimonadetes (4.1 %-6.5 %), Verrucomicrobia (3.8 %-6.3 %), and Chloroflexi (2.6 %-5.7 %) (Fig. 3a). Furthermore, Planctomycetia in the phylum Planctomycetes (10.3 %, 10.8 %, and 11.1 % in groups A, C, and E, respectively) was the dominant class, Alphaproteobacteria in the phylum Proteobacteria (10.4 %) was predominant in recently harvested farmland samples (group B), while Alphaproteobacteria represented 9.50 %, 6.90 %, 8.30 %, and 6.80 % in groups A, B, C, and E, respectively. A total of 6 distinct fungal phyla were detected across all fifteen samples and were dominated by the phylum Ascomycota (61.9 %-70.3 %), while the phyla Basidiomycota and Zygomycota represented only 7.6 %-8.6 % and 3.6 %-10.1 % reads, respectively (Fig. 3b). Furthermore, compared with group A, Sordariomycetes was decreased in group B and was gradually restored with a longer time to replant maize. Conversely, Leotiomycetes and Tremellomycetes were more abundant in group B than in group A and gradually decreased to the level of group A when the replanting maize time reached 6 years (Fig. 3d).
Relative abundance of primary bacterial phyla (average relative abundance ≥ 0.5 %) (a), fungal phyla (average relative abundance ≥ 0.5 %) (b), bacterial class (average relative abundance ≥ 0.5 %) (c) and fungal class (average relative abundance ≥ 0.5 %) (d) present in different soil groups of bacterial and fungal communities
At the bacterial genus level, the relative abundances of Chthoniobacter (p < 0.05), Opitutus (p < 0.01), Prosthecobacter (p < 0.05), Adhaeribacter (p < 0.05), Luteolibacter (p < 0.01), Agrobacterium (p < 0.05), Balneimonas (p < 0.05), Pontibacter (p < 0.05), Skermanella (p < 0.05), and Arthrobacter (p < 0.05) in group B were significantly lower, representing 1.5 % of the sequences (Fig. 4a, S5). Furthermore, two genera (Adhaeribacter and Luteolibacter) were gradually restored with a longer time to replant maize. Correspondingly, group B were significantly higher in DA101 (p < 0.01), Mycobacterium (p < 0.05), Rhodoplanes (p < 0.01), Pedobacter (p < 0.05), Rhodanobacter (p < 0.01) and Bacillus (p < 0.05). Except for DA101, which did not obviously change with a longer time to replant maize, other genera were gradually restored to the levels of group A (Fig. 4a, S6). At the fungal genus level, group B were significantly lower in Myrothecium (p < 0.05), Volutella (p < 0.05), Wardomyces (p < 0.05), Thelebolus (p < 0.01), Leptosphaeria (p < 0.05), Passalora (p < 0.05), Truncatella (p < 0.01), Protomyces (p < 0.01), Toninia (p < 0.05), Rachicladosporium (p < 0.05), Hydnum (p < 0.01), and Marasmiellus (p < 0.01) (Fig. 4b, S7), representing 1.5 % of the sequences. Furthermore, three genera (Myrothecium, Hydnum, and Rachicladosporium) were gradually restored as replanting maize time increased, whereas three genera (Marasmiellus, Leptosphaeria, and Volutella) did not obviously change with a longer time to replant maize. Additionally, three genera (Thelebolus, Wardomyces, and Passalora) were consistently lower in the farmlands of replanting maize. Only Thelebolus and Wardomyces were gradually restored in group E. Interestingly, Toninia was restored in group C but was again significantly lower in group D. Correspondingly, group B were significantly higher in Retroconis (p < 0.05), Chaetomium (p < 0.05), Pseudeurotium (p < 0.05), Hypomyces (p < 0.01), Phialemonium (p < 0.01), Rhinocladiella (p < 0.01), Leptodiscella (p < 0.05), Torrubiella (p < 0.01), Monodictys (p < 0.05), and Cladosporium (p < 0.01) and were gradually restored to the levels of group A with a longer time to replant maize. Although Retroconis and Chaetomium fluctuated by varying degrees during replanting maize periods, they also returned to group A levels (Fig. 4b, S8).
Heatmap of weighted Bray-Curtis with hierarchal clustering of some significantly different bacteria (a) and fungi (b) from the soil of farmland that never planted American ginseng (Group A) and recently harvested American ginseng (Group B) at the genus level. The relative abundance for each genus is depicted by color intensity in each field. Higher values are represented by red whereas lower values are represented by green. A, B, C, D, E represented the five groups
Bacterial and fungal Beta-diversity analysis and correlation with environmental parameters
The planting of American ginseng had a significant effect on the soil microbial community based on UniFrac distances. The Adonis analysis of the 16 S rRNA gene and ITS data using unweighted and weighted methods revealed that the bacterial and fungal community structures and compositions were significantly changed (Table 1). This phenomenon was also observed through PCoA and NMDS analyses (Fig. S3, S4). Significant variations between different groups were revealed by PCoA ordination for bacterial communities and fungi. PC1 (first principal component, 30.51 % contribution), PC2 (second principal component, 13.42 % contribution), and PC3 (third principal component, 11.39 % contribution) differentiated the bacterial communities (Fig. S3a, b, c); PC1 (first principal component, 17.88 % contribution), PC2 (second principal component, 12.98 % contribution), and PC3 (third principal component, 10.44 % contribution) differentiated the fungal communities (Fig. S3d, e, f). A similar phenomenon was observed via NMDS analysis (Fig. S4).
Redundancy analysis suggested that changes in soil chemistry played an important role in shaping the structure and composition of microbial communities. Changes in SM, NN, SS, SOC, and CN had a more obvious effect on the alteration of bacterial structures and compositions in different farmland soils; however, the changes in pH, TP, AN, AP, and TN had a relatively low impact (Fig. 5a). The changes in pH, SM, NN, SS, and CN had a more obvious effect on the change in fungal community structure and composition; however, the changes in AP, OC, AN, TN, and TP had a relatively low impact (Fig. 5b).
Redundancy analysis of dominant bacterial phyla (a), dominant fungal phyla (b) (average relative abundance > 0.5 %), significantly different bacterial genera (c) and significantly different fungal genera (d) across all of the farmland soil samples. Phyla and genera are indicated by blue vectors and physicochemical variables are represented by red vectors. The positions and lengths of the arrows indicate the directions and strengths, respectively, of the effects of variables on microbial communities. Samples were analyzed in triplicate plots. Blue right triangles represent never planted American ginseng farmland (Group A). Black stars represent recently harvested American ginseng farmland (Group B). Whereas orange diamonds, green circles and red squares represent replanted maize for 2 years (Group C) ,4 years (Group D) and 6 years (Group E) after planting American ginseng, respectively. Abbreviations in a, Aci, Acidobacteria, Act, Actinobacteria, Bac, Bacteroidetes, Chl, Chloroflexi, Cya, Cyanobacteria, Fir, Firmicutes, Gem, Gemmatimonadetes, Nit, Nitrospirae, Pla, Planctomycetes, Pro, Proteobacteria, Ver, Verrucomicrobia. Abbreviations in b, Ich, Ichtyosporea, Asc, Ascomycota, Bas, Basidiomycota, Bla, Blastocladiomycota, Chy, Chytridiomycota, Zyg, Zygomycota, uni, unidentified. Abbreviations in c, Ped, Pedobacter, Myc, Mycobacterium, Rhoda, Rhodanobacter, Rhodo, Rhodoplanes, Bac, Bacillus, Cht, Chthoniobacter, Opi, Opitutus, Pro, Prosthecobacter, Adh, Adhaeribacter, Lut, Luteolibacter, Agr, Agrobacterium, Ske, Skermanella, Art, Arthrobacter, Bal, Balneimonas, Pon, Pontibacter. Abbreviations in d, Ret, Retroconis, Pse, Pseudeurotium, Mon, Monodictys, Rhi, Rhinocladiella, Tor, Torrubiella, Phi, Phialemonium, Cla, Cladosporium, Cha, Chaetomium, Hyp, Hypomyces, Lep, Leptodiscella, Sta, Stagonosporopsis, Myr, Myrothecium, Ton, Toninia, Pas, Passalora, Vol, Volutella, The, Thelebolus, Rac, Rachicladosporium, Tru, Truncatella, Pro, Protomyces, War, Wardomyces, Lept, Leptosphaeria, Mar, Marasmiellus, Hyd, Hydnum
At the bacterial phylum level, all bacterial phyla with an average relative abundance greater than 0.5 % exhibited different degrees of negative correlations with changes in TN, CN and OC, particularly the phyla OD1, Verrucomicrobia, Gemmatimonadetes, Actinobacteria, and Acidobacteria. The phyla Nitrospirae, WS3, Firmicutes, Chloroflexi, Proteobacteria, Actinobacteria, and Acidobacteria possessed a strongly positive association with SS, AN, NN and AP, while Cyanobacteria, Planctomycetes, and Bacteroidetes were strongly negatively associated. Additionally, Nitrospirae, WS3, Firmicutes, Chloroflexi, Proteobacteria, Acidobacteria, Actinobacteria, and Gemmatimonadetes revealed a strongly negative association with pH and SM, while Cyanobacteria, Planctomycetes, and Bacteroidetes had a strongly positive association. Nitrospirae and WS3 were positively associated with TP, whereas Verrucomicrobia, Bacteroidetes, and Planctomycetes were strongly negatively correlated with TP (Fig. 5a). At the fungal phylum level, Ichtyosporea was negatively correlated with TP, pH, and CN, while there was a distinctly positive correlation with SS, TN, AN, NN and AP. Basidiomycota, Blastocladiomycota, and Chytridiomycota showed different degrees of negative correlations with TN, AN, OC, TP, AP, and SM, while Blastocladiomycota and Chytridiomycota were positively correlated with pH. Basidiomycota and Chytridiomycota were positively correlated with SS and NN, whereas Ascomycota was positively correlated with TN, AN, OC, TP, AP, and SM. Additionally, Zygomycota was negatively correlated with pH and TP, while there was a positive correlation with TN, AN, and AP (Fig. 5b).
According to the results of the RDA, the bacterial genera showed significant differences between groups A and B. Pedobacter, Rhodanobacter, Rhodoplanes, and Bacillus were strongly positively associated with SE, TN, AN, NN, and AP, whereas Opitutus, Prosthecobacter, Adhaeribacter, and Balneimonas demonstrated a strongly negative association. DA101, Pedobacter, Mycobacterium, Rhodanobacter, Rhodoplanes, and Bacillus were strongly negatively correlated with pH and CN, whereas Adhaeribacter, Luteolibacter, Skermanella, and Balneimonas were strongly positively correlated (Fig. 5c). Simultaneously, at the fungal genus level, all of the most significantly increased fungi in group B, in contrast to group A, were negatively correlated with pH, CN, TP, and SM to varying degrees. Furthermore, all significantly decreased fungi were positively associated with these variables, except for Monodictys, which was slightly positively associated with TP and had no relationship with CN. Conversely, all significantly decreased fungi in group B compared with group A were negatively correlated with SS, AN, and NN, whereas all significantly increased fungi were positively correlated. Furthermore, except for Retroconis, Chaetomium, Leptodiscella, and Toninia, which were negatively correlated with TN, all of the significantly changed fungi were positively associated with TN. In addition, Retroconis, Rhinocladiella, Phialemonium, Chaetomium, Leptodiscella, and Toninia were negatively correlated, while Torrubiella and Hypomyces had no relevance to SOC, and other significantly changed fungi were positively associated with SOC (Fig. 5d).
Discussion
Soil pH, salt content, and nitrogen content are important physicochemical factors affecting continuous cropping obstacle of American ginseng
Analysis of the changes in soil physicochemical properties showed that SE, SS, NN, and pH played extremely important roles in the continuous cropping problem of American ginseng. The obstacles to continuous cropping are likely attributable to the decrease in soil pH and the increase in SE, SS, and NN levels associated with the cultivation process.
Our results indicate that soil pH of group B was remarkably lower (p < 0.01) than that of group A and gradually recovered with the increasing length of replanting maize time (Fig. 1b). This is consistent with previous researches (Zhang et al. 2020; You et al. 2015) and suggest that the decrease in soil pH is a vital factor in continuous cropping obstacles of American ginseng. Furthermore, because previous studies have reported that the root weights of American ginseng in soils with continuous cropping 2-, 3- and 4-years decreased by 24 %, 17 % and 32 % respectively (Zhao 2009). The seedling survival rate of American ginseng after cropping rotation maize and wheat for 5 years increased from 30 % to about 85 % (Zhang 2013). We speculated that the decrease in soil pH may be an important reason for the decline in American ginseng production. Additionally, we believe that the soil pH of fields where replanting maize after harvesting American ginseng may gradually increase and approach the pH of group A as time length increases, because the soil pH had begun to rise significantly when the farmland was replanted maize for 4-years.
Soil pH has the capacity to impact all chemical, physical, and biological soil properties (Brady et al. 2008; Kemmitt et al. 2006). The present study revealed that the soil pH was similarly correlated with SOC, TN and CN (Pietri and Brookes 2008) and related to changes in the quantity of released NH4+ and NO3− in soil (Feizi et al. 2017; Nicol et al. 2008). Our work demonstrated that the soil pH was negatively associated with TN, AN and NN (Table S3). These results were consistent with a previous study (Curtin et al. 1998). Additionally, our experiments also demonstrated that TN (p < 0.01), AN (p < 0.05) and NN (p < 0.01) were significantly increased for recently planted American ginseng and were generally restored after replanting maize (Fig. 1f, g, h). These results indicated that changes in soil pH caused by planting American ginseng may cause changes in soil nitrogen content since primary transformations of N in soil-plant systems are affected by NH4+-N and NO3−-N concentrations (De Klein and Van Logtestijn 1994). Organic N mineralization contributes to changes in soil pH values by initially consuming H+ during the ammonification process, followed by the release of H+ during nitrification (Xu et al. 2006). Our research appeared to demonstrate this, as soil pH was negatively associated with NN. These results suggested that changes in TN, NN, and AN are material factors affecting the continuous cropping obstacles related to American ginseng. In association with the changes in TN, CN decreased significantly following the cultivation of American ginseng, which was generally restored to the control level after replanting maize (Fig. 1k). These results indicate that more attention should be paid to the application of N fertilizer when planting American ginseng. CN was also shown to be an important factor in regulating the composition of soil microbial communities and was related to soil pH (Bengtsson et al. 2003; Lovett et al. 2002). This explains the positive correlation between CN and soil pH (r = 0.632, p < 0.05) (Table S3).
Furthermore, in our study, the SE and SS of group B were remarkably higher than group A (p < 0.01) and were restored to the level of group A after replanting maize for 2 years (p < 0.01) (Fig. 1c, d). This result suggests that the increase in soil salt content associated with the continuous cropping obstacle of American ginseng and may be reversible. There have been numerous previous studies to illustrate that the soil salt content is important for plant growth (Omer 2004; Wang et al. 2008). The present study showed that crops may develop best when the surface soil salt content is < 0.5 g/kg. Crop growth is restricted when the soil salt content is between 0.5 and 4 g/kg, and little crop growth occurs when the soil salt content is greater than 4.0 g/kg (Jin et al. 2012). It was confirmed in our research that the cultivation of American ginseng contributes to a significant increase in the soil salt content, recording levels that ranged from less than 0.4 g/kg in soils of group A to close to 1.0 g/kg in soils of group B. The soil salt content decreased significantly to the level before planting American ginseng, and it was maintained at this level with a longer time to replant maize. This result indicates that the development of effective methods to reduce the soil salt content following the cultivation of American ginseng may assist in alleviating the continuous cropping problem of American ginseng.
Soil cellulase and alkaline phosphatase are important soil enzymes affecting continuous cropping obstacle of American ginseng
Biochemical parameters include multiple indicators, and soil enzyme activities are employed as the most important soil quality and fertility indicators (Bastida et al. 2008; Stott et al. 2010). In our study, neutral phosphatase activity (Fig. 2a) was significantly increased (p < 0.01) in the soil under the continuous cropping of American ginseng compared with the other groups. However, the alkaline phosphatase (Fig. 2b) and cellulase (Fig. 2d) activities were significantly decreased (p < 0.01).
The CN and the types of C and N sources, as well as their individual concentrations, are important parameters in cellulase production. Several studies have shown that the cellulase enzymes might influence microbial activity (Torres et al. 2016), the reduction in cellulase activity associated with continuous cropping obstacle is related to Myrothecium, Volutella, and Thelebolus, which are known to participate in the generation of cellulase activity (Kidder III and Goddard 1965; Singh et al. 2016). The results of our experiments revealed that relative abundance of Myrothecium, Volutella, and Thelebolus were significantly decreased in group B. Levels of Myrothecium and Thelebolus were gradually restored, although they did not recover to the level of group A (Fig. S7 a, d, g). This explained the positive correlation between cellulase activity and the relative abundance of Myrothecium (r = 0.579, p < 0.05), Volutella (r = 0.324, p = 0.240), and Thelebolus (r = 0.314, p = 0.255) (Table S4). Additionally, changes in the relative abundance of Chaetomium were significantly negatively correlated with cellulase activity (r = -0.653, p < 0.01). Chaetomium is an important fungus that is contributed to decompose cellulose and resides within various plant residues, cellulose-rich substrates and soils (Hubbard et al. 2011; Lee and Hanlin 1999). Here, the cellulase activity gradually recovered with increasing replanting maize time. This suggests that cellulase activity is a key factor that affects the continuous cropping problem of American ginseng, which was impacted by changes in soil microbial community.
Alkaline phosphatase (AlkP) activity was significantly decreased in group B compared with group A; AlkP declined in group C and then gradually recovered (Fig. 2b). Previous studies have found that AlkP increases as the soil pH increases (Dick et al. 2000). These reports articulated that alkaline phosphatase was positively correlated with soil pH (r = 0.910, p < 0.01) (Table S5). Combined with the influence on the soil microbial community (Acostamartinez et al. 2003), we believe that alkaline phosphatase is another crucial extracellular enzyme that affects the continuous cropping of American ginseng and is closely related to changes in microbial community structure and composition.
Changes in soil microbial community are closely related to changes in soil physicochemical and enzyme activities
For this study, we characterized the changes in microbial community structure and composition of farmlands soils from which planted with American ginseng and replanted maize after American ginseng. Our research suggests that the bacterial diversity of soil tends to be lower following the planting of American ginseng, while the fungal diversity and bacterial and fungal richness both tend to be higher. Furthermore, Pearson’s correlation analysis between the alpha diversity index and soil pH showed that soil pH was positively correlated with bacterial diversity (r = 0.535, p < 0.05), but it was negatively correlated with fungal diversity (r = − 0.080, p = 0.777) (Table S6). This is consistent with the findings of previous studies (Dong et al. 2017; Rousk et al. 2010; Zheng et al. 2019).
Specifically, at the bacterial phylum level, bacterial community composition was dominated by Bacteroidetes, Proteobacteria, Planctomycetes, Acidobacteria, Actinobacteria, etc. This was consistent with previous studies (Wolińska et al. 2017; Yu et al. 2018; Zheng et al. 2019). Acidobacteria is believed to have an extensive range of metabolic and genetic functions (Kielak et al. 2016). Soil pH is one of the key factors that influences the composition and structure of Acidobacteria communities (Sait et al. 2006; Zhang et al. 2014) and Acidobacteria exhibited a robust, inverse response to soil pH (Zheng et al. 2019). The relative abundance of Acidobacteria has been shown to increase with lower soil pH (Dimitriu and Grayston 2010; Rousk et al. 2010). According to the RDA, our study also demonstrated that Acidobacteria was strongly negatively correlated with soil pH (Fig. 5a). In comparison, Nitrospirae (0.8 %) and WS3 (0.9 %) significantly increased in soils planted with American ginseng (p < 0.01) and gradually recovered to pre-cultivation levels after replanting maize (Fig. 6). This result indicates that increases in Nitrospirae and WS3 are potentially important factors in the continuous cropping problem of American ginseng. Furthermore, our study reveals that the relative abundance of Nitrospirae is positively correlated with AN (r = 0.515, p < 0.05) and NN (r = 0.840, p < 0.01) (Table S7, Fig. 5a). These results are in alignment with a previous study, in which the higher abundance of Nitrospirae reinforces the idea of a community adapted to improved N mining in these environments (Carbonetto et al. 2014).
Significantly different bacterial phyla of recently harvested American ginseng farmland (Group B) compared with never planted American ginseng farmland (Group A). Data are means ± standard deviation (n = 3). Different letters (a, b, c) indicates a significant difference from each other at p ≤ 0.05, and different letters (A, B, C) indicates a remarkable difference from each other at p ≤ 0.01
At the bacterial genus level, the relative abundance of Bacillus was significantly increased following the planting of American ginseng. Most Bacillus members can inhibit the propagation of pathogenic microbes in soils; additionally, they can invade the roots of plants, reduce soil-borne diseases in plants, regulate and balance soil pH, adjust the ecological environments of plant roots, and form dominant colonies to remediate the damage inflicted on soils by chemical fertilizers, pesticides, and other harmful factors (Lin et al. 2014; Shafi et al. 2017). Consequently, increased Bacillus may be a response to changes in the physicochemical properties and microbial community structures and compositions of soils, particularly in terms of decreasing pH, increasing SS, and harmful bacteria and fungi. Opitutus is believed to contribute to the N cycle by reducing nitrate to nitrite, which other bacteria convert further (Chin et al. 2001). Our results also reveal that changes in the relative abundance of Opitutus were significantly negatively correlated with TN (r = -0.560, p < 0.05), AN (r = -0.580, p < 0.05), and NN (r = -0.718, p < 0.01). Our results show that some Proteobacteria, such as Agrobacterium, Balneimonas, and Skermanella, were significantly decreased. In addition, there were other beneficial bacterial genera that also significantly decreased following the planting of American ginseng. For example, Chthoniobacter was found to improve the health of soils (Sangwan et al. 2004).
At the fungal phylum level, fungal communities were dominated by Ascomycota, Basidiomycota, and Zygomycota, which is consistent with the findings of previous studies (Egidi et al. 2019; Huang et al. 2015; Tan et al. 2017). We found that almost all the significantly altered fungal genera in group B (compared with group A) were members of Ascomycota and Basidiomycota. Previous studies revealed that the members Ascomycota and Basidiomycota were primary soil fungal decomposers (Bastian et al. 2009). The decomposition of crop residues by soil microbial communities is a key step toward the release of inorganic nutrients from plant residues (Ma et al. 2013). We found that almost all significantly increased fungal genera in group B, in contrast to group A, were significantly positively correlated with SE, SS, AN, and NN (Table S8). Furthermore, the soil SE and SS were significantly positively correlated with Torrubiella (r = 0.889, p < 0.001), Cladosporium (r = 0.849, p < 0.001), Rhinocladiella (r = 0.874, p < 0.001), and Phialemonium (r = 0.904, p < 0.001), whereas the soil NN was significantly positively correlated with Torrubiella (r = 0.900, p < 0.001), Rhinocladiella (r = 0.907, p < 0.001), and Cladosporium (r = 0.896, p < 0.001).
A previous study reported that soil pH positively affected plant diversity in regions with predominately neutral soils but negatively affected plant diversity in regions where acidic soils dominated (Partel 2002). This confirmed that the most significantly changed fungi were strongly related to soil pH. As such, the soil pH was significantly positively correlated with Myrothecium (r = 0.748, p < 0.01), Truncatella (r = 0.820, p < 0.001), and Protomyces (r = 0.766, p < 0.001), while it was significantly negatively correlated with Chaetomium (r = -0.525, p < 0.05), Cladosporium (r = -0.583, p < 0.05), and Leptodiscella (r = -0.567, p < 0.05). The presence of Myrothecium is extensive in plants and soil, with most species having a strong capacity to decompose cellulose. Some species can produce antibiotics, which is related to a decrease in cellulase activity (Brian et al. 1948; Brian and Mcgowan 1946). Our results showed that changes in the relative abundance of Myrothecium were significantly positively correlated with cellulase activity (r = 0.579, p < 0.05). As an important cellulolytic fungus, Chaetomium plays a critical role in the carbon cycle and soil improvement in natural ecosystems, and many species have beneficial biocontrol potential in agricultural production. Changes in the relative abundance of Chaetomium were significantly negatively correlated with soil pH (r = -0.525, p < 0.05) and positively correlated with SS (r = 0.573, p < 0.05) in response to decreased soil pH and increased SS. Additionally, as a potential biocontrol fungus, changes in the relative abundance of Hydnum were significantly decreased in group B compared with group A, significantly negatively correlated with soil pH (r = -0.634, p < 0.05), and positively correlated with SS (r = 0.728, p < 0.01). A study has shown that members of Hydnum might be associated with the symptoms of root rot for a broad array of woody and herbaceous host plants (Bastos et al. 1981).
In summary, we believe that the change in bacterial and fungal community structure is an important factor that leads to the failure of continuous cropping for American ginseng; balancing soil pH may be an essential prerequisite to improve the diversity and abundance of fungal communities to resolve the continuous cropping problem of American ginseng. A number of effective measures could be performed in actual cultivation applications, such as the addition of organic amendments, which have proven to be a viable strategy for the regulation of soil pH, nitrogen availability, and microbial community composition (Bowles et al. 2014; Feizi et al. 2017). Furthermore, fertilization with wood ash and lime has the capacity to dramatically increase and improve the soil pH over time to levels that are suitable for plant growth (Demeyer et al. 2001; Fritze et al. 1994).
Conclusions
In this study, we collected soils from maize fields where American ginseng had never been planted, from fields where American ginseng had just been harvested, and from fields where maize had been planted for 2, 4 and 6 years after harvesting American ginseng. We analyzed the physical and chemical properties, enzymatic activities, and microbial community structure and composition. We found that a significant decrease in soil pH and a substantial increase in soil salt content were critical factors that led to the continuous cropping problem of American ginseng. According to our results, the modification of these physical and chemical properties caused significant changes in microbial community structures, as well as changes in soil bacterial and fungal diversity and composition following the cultivation of American ginseng. Furthermore, the relative abundance of some bacteria, such as Nitrospirae, Opitutus, Agrobacterium, Balneimonas, and Skermanella, were altered, as were the relative abundance of associative fungal communities, such as Myrothecium, Chaetomium, Volutella, and Thelebolus. Pearson’s correlation analysis shown that these changes were influenced by the alteration of soil physical and chemical properties. Soil cellulase and alkaline phosphatase activities were also changed following the cultivation of American ginseng.
In summary, changes in soil pH, soil salt content and nitrogen content lead to changes in microbial community structure and composition, as well as cellulase and alkaline phosphatase activities, these changes may cause the continuous cropping obstacles of American ginseng and may be improved by replanting maize.
References
Abarenkov K et al (2010) The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 186:281–285
Acostamartinez V, Zobeck TM, Gill TE, Kennedy AC (2003) Enzyme activities and microbial community structure in semiarid agricultural soils. Biol Fertil Soils 38:216–227
Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N (2015) Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029
Banerjee S, Bora S, Thrall PH, Richardson AE (2016) Soil C and N as causal factors of spatial variation in extracellular enzyme activity across grassland-woodland ecotones. Appl Soil Ecol 105:1–8
Bastian F, Bouziri L, Nicolardot B, Ranjard L (2009) Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol Biochem 41:262–275
Bastida F, Zsolnay A, Hernández T, García C (2008) Past, present and future of soil quality indices: a biological perspective. Geoderma 147:159–171
Bastos CN, Evans HC, Samson RA (1981) A new hyperparasitic fungus, Cladobotryum amazonense, with potential for control of fungal pathogens of cocoa. Trans Br Mycol Soc 77:273–278
Becker JC, Rodibaugh KJ, Hahn D, Nowlin WH (2017) Bacterial community composition and carbon metabolism in a subtropical riverscape. Hydrobiologia 792:209–226
Bengtsson G, Bengtson P, Månsson KF (2003) Gross nitrogen mineralization-, immobilization-, and nitrification rates as a function of soil C/N ratio and microbial activity. Soil Biol Biochem 35:143–154
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Blagodatskaya EV, Anderson T-H(1998) Interactive effects of pH and substrate quality on the fungal-to-bacterial ratio and qCO2 of microbial communities in forest soils. Soil Biol Biochem 30:1269–1274
Bowles TM, Acostamartinez V, Calderon FJ, Jackson LE (2014) Soil enzyme activities, microbial communities, and carbon and nitrogen availability in organic agroecosystems across an intensively-managed agricultural landscape. Soil Biol Biochem 68:252–262
Brady NC, Weil RR, Weil RR (2008) The nature and properties of soils vol 13. Prentice Hall, Upper Saddle River
Brian PW, Mcgowan JC (1946) Biologically active metabolic products of the Mould Metarrhizium glutinosum S. Pope. Nature 157:334–334
Brian PW, Hemming HG, Jefferys EG (1948) Production of antibiotics by species of Myrothecium Mycologia 40:363–368
Brockett BF, Prescott CE, Grayston SJ (2012) Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil biology biochemistry 44:9–20
Burns RG et al (2013) Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol Biochem 58:216–234
Buyer JS, Teasdale JR, Roberts DP, Zasada IA, Maul JE (2010) Factors affecting soil microbial community structure in tomato cropping systems. Soil Biol Biochem 42:831–841
Carbonetto B, Rascovan N, Alvarez R, Mentaberry A, Vázquez MP (2014) Structure, composition and metagenomic profile of soil microbiomes associated to agricultural land use and tillage systems in Argentine Pampas. PLoS One 9(6):e99949
Cederlund H et al (2014) Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl Soil Ecol 84:62–68
Chin K, Liesack W, Janssen PH (2001) Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int J Syst Evol Microbiol 51:1965–1968
Cramer M, Lewis O (1993) The influence of nitrate and ammonium nutrition on the growth of wheat (Triticum aestivum) and maize (Zea mays) plants. Ann Bot 72:359–365
Curtin D, Campbell C, Jalil A (1998) Effects of acidity on mineralization: pH-dependence of organic matter mineralization in weakly acidic soils Soil. Biology Biochemistry 30:57–64
Daliakopoulos I, Tsanis I, Koutroulis A, Kourgialas N, Varouchakis A, Karatzas G, Ritsema C (2016) The threat of soil salinity: A European scale review. Sci Total Environ 573:727–739
De Klein C, Van Logtestijn R (1994) Denitrification in the top soil of managed grasslands in The Netherlands in relation to soil type and fertilizer level. Plant Soil 163:33–44
Demeyer A, Nkana JCV, Verloo MG (2001) Characteristics of wood ash and influence on soil properties and nutrient uptake: an overview. Bioresour Technol 77:287–295
Deng S, Tabatabai M (1994) Cellulase activity of soils. Soil Biol Biochem 26:1347–1354
DeSantis TZ et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with. ARB Appl Environ Microbiol 72:5069–5072
Dick WA, Cheng L, Wang P (2000) Soil acid and alkaline phosphatase activity as pH adjustment indicators. Soil Biol Biochem 32:1915–1919
Dimitriu PA, Grayston SJ (2010) Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microbial Ecol 59:563–573
Dong L, Xu J, Zhang L, Yang J, Liao B, Li X, Chen S (2017) High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin Med 12:18
Drenovsky R, Vo D, Graham K, Scow K (2004) Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microb Ecol 48:424–430
Edgar RC (2010) Search and clustering orders of magnitude faster than. BLAST Bioinformatics 26:2460–2461
Egidi E et al (2019) A few Ascomycota taxa dominate soil fungal communities worldwide. Nat Commun 10:1–9
Eivazi F, Tabatabai M (1977) Phosphatases in soils. Soil Biol Biochem 9:167–172
Fageria N, Baligar V (2005) Enhancing nitrogen use efficiency in crop plants. Adv Agron 88:97–185
Feizi M, Jalali M, Renella G (2017) Available alkalinity and N mineralization are key factors regulating soil pH value of an organically amended Iranian agricultural soil. Arid Land Res Manag 31:140–158
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579
Flowers T, Bremner J (1991) A rapid dichromate procedure for routine estimation of total nitrogen in soils. Commun Soil Sci Plant Anal 22:1409–1416
Friedel J, Mölter K, Fischer W (1994) Comparison and improvement of methods for determining soil dehydrogenase activity by using triphenyltetrazolium chloride and iodonitrotetrazolium chloride. Biol Fertil Soils 18:291–296
Fritze H, Smolander A, Levula T, Kitunen V, Malkonen E (1994) Wood-ash fertilization and fire treatments in a Scots pine forest stand: Effects on the organic layer, microbial biomass, and microbial activity. Biol Fertil Soils 17:57–63
Gao Z et al (2019) Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front Microbiol 10:2269
He CN, Gao WW, Yang JX, Bi W, Zhang XS, Zhao YJ (2009) Identification of autotoxic compounds from fibrous roots of Panax quinquefolium L. Plant Soil 318:63–72
Henry HA (2013) Reprint of “Soil extracellular enzyme dynamics in a changing climate”. Soil Biol Biochem 56:53–59
Horchani F, Hajri R, Aschi-Smiti S (2010) Effect of ammonium or nitrate nutrition on photosynthesis, growth, and nitrogen assimilation in tomato plants. J Plant Nutr Soil Sci 173:610–617
Huang X, Liu L, Wen T, Zhu R, Zhang J, Cai Z (2015) Illumina MiSeq investigations on the changes of microbial community in the Fusarium oxysporum f. sp. cubense infected soil during and after reductive soil disinfestation. Microbiol Res 181:33–42
Hubbard J, Harman G, Eckenrode C (2011) Interaction of a biological control agent, Chaetomium globosum, with seed coat microflora Canadian. J Microbiol 28:431–437
Hui D, Mayes MA, Wang G (2013) Kinetic parameters of phosphatase: a quantitative synthesis. Soil Biol Biochem 65:105–113
Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560
Jangid K et al (2008) Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem 40:2843–2853
Jia W, Zhou H, Xie W, Guan C, Gao C, Shi Y (2008) Effects of long-term inorganic fertilizer combined with organic manure on microbial biomass C, N and enzyme activity in cinnamon soil . Plant Nutri Fertil Sci 14:700–705
Jian ZY, Wang WQ, Meng L, Zhang ZL (2008) Research progress on continuous cropping obstacles of medicinal plants of ginseng. Mod Chin Med 2008(06):3–5
Jin X, Vekerdy Z, Zhang Y-K, Liu J (2012) Soil salt content and its relationship with crops and groundwater depth in the Yinchuan plain (China) using remote sensing. Arid Land Res Manag 26:227–235
Kandeler E, Gerber H (1988)Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fertil Soils 6:68–72
Kaur J, Singh J (2014) Long-term effects of continuous cropping and different nutrient management practices on the distribution of organic nitrogen in soil under rice-wheat system . Plant Soil Environ 60:63–68
Kemmitt SJ, Wright D, Goulding KW, Jones DL (2006) pH regulation of carbon and nitrogen dynamics in two agricultural soils. Soil Biol Biochem 38:898–911
Kidder GW III, Goddard DR (1965) Studies on the inhibitor resistant respiration of the fungus Myrothecium verrucaria. Plant Physiol 40:552
Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE (2016) The ecology of Acidobacteria: moving beyond genes and genomes. Front Microbiol 7:744
Kitts DD, Wijewickreme AN, Hu C (2000) Antioxidant properties of a North American ginseng extract. Mol Cell Biochem 203:1–10
Koyama A, Wallenstein MD, Simpson RT, Moore JC (2014) Soil bacterial community composition altered by increased nutrient availability in Arctic tundra soils. Front Microbiol 5:516
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental Scale. Appl Environ Microbiol 75:5111–5120
Lee S, Hanlin RT (1999) Phylogenetic relationships of Chaetomium and similar genera based on ribosomal DNA sequences. Mycologia 91:434–442
Li X, Li Y, An S, Zeng Q (2016) Effects of stem and leaf decomposition in typical herbs on soil enzyme activity and microbial diversity in the south Ningxia loess hilly region of Northwest China . Ying Yong Sheng Tai Xue Bao 27:3182–3188
Lin Y, Du D, Si C, Zhao Q, Li Z, Li P (2014) Potential biocontrol Bacillus sp. strains isolated by an improved method from vinegar waste compost exhibit antibiosis against fungal pathogens and promote growth of cucumbers. Biol Control 71:7–15
Lovett GM, Weathers KC, Arthur MA (2002) Control of Nitrogen loss from forested watersheds by soil carbon: Nitrogen ratio and tree species composition. Ecosystems 5:0712–0718
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585
Ma K, Zhang L, Du Q, Song N (2010) Effect of potato continuous cropping on soil microorganism community structure and function. J Soil Water Conserv 24:e33
Ma A, Zhuang X, Wu J, Cui M, Lv D, Liu C, Zhuang G (2013) Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS One 8(6):e66146
Meng P-p et al (2012) Fungal population structure and its biological effect in rhizosphere soil of continuously cropped potato. Ying Yong Sheng Tai Xue Bao 23(11):3079–86
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Omer LS (2004) Small-scale resource heterogeneity among halophytic plant species in an upper salt marsh community. Aquat Bot 78:337–348
Pan Y et al (2014) Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol Ecol 90:195–205
Partel M (2002) Local plant diversity patterns and evolutionary history at the regional scale. Ecology 83:2361–2366
Pathak H, Rao D (1998) Carbon and nitrogen mineralization from added organic matter in saline and alkali soils. Soil Biol Biochem 30:695–702
Pietri JA, Brookes P (2008) Relationships between soil pH and microbial properties in a UK arable soil. Soil Biol Biochem 40:1856–1861
Pu P, Zhang M, Zhang LN (2014) A study on temperature and time conditions of colorimetric method in measuring soil available phosphorus. In: Advanced Materials Research. Trans Tech Publ, Bäch, pp 2047–2051
Qi J, Zhao X, Zhou L, Sun P, Zhang X, Li X (2010) Soil microbial community composition and diversity in Panax quinquefolius rhizosphere. Zhongguo Zhong Yao Za Zhi 35:2378–2382
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160
Reeves D (1997) The role of soil organic matter in maintaining soil quality in continuous cropping systems. Soil Tillage Res 43:131–167
Rousk J et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Sahrawat K, Prasad R (1975) A rapid method for determination of nitrate, nitrite, and ammoniacal nitrogen in soils. Plant Soil 42:305–308
Sait M, Davis KE, Janssen PH (2006) Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl Environ Microbiol 72:1852–1857
Sangwan P, Chen X, Hugenholtz P, Janssen PH (2004) Chthoniobacter flavus gen. nov., sp. nov., the First Pure-Culture Representative of Subdivision Two, Spartobacteria classis nov of the Phylum Verrucomicrobia . Appl Environ Microbiol 70:5875–5881
Schnecker J et al (2015) Microbial community composition shapes enzyme patterns in topsoil and subsoil horizons along a latitudinal transect in Western Siberia. Soil Biol Biochem 83:106–115
Sen S, Chen S, Feng B, Wu Y, Lui E, Chakrabarti S (2012) Preventive effects of North American ginseng (Panax quinquefolium) on diabetic nephropathy. Phytomedicine 19:494–505
Shafi J, Tian H, Ji M (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotechnol Equip 31:446–459
Shrivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci 22:123–131
Singh P, Roy U, Tsuji M (2016) Characterisation of yeast and filamentous fungi from Brøggerbreen glaciers Svalbard. Polar Rec 52:442–449
Sinha AK (1972) Colorimetric assay of catalase. Anal Biochem 47:389–394
Sinsabaugh RL et al (2014) Extracellular enzyme kinetics scale with resource availability. Biogeochemistry 121:287–304
Sinsabaugh RL, Gallo ME, Lauber C, Waldrop MP, Zak DR (2005) Extracellular enzyme activities and soil organic matter dynamics for northern hardwood forests receiving simulated nitrogen deposition. Biogeochemistry 75:201–215
Stott D, Andrews S, Liebig M, Wienhold BJ, Karlen D (2010) Evaluation of β-glucosidase activity as a soil quality indicator for the soil management assessment framework. Soil Sci Soc Am J 74:107–119
Tabatabai M (1994) Soil enzymes. In: Weaver RW et al (ed) Methods of soil analysis, Part 2. Microbiological and Biochemical Properties. SSSA Book Ser. 5. SSSA, Madison, WI, pp 801–834
Tabatabai M, Bremner J (1972) Assay of urease activity in soils. Soil Biol Biochem 4:479–487
Tan Y, Cui Y, Li H, Kuang A, Li X, Wei Y, Ji X (2017) Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol Res 194:10–19
Torres I, García C, Ruiz-Navarro A, Hernández T, Bastida F (2016) The enzymatic and physiological response of the microbial community in semiarid soil to carbon compounds from plants. Eur J Soil Sci 67:456–469
Van Wyk DA, Adeleke R, Rhode OH, Bezuidenhout CC, Mienie C (2017) Ecological guild and enzyme activities of rhizosphere soil microbial communities associated with Bt-maize cultivation under field conditions in North West Province of South Africa. J Basic Microbiol 57:781–792
Wang Y, Li Y, Xiao D (2008) Catchment scale spatial variability of soil salt content in agricultural oasis Northwest China. Environ Geol 56:439–446
Wang H-w, Wang X-x, Lv L-x, Xiao Y, Dai C-c (2012) Effects of applying endophytic fungi on the soil biological characteristics and enzyme activities under continuously cropped peanut. Ying Yong Sheng Tai Xue Bao 23(10):2693–700
Wang L, Yao Y, Sang W, Yang X, Ren G (2015a) Structural features and immunostimulating effects of three acidic polysaccharides isolated from Panax quinquefolius. Int J Biol Macromol 80:77–86
Wang L, Yu X, Yang X, Li Y, Yao Y, Lui EMK, Ren G (2015b) Structural and anti-inflammatory characterization of a novel neutral polysaccharide from North American ginseng (Panax quinquefolius). Int J Biol Macromol 74:12–17
Wang C, Zheng M, Song W, Wen S, Wang B, Zhu C, Shen R (2017) Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol Biochem 113:240–249
Wei L-h (2009) Comparision of decomposition and colorimetry methods in determination of soil total phosphorus. J Liaoning Agric College 11(2):1–3
White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:e1000352
Wichern J, Wichern F, Joergensen RG (2006) Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 137:100–108
Wolińska A, Kuźniar A, Zielenkiewicz U, Izak D, Szafranek-Nakonieczna A, Banach A, Błaszczyk M (2017) Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl Soil Ecol 119:128–137
Wu W, Tang X-P, Yang C, Liu H-B, Guo N-J (2013) Investigation of ecological factors controlling quality of flue-cured tobacco (Nicotiana tabacum L.) using classification methods. Ecol Informatics 16:53–61
Wu Y, Qin C, Lu X, Marchiori J, Feng Q (2016) North American ginseng inhibits myocardial NOX2-ERK1/2 signaling and tumor necrosis factor-α expression in endotoxemia. Pharmacol Res 111:217–225
Xu J, Tang C, Chen ZL (2006) The role of plant residues in pH change of acid soils differing in initial pH. Soil Biol Biochem 38:709–719
Yeomans JC, Bremner JM (1988) A rapid and precise method for routine determination of organic carbon in soil. Commun Soil Sci Plant Anal 19:1467–1476
Ying Y, Ding W, Zhou Y, Li Y (2012) Influence of Panax ginseng continuous cropping on metabolic function of soil microbial communities. Chin Herbal Med 4:329–334
You J, Liu X, Zhang B et al (2015) Seasonal changes in soil acidity and related properties in ginseng artificial bed soils under a plastic shade. J Ginseng Res 39(1):81–88
Yu X, Yang X, Cui B, Wang L, Ren G (2014) Antioxidant and immunoregulatory activity of alkali-extractable polysaccharides from North American ginseng. Int J Biol Macromol 65:357–361
Yu D, Wen Z, Li X, Song X, Wu H, Yang P (2018) Effects of straw return on bacterial communities in a wheat-maize rotation system in the North China Plain. PLoS One 13(6):e0198087
Zhalnina K et al (2015) Soil pH determines microbial diversity and composition in the park grass experiment. Microb Ecol 69:395–406
Zhang XS (2013) Abatement effect of crop rotation on replant problem of Panax quinquefolium L. (Doctoral dissertation). Chinese Academy Of Medical Sciences & Peking Union Medical College, Beijing
Zhang D (2019) Global and Chinese American ginseng overall scale analysis. In: Zhang D (ed) 2019 American ginseng global and Chinese market deep research report. QYResearch, Beijing, pp 94–96
Zhang Y et al (2014) Community structure and elevational diversity patterns of soil Acidobacteria. J Environ Sci 26:1717–1724
Zhang ZY, Lin WX (2009) Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants . Chin J Eco Agric 17:189–196
Zhang X-l, Pan Z-g, Zhou X-f, NI W-z (2007) Autotoxicity and continuous cropping obstacles: A review . Chin J Soil Sci 4:33
Zhang G-K, Song X-Y, Liu S-T, Nan Z-W, Jiang W (2015) Effect of long-term application on nitrogen utilization and yield quality of wheat and maize. Acta Agric Boreal Sin 30:157–161
Zhang J, Fan S, Qin J, Dai J, Zhao F, Gao L, Lian X, Shang W, Xu X, Hu X (2020) Changes in the microbiome in the soil of an American ginseng continuous plantation. Front Plant Sci 11:572199
Zhao XM (2009) The effect of continous cropping on the growth of American ginseng and allelochemieal (Doctoral dissertation). Chinese Academy Of Medical Sciences & Peking Union Medical College, Beijing
Zheng Q et al (2019) Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol Biochem 136:107521
Zydlik Z, Zydlik P (2013) The effect of microbiological products on soil properties in the conditions of replant disease. Zemdirbyste-Agriculture 100:19–24
Funding
This work was supported by National Natural Science Foundation of China [grant numbers 31670299]; the Fundamental Research Funds for the Central Universities [grant numbers GK202103065 and GK201806006]; the National Key Technologies R & D Program for Modernization of Traditional Chinese Medicine [grant numbers 2017YFC1701300 and 2017YFC1700706]; and the Key R&D Program of Shaanxi Province [grant numbers 2019SF-307].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Conceptualization: Shuai Liu, Jun-feng Niu and Shi-qiang Wang; Methodology: Shuai Liu and Shu-ke Zhang; Writing-Original Draft: Shuai Liu; Formal analysis: Shuai Liu, Zhan-yu Wang and Kai-Kai Dang; Resources: Shuai Liu and Shi-qiang Wang; Data Curation: Zhan-yu Wang, Kai-Kai Dang and Shu-ke Zhang; Investigation: Zhan-yu Wang and Kai-Kai Dang; Funding acquisition: Jun-feng Niu, Shi-qiang Wang and Zhe-zhi Wang; Writing- Reviewing and Editing: Jun-feng Niu and Zhe-zhi Wang. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Sven Marhan.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Fig. S1 Rarefaction curve (a, b) and rank abundance curve (c, d) of bacterial (a, c) and fungal (b, d) OUT of 15 soil samples. Fig. S2 Changes in alpha diversity of microbial community. Bacterial chao 1 (a), bacterial ACE (b), bacterial Shannon index (c), bacterial Simpson index (d), fungal chao 1 (e), fungal ACE (f), fungal Shannon index (g), fungal Simpson index (h). Data are means ± standard deviation (n=3). Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a significant difference at p ≤ 0.01. Fig. S3 Principal coordinates analysis (PCoA) analysis based on Bray-Curtis. Dissimilarity displaying sites using unweighted UniFrac analysis for bacterial (a, b, c) and fungal (d, e, f) community composition in five group soils. Fig. S4 Nonmetric multidimensional scaling (NMDS) analysis based on Bray-Curtis. Dissimilarity displaying sites using unweighted UniFrac analysis for bacterial (a) and fungal (b) community composition in the five soil groups. Black up triangles and orange squares represent never planted American ginseng farmland (group A), red crosses and red circles represent recently harvested American ginseng farmland (group B), green X marks and blue up triangles represent replanted maize for 2 years after harvesting American ginseng (group C), blue diamonds and green diamonds represent replanted maize for 4 years after harvesting American ginseng (group D), whereas blueness down represent and circles represent replanted maize for 6 years after harvesting American ginseng (group E) in Fig. a and b, respectively. Fig. S5 Significantly decreased bacterial genera of recently harvested American ginseng farmland (Group B) compared with never planted American ginseng farmland (Group A). Data are means ± standard deviation (n=3). Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a significant difference at p ≤ 0.01. Fig. S6 Significantly increased bacterial genera of recently harvested American ginseng farmland (Group B) compared with never planted American ginseng farmland (Group A). Data are means ± standard deviation (n=3). Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a significant difference at p ≤ 0.01. Fig. S7 Significantly decreased fungal genera of recently harvested American ginseng farmland (Group B) compared with never planted American ginseng farmland (Group A). Data are means ± standard deviation (n=3). Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a significant difference at p ≤ 0.01. Fig. S8 Significantly increased fungal genera of recently harvested American ginseng farmland (Group B) compared with never planted American ginseng farmland (Group A). Data are means ± standard deviation (n=3). Different lowercase letters (a, b, c) indicate a significant difference at p ≤ 0.05, whereas different uppercase letters (A, B, C) indicate a significant difference at p ≤ 0.01. (DOCX 930 KB)
ESM 2
(DOCX 45.4 KB)
Rights and permissions
About this article
Cite this article
Liu, S., Wang, Z., Niu, J. et al. Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 463, 427–446 (2021). https://doi.org/10.1007/s11104-021-04911-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-021-04911-2