Abstract
Background and aims
Diazotrophic bacteria, including those of the genus Azospirillum and Nitrospirillum, colonize Brachiaria genotypes and contribute to plant development through nitrogen fixation, production of phytohormones and bioavailability of nutrients. This study aimed to determine the phylogenetic positioning and evaluate the functional abilities of diazotrophic bacteria isolated from Brachiaria genotypes.
Methods
Diazotrophic bacterial counting and isolation were carried out with rhizosphere soil and root samples from 20 Brachiaria genotypes after inoculation in nitrogen-free semi-solid NFb and LGI media. The isolates were analyzed using 16S rRNA and nifH sequences, and tested for their functional abilities to produce auxin and siderophores, to solubilize phosphate and zinc, and to degrade cellulose.
Results
The diazotrophic population ranged from 102 to 108 g−1 rhizosphere soil or roots. Sequencing of 16S rRNA from 213 isolates confirmed the presence of genera Azospirillum and Nitrospirillum, and revealed the presence of 14 other diazotrophic genera. The genus Nitrospirillum was detected colonizing all niches of most Brachiaria species. A PCA analysis showed a positive correlation between the ability to produce siderophores with the ability to produce IAA; and between phosphate and zinc solubilisation.
Conclusions
The results showed a high diversity of diazotrophic bacterial species colonizing 20 Brachiaria genotypes and revealed the presence of bacteria with variable growth-promoting characteristics, highlighting their potential as good candidates for the development of biofertilizers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pasturelands represent one of the largest ecosystems in Brazil (Guarda and Guarda 2014), with the genus Brachiaria occupying about 260 million ha, equivalent approximately to 80% of tropical and subtropical regions of the Brazilian pastures (Valle et al. 2010). This forage grass, constituted by around 100 species, is responsible for most of the 221 million cattle that supply the domestic demand for meat and the exportation to different countries (USDA 2018). Despite its economic importance, it is estimated that between 50 to 70% of pastures in Brazil present different degrees of degradation (Dias-Filho 2011). This phenomenon can mainly be explained by the low natural soil fertility of tropical regions and the lack of nutrient replacements (Macedo 2005) such as nitrogen, which is considered to be the most limiting element to productivity of Brachiaria spp. (Boddey et al., 2004). However, the use of nitrogen fertilizers greatly affects the cost of production, and due to the increasing demand for food, sustainable alternatives such as Biological Nitrogen Fixation (BNF) have to be used. BNF is known to be performed by diazotrophic bacteria that can colonize the rhizosphere, the surface and/or the interior of plant tissues (Compant et al. 2010).
In a review published by Baldani and Baldani (2005) related to the BNF process in forage grasses, the authors indicate that BNF is a renewable and continuous source of nitrogen. In this way, BNF in pastures may play an important role in places where the practice of inoculation with specific associative diazotrophic bacteria may substitute, at least partially, the chemical nitrogen source in intensive systems. Azospirillum spp. and Nitrospirillum amazonense (older Azospirillum amazonense), are among the most studied groups of diazotrophic bacteria that have been found to be associated with several plant families (Reis et al. 2015), presenting a wide distribution, as they are found in the soil, roots, leaves and stems of plants (Baldani et al. 2014). These genera have been isolated from several forage grasses such as Brachiaria sp. (Reis et al. 2001; Reis Junior et al. 2004; Brasil et al. 2005; Silva et al. 2013), Panicum maximum (Santos et al. 2013), Axonopus purpusii, Hymenachne amplexicaulis and Mesosetum chaseae (Souza et al. 2017), Miscanthus sinensis cv. giganteus (Eckert et al. 2001) and Pennisetum purpureum (Videira et al. 2012). Other diazotrophic bacterial genera have also been isolated from Brachiaria grown in Northeast region (Caatinga biome) of Brazil, and these include Burkholderia sp., Bacillus sp., Enterobacter sp., Klebsiella sp., Sphingomonas sp., Pantoea sp. and Rhizobium sp. (Oliveira et al. 2018). Similarly, a high diverse non-nitrogen-fixing bacterial community belonging to genera Pseudomonas, Pantoea, Acinetobacter and Enterobacter was detected in different Brachiaria plants naturally grown in soil in Nairobi, Kenya (Mutai et al. 2017).
Diazotrophic bacteria can play an important role in the rehabilitation and sustainability of pastures, since they incorporate N through the biological fixation process in amounts that vary from 25 to 50 kg ha−1. These amounts have been measured in Brachiaria species and Panicum maximum genotypes grown in cylinders filled with 15N labelled soil maintained outside the greenhouse (Boddey and Victoria 1986; Miranda et al. 1990). In addition, diazotrophic bacteria possess the capacity to produce and release plant growth regulators, such as auxins (Reis Junior et al. 2004; Videira et al. 2012), solubilize inorganic phosphate (Rodriguez et al. 2004; López-Ortega et al. 2013), and produce siderophores (Tortora et al. 2011). Further studies on the potential of these diazotrophic bacteria in association with forage grasses are therefore needed in order to understand the role of these microorganisms in plant growth-promoting activities. Recently, a commercial inoculant containing two variants (Abv5 and Abv6) of Azospirillum brasilense strain Sp7 was applied with success in maize (Hungria et al., 2010) and later its use was extended to Brachiaria pastures. However, these increments occur mostly when inoculant is applied together a half dose of nitrogen fertilizer required by the crop (Hungria et al. 2016), suggesting that other mechanisms than the BNF was acting in the plant-bacteria interaction. Despite of that, it should be chased the development of an inoculant containing efficient nitrogen-fixing strains in addition to other biotechnological functionalities. In this sense, we searched for the occurrence of diazotrophic bacteria (associative and endophytic) in twenty Brachiaria genotypes, expecting to find out high diversity of bacterial species/strains containing these functionalities and that could be more efficient when applied in future inoculation studies with some of these Brachiaria genotypes that occupy around 80% of the Brazilian pastures. Our hypothesis is that Brachiaria genotypes could establish a more specific diazotrophic association considering that the plants have been maintained in the same Germoplasm Active Bank for around ten years.
Material and methods
Origin of the genotypes
The study comprised the analysis of 20 Brachiaria genotypes collected at the Germplasm Active Bank (GAB) of the Centro Nacional de Pesquisa de Gado de Corte (CNPGC), part of the Embrapa project number 02.13.08.004.00.00. The genotypes were B. decumbens (Basilisk, D24 / 27, D24 / 45, D24 / 2, 79–10, X009, B001), B. brizantha (Marandu, Paiaguás, Piatã, Xaraés and B140), B. humidicola (Common, Tupi, Ipyporã, H47), B. ruziziensis (Common), B. arrecta (A2) and Brachiaria spp. (Llanero and Mulato II). The GAB has been maintained for up to 10 years at the same place (geographic coordinates 20°27’ S and 54°37’ W) in a Typical Red Clay Latosol. The GAB is fertilized annualy with phosphorus and potassium to maintain the equivalent to 4–5 mg P dm3 and 50–60 mg K dm3), respectively. In addition, nitrogen (70 Kg N ha−1), sulfur (30 kg S ha−1), micronutrients (40 kg FTE-BR10 ha−1) and dolomitic lime (3 ton ha−1) were applied every year to maintain the nutrient plant demand.
Counting and isolation of diazotrophic bacteria
The plant samples containing rhizosphere soil of the 20 genotypes were transported to Embrapa Agrobiologia, and then properly labeled and transplanted to 6 kg pots containing sterile sand and vermiculite (1:1) and kept in a greenhouse. Nutrient fertilization was performed every 15 days with 30 mL of KH2PO4 (35 g L−1); 5 mL of CaCl2.2H2O (150 g L−1) and 5 mL of MnSO4.H2O (5 g L−1), totalizing six fertilizations, and the pots remained in the greenhouse for up three months.
Counting and isolation followed the methodology described by Baldani et al. (2014) using nitrogen-free LGI and NFb semi-solid media. The NFb (Azospirillum species) and LGI (Nitrospirillum amazonense) semi-solid media were chosen based on the widespread occurrence of these diazotrophic species in high numbers in association with Brachiaria species in Brazil (see introduction). Briefly, the bulk soil close to roots was removed and the rhizospheric soil (soil adherent to roots) was taken for analysis. The roots were washed in tap water to remove the remained soil residues and processed with or without surface disinfection. For the root disinfection process, a concentration of 10 g L−1 of Chloramine-T (CH3C6H4SO2NNaCl.3H2O) was used for 5 min. After disinfection time, the roots were placed in sterile distilled water for 1 min and 30 s, and then transferred to phosphate buffer solution (50 mM, pH 7.0) for 1 min and 30 s, and again in distilled water for 5 min. The disinfected roots were used for the isolation of diazotrophic bacteria considered endophytic. For non-disinfected roots, the samples (10 g fresh tissues) were kept in distilled water for the same time as the one used in the disinfection process. The samples (10 g umid soil or fresh roots) were mixed with 90 mL saline solution, agitated to homogenize it for 5 to 10 min on a rotary shaker (~150 rpm) and then diluted up to 10−7, inoculated in both nitrogen free semi-solid media and incubated at 30 °C for 5 days. Flasks (1 to 3) of the highest counting dilution/repetition, showing subsurface veil pellicles characteristic of diazotrophic bacteria, were transferred to new semi-solid media and purified following the indicated methodology (see Baldani et al. 2014). The final number of diazotrophic bacterial isolates varied according to the richness of the target bacteria in the veil pellicle and the success during the steps of purification due to the presence of scavenger bacteria. Purified cultures were then stored in duplicates in appropriate slant media under mineral oil.
Phylogenetic positioning of the isolates
Amplification of the 16S rRNA gene fragment with the primers 27f (5′-AGA GTTTA TCC TG CTG AG-3′) (Furushita et al. 2003) and Amp2 (5′-AAG GAG GT ATC CAT CCG CA-3′) (Wang et al. 1996) was carried out as described by Videira et al. (2012). For amplification of the nifH gene the primers PolF (5’-TC GAY CCS AAR GCB GAC TC-3′) and PolR (5’-ATS GCC ATC ATY TCR CCG GA-3′) were used (Poly et al. 2001). PCR products were purified with the Exo-SAP kit, according to the manufacturer’s description.. PCR products were sequenced on the ABI PRISM 3500 Genetic Analyzer automatic sequencer (Applied Biosystems) using the commercial Big Dye® Terminator Cycle Sequencing v3.1 kit from Thermo Fisher Scientific (Cat. No.4337456).
The amplicons were sequenced at both ends (forward and reverse), using the same PCR pair of primers. Sequences of the 16S rRNA and nifH genes were analyzed and assembled using the program BioEdit version 7.1.9 (Hall 1999) to check the quality of the amplified sequencing products. A visual inspection of the chromatograms was carried out to identify the sequence quality of the fragments and those with lower quality were deleted. The sequences were then aligned and compared with those deposited in the GenBank database using the BLAST program (Altschul et al. 1997). Phylogenetic trees with 16S rRNA (~1400 bp) and nifH (~ 360 bp) sequences were constructed to verify their phylogenetic position in relation to the species type of each identified strain. Multiple sequence alignment was done using the ClustalX program (Tuimala, 2004) and the sequences were processed by the MEGA6 program (Tamura et al. 2013). The consensus tree was constructed by the Neighbor-Joining method, considering a bootstrap of 1000 replicates. Sequencing of the nifH amplified fragments was performed for the 32 strains selected according to their variable functional characteristics and the representative taxonomical species.
Functional capacity of the strains
Biological nitrogen fixation- qualitative evaluation
The ability of the isolates to perform the BNF process was evaluated qualitatively in penicillin flasks containing 7 mL of nitrogen free NFb or LGI semisolid media. The inocula cultures were grown in their respective semi-solid medium. The flasks were incubated for 72 h at 30 °C in the dark followed by analysis of the growth pellicle formed near the surface of the bottles. Three replicates were performed for each bacterial isolate, and this procedure was repeated at least three times.
Solubilization of inorganic phosphate and zinc oxide
The solubility of P by the isolates was evaluated in a NBRIP solid medium (Nautiyal 1999), containing insoluble β-tricalcium phosphate [Ca3(PO4)2] or zinc oxide (ZnO), as the sole source of phosphorus and zinc, respectively. A total of 10 μL of bacterial cultures was inoculated onto NBRIP agar plates. The appearance of a clear halo around the colonies on the 12th and 15th days after inoculation was used to measure the phosphate and zinc oxide solubilization, respectively. The criteria used to calculate the solubilization index (SI) were those described by Kumar and Narula (1999). Each isolate was tested three times in NBRIP agar medium to confirm the phenotype of phosphate and zinc solubilization. The strain Gluconacetobacter diazotrophicus PAL5 (BR11281) was used as positive control.
Production of siderophores
The production of siderophores was evaluated using the chromium azurol S (CAS) method adapted from Schwyn and Neilands (1987). A characteristic colony of each isolate was inoculated in a test tube containing 5 mL of NFB or LGI liquid medium, without Fe3+ iron and 1.0 g L−1 of (NH4)2SO4 for induction of the production of siderophores or chelating compounds of iron. The tubes were incubated at 180 rpm, 30 °C for 48 h. After incubation, three aliquots of 10 μL each were inoculated onto NFb or LGI plate [without Fe3+ iron and 1.0 g L−1 of (NH4)2SO4] containing 1% CAS. Bacteria were incubated for 3 days at 30 °C. The intensity of the production of siderophores was classified as low, medium or high according to the size of the halo. It was also evaluated the type of siderophores produced by the isolates, being the blue/yellow color for catecholate type and blue/pink color for hydroxamate type (Schwyn and Neilands 1987).
Cellulolytic activity
The cellulolytic activity of the bacterial strains was evaluated according to Kasana et al. (2008). Ten μl culture was inoculated in triplicate onto a plate containing CMC agar medium [(0.2% NaNO3, 0.02% K2HPO4, 0.05% MgSO4, 0.5% KCl, 0.2% sodium CMC (carboxymethylcellulose), 0.02% peptone and 1.7% agar)]. Plates were incubated for four days at 30 °C. After incubation, the plates were flooded with iodine solution (2.0 g KI, 1.0 g iodine diluted in 300 mL distilled water) for five minutes. The excess solution was drained and the presence of halo around the bacterial colonies indicated a positive result for the cellulolytic activity. The halo was measured with the aid of a digital caliper and the cellulolytic activity index (CI) was calculated as proposed by Hankin and Anagnostakis (1975) and classified according to Dantur et al. (2015).
Production of indole compounds
Bacterial strains were analyzed for the synthesis of indole compounds according to the microplate method proposed by Sarwar and Kremer (1995). Each bacterial isolate was cultured in a tube containing 5 mL of liquid DYGS for 24 h under 30 °C. Subsequently, a 20 μL aliquot of each tube was transferred to another tube containing liquid NFb or LGI culture medium, both without the pH indicator, and L-tryptophan (100 μg mL−1) was added and incubated under shaking at 180 rpm for a period of 72 h, in an environment without light and with an ambient temperature of 30 °C. Aliquots (1 mL) were then withdrawn from each tube and centrifuged at 12,000 rpm for 5 min. After that, 150 μL of the supernatant from each sample was added to three wells (96 wells) and mixed with 100 μL of the Salkowski reagent. The samples were incubated in the dark for 30 min at room temperature and after this period, readings were taken using a spectrophotometer (Labsystems iEMS Reader MF) at a wavelength of 540 nm and the absorbance data were processed by the program Ascent for iEMS Reader MF. The strain Azospirillum brasilense Sp245 was used as a positive control. The readings were standardized by means of a standard curve previously obtained with known concentrations of 3-indole-acetic acid (25 to 600 μg mL−1). All samples were analyzed in triplicate and the result was an average of three readings.
Statistical analysis
The functional information was used to calculate a Principal Component Analysis (PCA) displaying how similar the different isolates are in their functional potential. The characteristics included in the analysis were the production of siderophores, production of IAA, Zn solubilization, and the ability of consuming CMC. The PCA was calculated with the correlation matrix since the variables are expressed in different units. The correlations between the different variables and the coordinate axes was displayed in the form of arrows in the PCA plot. All analyses were done with the package vegan Oksanen et al. (2019) implemented in the R platform (R Core Team 2018).
Results
MPN count and isolation of diazotrophic bacteria in semi-solid NFb and LGI media
The estimated number of diazotrophic bacteria present in the rhizosphere soil and in the roots of the plant genotypes differed according to the sample used for isolation [non-disinfected root (NDR), surface disinfected roots (DR) and rhizosphere soil (RS)], as well as the semi-solid medium (NFb and LGI). In general, the population size of viable diazotrophic bacteria colonizing the 20 Brachiaria genotypes ranged from 102 to 108 bacteria per gram of fresh soil or fresh root tissues. The largest population of diazotrophic bacteria was observed in the B. decumbens genotype Basilisk counted in NFb medium and B. humidicola genotype H47 counted in the LGI medium (Table 1). In contrast, lower diazotrophic population or no detection (below the minimum detection level) was observed in some genotypes, mainly in the B. ruziziensis R134 and B. humidicola genotype Tupi counted in both semisolid media. Concerning the niche occupation in the brachiaria forages, the bacterial counting showed a tendence of similar bacterial population in the non-disinfected and disinfected roots, except for disinfected roots that were slightly higher for Basilisk and H47. The rhizospheric soil diazotrophic population was lower as compared to roots and in some cases, non-detectable population was observed for some genotypes counted in both culture media (Table 1).
A total of 213 diazotrophic isolates (110 in NFb medium and 103 in LGI medium) were obtained from the different niches (rhizospheric soil, surface disinfected root and non-disinfected root) of the 20 brachiaria genotypes (Table 1). The highest percentage (22.53%) was obtained from NDR followed by 18.78% for RS and 10.32% for the DR isolated in semi-solid NFb medium, while in the LGI medium the percentage of isolates was 20.65% for NDR, 14.55% for DR and 13.14% for RS. The sum of isolates obtained in both media also varied with the forage grass genotype, with the genotypes B. decumbens cv. X009 (25), cv. 24 /27 (20), cv 79–10 (19) and cv. D24/45 (18) showing the highest number of isolates. In contrast, Brachiaria ruziziensis cv. R134 (1), B. decumbens cv. D24/2 (3) and B. humidicola cv. Tupi (4) presented the lowest number of isolates (Table 1). The number of bacterial isolates also varied with Brachiaria species, with an average quite similar (14.6 and 13), for B. decumbens and B. arrecta, respectively followed by Brachiaria spp, B. brizantha and B. humidicola with an average of 9.5, 9.4 and 7.8, respectively while only one representing bacterial isolate was obtained from B. ruziziensis (Table 1).
A Venn dendrogram analysis involving the niche of the diazotrophic bacterial genera isolation and each group of Brachiaria genotypes (B. brizantha, B. humidicola, B. decumbens and Brachiaria spp plus B. arrecta) showed the occurrence of the bacterial genera randomized among the Brachiaria species (Fig. S1). The genera Nitrospirillum and Azospirillum were common to to all three niches of the B. brizantha and B. decumbens while Nitrospirillum genus was also common to the three niches of the other Brachiaria species. The B. decumbens presented the highest number of genera colonizing the three niches while no single genus was found colonizing only the rhizosphere soil of B. brizantha and B. decumbens. The genus Pseudomonas seems to be more commonly found colonizing the root interior of Brachiaria species, except for the Brachiaria spp. The genus Rhizobium was also found colonizing the root interior of the Brachairia species: B. decumbens, B. humidicola and Brachiaria spp. In contrast, the genus Herbaspirillum was detected associated with the rhizosphere soil of B. brizantha while the Lysinobacillus was associated with B. humidicola (Fig. S1).
Molecular characterization and diazotrophic bacterial diversity analysis
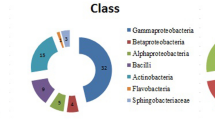
Sequences of the 16S rRNA subunit from the bacterial isolates showed a high level of similarity (97% and 99%) to sequences deposited in the NCBI database. It was possible to taxonomically identify 213 isolates at the genus level, indicating a high bacterial diversity constituted by 16 genera, belonging to the Proteobacteria classes (alpha, beta and gamma) and Bacilli, therefore confirming the semi-selectivity of the semisolid NFb and LGI culture media (Fig. 1). The genus Nitrospirillum (36.3%), followed by Azospirillum (33.0%), Bacillus and Stenotrophomonas (6.5%), respectively were highly represented among the isolates of the 20 Brachiaria genotypes at harvest time. Other genera with lower number of isolates were also found associated with these Brachiaria genotypes including Rhizobium, Pseudomonas, Paraburkholderia, Gluconacetobacter, Bosea, Kosakonia, Zoogloea, Herbaspirillum, Lysinibacillus, Ochrobactrum, Phytobacter and Sphingobium (Fig. 1). The sequences of these 213 strains were submitted to the Genbank® database (National Center for Biotechnology Information - NCBI) under accession numbers MK542907 to MK543119.
Phylogenetic analyses indicated the presence of 78 strains closely related to Nitrospirillum amazonense species, 8 strains to Azospirillum brasilense, 27 strains to A. lipoferum, 24 strains to A. formosense, 6 strains to A.oryzae and 3 strains to A. melinis (Fig. 2). In addition to these phylogenetically related species, sequences close to Paraburkholderia tropica, Paraburkholderia silvatlantica, Gluconacetobacter diazotrophicus, Herbaspirillum seropedicae and Stenotrophomonas pavanii were also identified (Fig. 2). Other bacteria phylogenetically close to the species Stenotrophomonas maltophilia, Phytobacter diazotrophicus and Ochrobactrum anthropi were identified among the diazotrophic isolated strains. Strains taxonomically related to genera Bacillus, Lysinibacillus, Pseudomonas, Rhizobium, Sphingobium, Bosea, Kosakonia and Zoogloa were also identified (Fig. 2 and Table S1).
Phylogenetic tree based on 16S rRNA gene sequences (~ 1450 bp), including the bacterial type strain deposited in the NCBI database. Phylogeny was based on clustering of sequences according to the Neighbor-joining algorithm and Kimura model using the MEGA6.1 program. Numbers located in the branches indicate the percentage of 1000 sub-samples (bootstrap). The scale represents the number of mutations per nucleotide position. The Frankia alni 16S rRNA gene sequence was used for tree rooting
Although all strains had been isolated using the nitrogen-free semi-solid media and presumily considered diazotrophic bacteria, the 213 strains were subjected to amplification of the nifH gene using primers described by Poly et al. (2001). The PCR reaction showed amplified product of the expected size (~ 360 bp) for most of the strains, except for 33 strains that did not show the target amplified gene (data not shown). Analysis of the nifH gene fragments of 32 representative strains showed that only 2 (Pseudomonas nitritireducens strain NRB021 and Bacillus aryabhattai strain NRB108) did not present the band of the expected size. Sequencing of strains previously identified based on the 16S rRNA gene indicated that 8 strains present 100% similarity with nifH gene of species Nitrospirillum amazonense (Fig. 3a). This high identity was also observed for a strain from Burkholderia silvatlantica, Stenotrophomonas maltophilia, Phytobacter sp. and Pseudomonas kuykendallii (Fig. 3), thus corroborating the results obtained in the analysis of the 16S rRNA gene (Fig. 3b). In addition, sequencing of the nifH gene from 6 strains showed that they are closely related to species A. brasilense and/or A. formosense as identified by the 16S rRNA. The other strains showed similarity with the nifH gene of A. melinis, A. lipoferum and A. oryzae and to that observed for the 16S rRNA gene, but also presented distinct clusters (Fig. 3a, b).
Comparison of the phylogenetic trees based on sequences of the nifH gene (~ 360 bp) (a) and the 16S rRNA (b) of a group of strains selected based on their physiological and phylogenetically positioning among the 213 characterized strains. Phylogeny was based on clustering of sequences according to the Neighbor-joining algorithm and Kimura model using the MEGA6.1 program. Numbers located in the branches indicate the percentage of 1000 sub-samples (bootstrap). The scale represents the number of mutations per nucleotide position. The Frankia alni nifH and 16S rRNA gene sequences were used for tree rooting
Evaluation of the functional capacity of strains
Plant growth-promoting characteristics (solubilization of Pi and Zn, cellulolytic capacity and, siderophore and indole production) were tested among the 213 diazotrophic strains aiming to identify strains with the potential to be used as biofertilizers in some Brachiaria genotypes studied here.
Solubilization of inorganic phosphate and zinc oxide
Out of the 213 strains, 84 (39.44%) presented the capacity to solubilize inorganic phosphate (Fig. 4a). According to Madhaiyan et al. (2004), the phosphate solubilization index (SI) can be classified into levels, high (SI > 3), medium (2 < SI <3) and low (SI <2). Out of the strains that showed the capacity to solubilize inorganic phosphate, 8.33% presented a medium level and the rest (91.67%) showed a low level of Pi solubilization (Fig. 4a). Among the strains classified as medium SI, the strain closely related to A. formosense NRB004 presented the highest SI (2.6) followed by Rhizobium pusense NRB010, Pseudomonas nitritereducens NRB075, Bosea thiooxidans NRB076, A. lipoferum NRB085, N. amazonense NRB180 and A. formosense NRB214 that presented a SI greater than 2.0. (Fig. 4b, Table S1). In contrast, the R. pusense NRB009 (1.01) and Bacillus aerius NRB031 (1.12) showed the smallest SI Few strains phylogenetically related to species A. brasilense, A. melinis, A. oryzae, Stenotrophomonas maltophilia, Phytobacter diazotrophicus and P. silvatlantica also presented low SI (Fig. 4b, Table S1).
Diazotrophic bacteria isolated from 20 Brachiaria genotypes showing different functional characteristics. Bacteria with phosphate solubilization activity (a), ZnO activity (c), siderophore production (e), cellulolytic activity (g), indol acetic acid activity (i) and the closely related bacterial species for each function (b, d, f, h, j), respectively)
In relation to the ability of a strain to solubilize zinc oxide, only 20 strains showed this functional activity, of which 10 strains showed high ability, 9 presented low and 1 showed a medium ability to solubilize ZnO (Fig. 4c). Among the bacterial species, the highest number of strains was observed for the phylogenetically related to A. formosense species followed by A. lipoferum, A. brasilense, N. amazonense and Stenotrophomonas maltophilia (Fig. 4d). Many of these strains also have the capacity to solubilize inorganic phosphate. The strain phylogenetically related to A. formosense NRB082 showed the highest zinc oxide solubility index (5.13), superior to strain PAL5 (4.51) used as positive control. An example of in vitro halo formation characteristic of solubilization of inorganic phosphate (A) and zinc oxide (B) by strains NRB081 and NRB082 grown in NBRIP medium containing insoluble β-tricalcium phosphate (Ca3(PO4)2) or zinc oxide (ZnO) is shown in Fig. S2.
Production of siderophores
Out the 213 strains, 179 strains (84.04%) showed a change in blue/yellowish color (Fig. S3) or blue/pink (Fig. S4) on the CAS plate medium indicating the production of siderophores. One hundred and twenty-two strains (68.16%) presented low intensity, forty-eight (26.82%) showed medium intensity, and nine (5.03%) indicated high production of siderophores (Fig. 4e). Among the bacterial species, the majority of strains was closely related to N. amazonense species followed by A. formosense and A. lipoferum species (Fig. 4f). Many other strains phylogenetically related to other genera such as Zoogloea, Stenotrophomonas, Sphingobium, Rhizobium, Pseudomonas, Phytobacter, Paraburkholderia, Ochrobactrum, Lysinibacillus, Kosakonia, Herbaspirillum, Bosea, Bacillus were also detected as siderophore producer (Fig. 4f).
The highest catecholate type siderophore production intensities were observed with phylogenetically related to strains of P. nitritireducens, A. lipoferum, R. pusense, P. silvatlantica, N. amazonense, Zoogloea oryzae and A. formosense (Fig. 4f and Table S1). The production of siderophore was so intense in A. lipoferum strain NRB034 that it completely altered the color of the agar plate culture medium (Fig. S3). On the other hand, all strains obtained using the semi-solid LGI medium produced hydroxamate -type siderophores since the final color of the plate medium was pink (Schwyn and Neilands 1987) (Fig. S4).
Determination of cellulolytic activity
Out of the 213 tested strains, 69 (32.39%) presented cellulolytic capacity, including representatives phylogenetically related to genera Azospirillum (38), Nitrospirillum (10), Bacillus (6), Pseudomonas (5), Rhizobium (3), Stenotrophomonas (3), Paraburkholderia (2), Ochrobactrum (1) and Bosea (1) (Fig. 4g). Among the 38 strains belonging to Azospirillum genus, one strain presented high activity, 15 showed medium activity while 25 had low cellulolytic capacity (Fig. 4h). The NRB011 strain (A. formosense) presented the highest cellulolytic activity with a cellulolytic index (CI) of 6.8, followed by the strains NRB033 (A. lipoferum) and NRB214 (A. formosense), which presented a CI of 4.92 and 4.62, respectively. However, only the strain NRB011 showed a CI higher than the positive control G. diazotrophicus PAL5 (5.38). The strains with the lowest rates of cellulose degradation were closely related to the genus Azospirillum (NRB027) and Stenotrophomonas (NR032 and NRB008). Concerning to the genus Nitrospirillum, seven strains showed medium cellulolytic capacity and three with low, with a CI ranging from 1.21 to 3.88. Examples of in vitro cellulolytic activity of strains isolated from Brachiaria genotypes are presented in Fig. S5.
Production of indole acetic acid (IAA)
Among the 213 strains tested for their ability to produce this phytohormone, it was observed that only 17.37% were able to produce indoles (Fig. 4i), mainly strains related to the Rhizobium pusense, A. lipoferum and A. brasilense species. Other species closey related to genera Stenotrophomonas, Bacillus, Pseudomonas and Ochrobactrum were also detected producing indols (Fig. 4j). The intrinsic ability to produce these substances in the presence of tryptophan varied greatly among strains. Values ranged from 13.96 to 470.53 μg IAA mg−1 protein, with an average of 137.87 for Azospirillum strains, 187.48 for Rizobium, 327.75 for Bacillus, 113.45 for Stenotrophomonas, 84.75 for Pseudomonas and 13.96 μg for Ochrobactrum strain (Table S1).
Overall, analysis of of the functional characteristics of the 213 diazotrophic bacterial strains showed that 39% were able to solubilize inorganic phosphate, 9% solubilized zinc oxide, 84% produced siderophores, 32% had cellulolytic activity, 18% were capable of producing indoles and 85% presented the fragment of the nifH gene of the expected size (Table S1). There was no clear relationship between the phylogenetic positioning of the isolate, from where it was isolated, and the presence of a given function. However, the PCA allowed us to identify some patterns such as the positive correlation between the ability to produce siderophores with the ability to produce IAA; and between phosphate and Zinc solubilisation Fig. 5). This correlation is indicated by the small angle between the arrows in the PCA biplot. A group of isolates with NRB023 (R. pusense), NRB028, NRB029 and NRB030 (B. aerius), NRB037 (A. oryzae), NRB033 and NRB097 (A. lipoferum), are among those that produced more IAA, and all of them were able to produce siderophores, either in a small or intermediate level. Some were able to solubilise phosphate and some able to degrade CMC. The isolate NBR085, an A. lipoferum isolated from the rhizosphere of B. humidicola, stands out among the isolates as possessing the ability to carry out all functions measured, with high rates of phosphate and zinc solubilization, CMC degradation, and siderophore production. It was also able to produce IAA in intermediate doses among the isolates and to fix N in semi-solid media (nifH present). The isolate NRB074, an A. brasilense isolated from B. brizantha, was also able to produce considerable amounts of siderophores, fix nitrogen, and solubilize zinc and phosphate. However, it was not able to degrade CMC.
Principal component analysis (PCA) of five functional activity (Pi and ZnO solubilisation, siderophore and indol acetic acid production, cellulolytic activity) of the 213 diazotrophic strains isolated from 20 different Brachiaria genotypes. Each number represents an individual strain (a) while the symbols represent the Brachiaria species the strains was isolated (b). The arrows indicated the functional activity showing the positive correlation between them
Discussion
Diazotrophic bacterial population and genetic diversity analysis
The results of the present study showed that the diazotrophic population associated with the 20 Brachiaria genotypes, maintained in a Germoplasm Active Bank (GAB), ranged from 102 to 108 bacteria per gram of rhizosphere soil/fresh tissue sample, with the largest population found colonizing the roots (surface or interior). Reis Junior et al. (2004) observed similar results in the roots and rhizosphere soil of three Brachiaria genotypes cultivated in soils of Goiás and Minas Gerais states (Cerrado Biome), as well as in the study carried out by Santos et al. (2013) on forage grasses grown in the semi-arid Northeast region (Caating Biome) of Brazil. Large diazotrophic bacterial population (102 to 106 bacteria g−1 fresh tissues) has also been observed in two elephant grass genotypes grown in soil of Rio de Janeiro state (Atlantic forest biome) (Videira et al. 2012) and, in the study with the tropical molasses grass (Melinis minutiflora) grown as pioneer plants in poor soils in a tropical region of China (Peng et al. 2006). Therefore, the results indicate that, independently of soil type, there is a direct relationship between the Poaceae and these diazotrophic bacteria (associative and endophytic) as attributed by Baldani and Baldani (2005).
The bacterial colonization of the root system is normally influenced by the release of exudates (Baudoin et al. 2003; Mommer et al. 2016) and variations that occur in the microbial population of the rhizosphere are highly dependent on the plant species and genotype (Barea et al. 2005; Richardson et al. 2009). Therefore, an important aspect to be considered in the selection of promising strains is the more tight relationship between plant and bacteria (Baldani and Baldani 2005). In our study, the GAB has been maintained in the same soil and place for around 10 years and therefore a structural native diazotrophic bacterial population could have been established and possibly modulated by the different Brachiaria species/genotypes. The highest diazotrophic population detected in the B. decumbens genotype Basilisk counted in NFb medium could be an indicative of this more specific type of association. The genotype Basilisk was the first Brachiaria grass introduced in Brazil in the early 70 (Mateus et al. 2013) and therefore could have developed a good interaction with the diazotrophic bacterial community along the years. In contrast, the diazotrophic bacteria population present in the B. ruziziensis genotype R134 was below the minimum detection level of the method (MPN) for both nitrogen-free semi-solid media. No clear explanation why the diazotrophic bacteria was so low in that genotype that could not be detected by the method despite the genotype has been maintained in the same GAB. Further studies applying molecular methods should be done to respond the missing information. Concerning the other genotypes, the relationship between the diazotrophic bacterial populations varied with species and the Brachiaria genotypes and could be detected in most of the genotypes independently of the semi-solid media.
Analyzes based on partial 16S rRNA and nifH gene sequences confirmed the presence of the genera Azospirillum and Nitrospirillum colonizing roots (surface and interior) and rhizospheric soil of 20 Brachiaria genotypes. Interesting, 15 other diazotrophic bacterial genera, belonging to the Proteobacteria classes (alpha, beta and gamma) and Firmicutes, were also identified using the nitrogen-free semi-specific semisolid NFb and LGI media. Among them, we highlight strains phylogenetically related to species Paraburkholderia silvatlantica, Stenotrophomonas maltophilia, Herbaspirillum seropedicae, Gluconacetobacter diazotrophicus, Stenotrophomonas pavanii, Phytobacter diazotrophicus, Ochrobactrum anthropi, Zoogloea oryzae, Rhizobium pusense. The species O. anthropi, P. diazotrophicus and Z. oryzae were found, for the first time, to be associated with Brachiaria genotypes. These results corroborate the studies carried out by Chalita et al. (2013) who also observed an enormous diversity of diazotrophic bacteria associated with B. brizantha and B. humidicola grown in Cerrado soils. Many of these diazotrophic bacterial species were also identified associated with B. decumbens and B. humidicola grown in the Northeast arid region of Brazil (Oliveira et al. 2018). In contrast, the use of rich media allowed the isolation of many bacterial strains belonging to different genera, mainly of Pseudomonas, Pantoea, Acinetobacter and Enterobacter species, from native Brachiaria grass grown in Kenya (Mutai et al. 2017). In addition to the interdiversity of the isolates, an intradiversity was also observed between the Azospirillum and Nitrospirillum strains (Figs. S6, S7, S8, S9 and S10). This fact corroborates the studies carried out by Reis Junior et al. (2006) and Azevedo et al. (2005), that found an intradiversity among Nitrospirillum amazonense (older Azospirillum amazonense) strains isolated from Brachiaria and rice, sorghum and maize, respectively.
The strains NRB074, NRB109, NRB194, NRB197, NRB200 and NRB214 are closely related to the species A. brasilense and/or A. formosense based on the partial 16S rRNA and nifH gene sequences, but each phylogenetic tree presented a different topology (Fig. S8). High similarity of the 16S rRNA and nifH genes was observed between the species A. brasilense and A. formosense (Lin et al. 2012). In contrast, other strains isolated from Brachiaria genotypes showed similarity with the nifH gene of A. melinis, A. lipoferum and A. oryzae, as well as for the 16S rRNA gene, but formed distinct clusters. This is due to the lack of evolutionary synchronism between the analyzed genes in the different species of Azospirillum, despite the high level of similarity of these genes (Liu et al. 2012). Gaby and Buckley (2014) also reported that the genetic divergence of the nifH and 16S rRNA genes generally produces different results. This event should be further investigated by analysing the complete clusters formed by the A. formosense and A. brasilense strains isolated from Brachiaria genotypes, considering that only one strain (CC-Nfb-7) was used in the description of A. formosense species by Lin et al. (2012). There is no clear explanation why the specific band of the nifH gene was not amplified in 33 strains isolated from both nitrogen free semisolid media and that clustered together to species already reported to contain nitrogen-fixing strains such as Bacillus aerius, B. cereus, Bosea thiooxidans, Nitrospirillum amazonense, Stenotrophomonas maltophilia and S. panacihumi. Except to the N. amazonense species, that is a widespread diazotrophic bacterium found associated with many grasses and other plants (Reis et al. 2015), the other species are constituted mostly by non-nitrogen-fixing strains and therefore additional studies are required including the use of degenerated nif primers.
The fact that the genera Nitrospirillum and Azospirillum presented the highest number of isolates is probably due to the use of the semi-selective LGI and NFb media for these diazotrophic species, respectively. In addition, their wide ecological distribution among various forage grasses (Reis Junior et al. 2004; Brasil et al. 2005; Videira et al. 2012; Santos et al. 2013; Silva et al. 2013; Souza et al. 2017) have also contributed to the frequency of isolation. Interesting was the occurrence of strains closely related to other Azospirillum species not yet reported in association with Brachiaria cultivated in Brazil, therefore expanding the possibility to explore the potential of these strains in association with Brachiaria. Additionally, it is was interesting the occurrence of a group of bacteria phylogenetically related to Rhizobium, a saprophytic bacteria that live in symbioses with legumes (Masson-Boivin et al. 2009). Oliveira et al. (2018) have also isolated strains of the genus Rhizobium from B. decumbens grown in the Northeast region of Brazil and reported a specificity between both partners. However, no further study was done to demonstrate this event. It seems that bacteria belonging to genus Rhizobium are quite commonly found colonizing grass plants such as observed for rice (Singh et al. 2006; Peng et al. 2008) and maize (Gutiérrez-Zamora and Martınez-Romero 2001; Ilyas et al. 2008).
Growth promoting characteristics
Our study showed that 84 strains were able to solubilize inorganic phosphate, 20 strains presented the capacity to solubilize zinc oxide while 10 strains showed both functional capacities. These strains were identified as belonging to the genera Azospirillum, Nitrospirillum, Paraburkholderia, Pseudomonas, among others. The ability of these genera to solubilize P and ZnO in vitro has been demonstrated by several authors (Fasim et al. 2002; Rodriguez et al. 2004; Estrada et al. 2013; Goteti et al. 2013; López-Ortega et al. 2013; Gupta et al. 2016; Nandal and Solanki 2017). These functional characteristic has been attributed to the release of several organic acids, with gluconic acid being the major one responsible (Rodriguez et al. 2004; Lin et al. 2006; Saravanan et al. 2007). It is known that phosphorus is one of the main nutrients that limit the development of roots and, consequently, the growth of plants, particularly in tropical soils (Cerrado biome) such as those cultivated with Brachiaria that occupy approximately 76 million ha (ABIEC, 2018).
The production of siderophores by associative diazotrophic bacteria can improve plant growth, either by increasing the availability of nutrients through iron uptake or by preventing the growth of soil pathogens due to iron limitation (Miethke and Marahiel 2007; Chaiharn et al. 2009; Sayyed and Chincholkar 2009). Our study showed that besides Azospirillum (catecholate type) and Nitrospirillum (hidroxamate type), other species have also shown the ability to produce siderophores, mainly the catecholate type. The production of siderophores by strains within the genus Azospirillum has also been observed by other authors (Saxena et al. 1986; Pedraza et al. 2004; Manivannan and Tholkappian 2013; Tortora et al. 2011). In the case of Nitrospirillum, there are no reports in the literature on siderophore production within this genus; however, Schwab et al. (2018) reported the presence of 2 copies of tonB and more than 50 genes encoding TonB dependent receptors, including 11 which are probably involved in iron uptake in Nitrospirillum amazonense strain CBAmC. The detection of siderophores among the other diazotrophic species identified in the present study indicated the presence of both types of siderophores with a high production of catecholate type by bacteria closely related to P. tropica, B. safensis, B. aerius, S. maltophilia, O. anthropi, P. nitritireducens strain and oxalate type by P.silvatlantica, H. seropedicae, P. diazotrophicus, B. cereus, S. panacihumi, Z. oryzae strains. The production of siderophores has also been observed by other authors using some bacteria of similar genera, such as Burkholderia (Lewenza et al. 1999), Pseudomonas (Saranraj et al. 2013; Kamran et al. 2017), Rhizobium (Carrillo-Castañeda et al. 2002) and Kosakonia (Kamran et al. 2017).
Differences among the isolates regarding the ability to produce siderophores, should be taken into account in the selection of strains for plant growth-promoting experiments. Strains with high siderophore production activity (e.g. NRB034, NRB021, NRB148, NRB158, NBR230) should be further investigated for their ability to confer resistance to plant diseases. The production of siderophores by associative bacteria can improve plant growth, both by increasing the availability of nutrients through iron uptake and by preventing the growth of soil pathogens due to iron limitation (Chaiharn et al. 2009; Sayyed and Chincholkar 2009). Thus, of the bacterial strains capable of producing siderophores should be further evaluated for their contribution to the control of phytopathogens and the development of plants in agriculture.
Bacterial strains that present cellulolytic activity can play a very important role in the endophytic colonization of the plant tissues, thus presenting an important ecological role in the interaction with plants (Hurek et al. 1994). Corroborating our study, Mostajeran et al. (2007) and Mehdipour-Moghaddam et al. (2010) also observed the cellulolytic capacity in strains of Azospirillum. In addition to this genus, cellulolytic capacity was also detected in strains of the genus Bacillus and Pseudomonas, and this capacity in both genera has been reported in the literature (Verma et al. 2001; Prakamhang et al. 2009; Reetha et al. 2014). Furthermore, the ability to degrade cellulose was also observed in some N. amazonense strains, however there is no report in the literature showing this capacity in the genus Nitrospirillum, including the sequenced genome of N. amazonense strain CBAmC (Schwab et al. 2018) and in a strain isolated from rice (Elbeltagy et al. 2000).
The closely related A. formosense strains NRB011 and NRB214 and A. lipoferum strain NRB033 should be further investigated due to their high biotechnology potential in the control of Fusarium, Rhizoctonia, and Pythium, causal agents of collection rot in Brachiaria (Duarte et al. 2007). In addition, these strains also presented the highest rates of cellulolytic activity and the ability to produce siderophores. It is known that bacteria producing cellulase-like enzymes and other hydrolytic enzymes may benefit their host by inhibiting fungal pathogens by degrading cell wall cells with the action of enzymes such as β-1,4-glycanases, leading to lysis of the cell wall and consequently the death of this pathogen (Sturz et al. 2000; Dobbelaere et al. 2003).
Our study also showed that the indole production was detected in strains belonging to the genera Azospirillum, Rhizobium, Bacillus, Stenotrophomonas, Pseudomonas and Ochrobactrum. Similar results were observed by Roesch et al. (2007) and Santos et al. (2015) working with different strains of the genus Azospirillum.. Pedraza et al. (2004) working with different diazotrophic bacteria observed that all bacterial strains produced IAA, but the Azospirillum strains presented higher values, with A. brasilense UAP14 producing the highest level (27.36 μg IAA mg protein−1). There is no explanation why the N. amazonense strains (total of 78) isolated from Brachiaria genotypes did not show indole acetic acid activity, although this characteristic has already been demonstrated in N. amazonense strains isolated from Brachiaria (Reis Junior et al. 2004) as well as from rice (Rodrigues et al. 2008).
It has been reported that IAA production by bacteria that colonize the rhizosphere, including fre-living and associative, is responsible for the stimulation of the growth and proliferation of the secondary roots, and for the pathogenesis in several plants (Glick 2012). Shigenaga and Argueso (2016), in a review, reported that auxins could act in the signaling of the plant defense process, as is already known for jasmonic acid and salicylic acid. Therefore, the use of diazotrophic bacteria that present the ability to synthesize IAA in the formulation of inoculants may play an important role in the control of diseases (Kazan and Manners 2009; Tian et al. 2017), besides stimulating the germination of the seeds and promoting the growth of secondary roots.
In conclusion, the present study showed a large cultivable population of diazotrophic bacteria colonizing rhizospheric soil and roots (associative and endophytic) of the twenty genotypes of Brachiaria, varying according to plant genotype, the sample and the culture medium used for isolation. There was no specific association of a bacterium genus and a Brachiaria genotype as for example the genus Nitrospirillum was detected associated with all Brachiaria species and colonizing the three sampled niches. In addition, it was observed that many of the identified genera were associated with the B. decumbens species and colonizing either the root (surface or interior) or the rhizosphere soil. The molecular analysis of the 213 bacterial strains confirmed the presence of diazotrophic bacteria closely related to the species Nitrospirillum amazonense and several species belonging to the genus Azospirillum. In addition, the presence of Paraburkolderia, Phytobacter, Gluconacetobacter, Herbaspirillum, Bacillus, Pseudomonas, Stenotrophomonas and others in the 20 Brachiaria genotypes was detected. Our study revealed, for the first time, the occurrence of the genera Phytobacter and Ochrobactrum colonizing this forage grass. There was no clear relationship between the phylogenetic positioning of the isolate, from where it was isolated, and the presence of a given function. However, the PCA allowed us to identify some patterns such as the positive correlation between the ability to produce siderophores with the ability to produce IAA; and between phosphate and zinc solubilisation. In addition, the functional activities of the bacterial strains revealed the presence of one strain (A. lipoferum strain NRB085) showing all the functional characteristics and twelve strains with five functional characteristics, thus indicating that these strains are potentially good candidates for field inoculation tests envisaging the development of biofertilizers. Furthermore, the results should assist in the development of ecologically efficient and sustainable agricultural practices.
References
ABIEC 2018- Associação Brasileira da Indústria Exportadora de Carnes. Exportações de carne bovina do Brasil. Available in: http://abiec.siteoficial.ws/images/upload/sumario-pt-010217.pdf. Accesses 27 July 2018
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. https://doi.org/10.1093/nar/25.17.3389
Azevedo MS, Teixeira KRDS, Kirchhof G, Hartmann A, Baldani JI (2005) Influence of soil and host plant crop on the genetic diversity of Azospirillum amazonense isolates. Pedobiologia 49(6):565–576. https://doi.org/10.1016/j.pedobi.2005.06.008
Baldani JI, Baldani VLD (2005) History on the biological nitrogen fixation research in graminaceous plants: special emphasis on the brazilian experience. An Acad Bras Cienc 77(3):549–579. https://doi.org/10.1590/S0001-37652005000300014
Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VLD (2014) The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384(1–2):413–431. https://doi.org/10.1007/s11104014-2186-6
Barea JM, Pozo MJ, Azcón R, Azcón-Aguilar C (2005) Microbial co-operation in the rhizosphere. J Exp Bot 56(417):1761–1778. https://doi.org/10.1093/jxb/eri197
Baudoin E, Benizri E, Guckert A (2003) Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil biology and biochemistry 35:1183-1192. Available online at www.sciencedirect.com
Boddey RM, Victoria RL (1986) Estimation of biological nitrogen fixation associated with Brachiaria and Paspalum grasses using 15N – labelled organic matter and fertilizer. Plant and Soil 90:265–292
Boddey RM, Macedo R, Tarré RM, Ferreira E, De Oliveira OC, Rezende CDP, Cantarutti RB, Pereira JM, Alves BJR, Urquiaga S (2004) Nitrogen cycling in Brachiaria pastures: the key to understanding the process of pasture decline. Agric Ecosyst Environ 103(2):389–403. https://doi.org/10.1016/j.agee.2003.12.010
Brasil MDS, Baldani JI, Baldani VLD (2005) Ocorrencia e diversidade de bactérias diazotróficas associadas a gramíneas forrageiras do pantanal sul matogrossense. R Bras Ci Solo 29(2):179–190. https://doi.org/10.1590/s0100-06832005000200003
Carrillo-Castañeda G, Juárez Muñoz J, Ramón Peralta-Videa J, Gomez E, Gardea-Torresdey JL (2002) Plant growth-promoting bacteria promote copper and iron translocation from root to shoot in alfalfa seedlings. J Plant Nutr 26(9):1801–1814. https://doi.org/10.1081/PLN-120023284
Chaiharn M, Chunhaleuchanon S, Lumyong S (2009) Screening siderophore producing bacteria as potencial biological control agente for fungal rice pathogens in Thailand. World Jornal Microbiol Biotecnol 25(11):1919–1928. https://doi.org/10.1007/s11274-009-0090-7
Chalita PB, Farias E, Zilli J, Silva KD (2013) Avaliação da diversidade genotípica de bactérias diazotróficas isoladas de plantas de Brachiaria brizantha e Brachiaria humidicola no estado de Roraima. In: Embrapa Roraima-Artigo em anais de congresso (ALICE). In: CONGRESSO BRASILEIRO DE CIÊNCIA DO SOLO, 34., 2013, Florianópolis. Ciência do solo: para quê e para quem: anais. Viçosa, MG: Sociedade Brasileira de Ciência do Solo
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42(5):669–678. https://doi.org/10.1016/j.soilbio.2009.11.024
Dantur KI, Enrique R, Welin B, Castagnaro AP (2015) Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 5(1):1–15. https://doi.org/10.1186/s13568-015-0101-z
Dias-Filho MB (2011) Degradação de pastagens: processos, causas e estratégias de recuperação. Documentos 411, 25 p. Embrapa Amazônia oriental, ISSN 1983-0513
Dobbelaere S, Vanderleyden J, Okon Y (2003) Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22(2):107–149. https://doi.org/10.1080/713610853
Duarte MDLR, Albuquerque FC, Sanhueza RMV, Verzignassi JR, Kondo N (2007) Etiology of collar rot of Brachiaria brizantha cv. Marandu in Amazonian pastures. Fitopatol Bras 32(3):261–265. https://doi.org/10.1590/S0100-41582007000300013
Eckert B, Weber OB, Kirchhof G, Halbritter A, Stoffels M, Hartmann A (2001) Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int J Syst Evol Microbiol 51(1):17–26. https://doi.org/10.1099/00207713-51-1-17
Elbeltagy A, Nishioka K, Suzuki H, Sato T, Sato Y, Morisaki H, Mitsui H, Minamisawa K (2000) Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci Plant Nutr 46(3):617–629. https://doi.org/10.1080/00380768.2000.10409127
Estrada GA, Baldani VLD, de Oliveira DM, Urquiaga S, Baldani JI (2013) Selection of phosphate-solubilizing diazotrophic Herbaspirillum and Burkholderia strains and their effect on rice crop yield and nutrient uptake. Plant Soil 369(1–2):115–129. https://doi.org/10.1007/s11104-012-1550-7
Fasim F, Ahmed N, Parsons R, Gadd GM (2002) Solubilization of zinc salts by a bacterium isolated from the air environment of a tannery. FEMS Microbiol Lett 213:1–6. https://doi.org/10.1111/j.15746968.2002.tb11277.x
Furushita M, Shiba T, Maeda T, Yahata M, Kaneoka A, Takahashi Y, Torii K, Hasegawa T, Ohta M (2003) Similarity of tetracycline resistance genes applied isolated from fish farm bacteria to those from clinical isolates. Appl Environ Microbiol 69(9):5336–5342. https://doi.org/10.1128/AEM.69.9.5336-5342.2003
Gaby JC, Buckley DH (2014) A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fxing bacteria. Database (Oxford) 2014:bau001. https://doi.org/10.1093/database/bau001
Glick BR (2012) Plant growth-promoting Bacteria: mechanisms and applications. Scientifica 2012:1–15. https://doi.org/10.6064/2012/963401
Goteti PK, Emmanuel LDA, Desai S (2013) Shaik MHA. Prospective zinc solubilising bacteria for enhanced nutrient uptake and growth promotion in maize (Zea mays L) Int J Microbiol 2013:1–8. https://doi.org/10.1155/2013/869697
Guarda VDA, Guarda RDA (2014) Brazilian tropical grassland ecosystems: distribution and research advances. Am J Plant Sci 5:924–932
Gupta D, Pandya H, Mistry S, Patel A, Shelat H, Vyas R (2016) Isolation of Azospirillum and compatibility testing with micronutrients, pgpr activity and efficacy on tomato. The Bioscan 11(2):891-896. Available online at: https://www.researchgate.net/publication/309406618
Gutiérrez-Zamora ML, Martınez-Romero E (2001) Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J Biotechnol 91(2–3):117–126. https://doi.org/10.1016/s0168-1656(01)00332-7
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hankin L, Anagnostakis SL (1975) The use of solid media for detection of enzymes production by fungi. Mycologia 67(3):597–607. https://doi.org/10.2307/3758395
Hungria M, Campo RJ, Souza EM, Pedrosa FO (2010) Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant and Soil, 331(1-2):413-425. https://doi.org/10.1007/s11104-009-0262-0
Hungria M, Nogueira MA, Araujo RS (2016) Inoculation of Brachiaria spp. with the plant growth-promoting bacterium Azospirillum brasilense: an environment-friendly component in the reclamation of degraded pastures in the tropics. Agric Ecosyst Environ 221:125–131. https://doi.org/10.1016/j.agee.2016.01.024
Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E (1994) Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol 176(7):1913–1923. https://doi.org/10.1128/jb.176.7.1913-1923.1994
Ilyas N, Bano A, Iqbal S (2008) Variation in Rhizobium and Azospirillum strains isolated from maize growing in arid and semiarid areas. Int J Agric Biol 10(6):612–618
Kamran S, Shahid I, Baig DN, Rizwan M, Malik KA, Mehnaz S (2017) Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front Microbiol 8:2593. https://doi.org/10.3389/fmicb.2017.02593
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) Rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr Microbiol 57(5):503–507. https://doi.org/10.1007/s00284-008-9276-8
Kazan K, Manners JM (2009) Linking development to defense: auxin in plant pathogen interactions. Trends Plant Sci 14(7):373–382. https://doi.org/10.1016/j.tplants.2009.04.005
Kumar V, Narula N (1999) Solubilization of inorganic phosphates and growth emergence of wheat as affected by Azotobacter chroococcum mutants. Biol Fertil Soils 28(3):301–305. https://doi.org/10.1007/s003740050497
Lewenza S, Conway B, Greenberg EP, Sokol PA (1999) Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol 81(3):748–756
Lin SY, Shen FT, Young LS, Zhu ZL, Chen WM, Young CC (2012) Azospirillum formosense sp. nov., a diazotroph from agricultural soil. Int J Syst Evol Microbiol 62(5):1185–1190. https://doi.org/10.1099/ijs.0.030585-0
Lin TF, Huang HI, Shen FT, Young CC (2006) The protons of glucônico acid are the major factor responsible for the dissolution of tricalcium phosphate by Burkholderia cepacia CC-A174. Bioresource Tecnology 97(7):957–960. https://doi.org/10.1016/j.biortech.2005.02.017
Liu J, Peng M, Li Y (2012) Phylogenetic diversity of nitrogen-fxing bacteria and the nifH gene from mangrove rhizosphere soil. Can J Microbiol 58(4):531–539. https://doi.org/10.1139/w2012-016
López-Ortega MDP, Criollo-Campos PJ, Gómez-Vargas RM, Camelo-Rusinque M, Estrada-Bonilla G, Garrido-Rubiano MF, Bonilla-Buitrago R (2013) Characterization of diazotrophic phosphate solubilizing bacteria as growth promoters of maize plants. Rev. col. Biotechnol 15(2):115–123. https://doi.org/10.15446/rev.colomb.biote.v15n2.36303
Macedo MCM (2005) Pastagens no ecossistema Cerrados: evolução das pesquisas Para o desenvolvimento sustentável. Reunião anual da Sociedade Brasileira de Zootecnia 42:56–84
Madhaiyan M, Saravanan VS, Jovi DBSS, Lee H, Thenmozhi R, Hari K, Sa T (2004) Occurrence of Gluconacetobacter diazotrophicus in tropical and subtropical plants of Western Ghats, India. Microbiol Res 159(3):233–243. https://doi.org/10.1016/j.micres.2004.04.001
Manivannan M, Tholkappian P (2013) Prevelence of Azospirillum isolates in tomato rhizosphere soils of coastal areas of Cuddalore district, Tamil Nadu. Int J recent Sci res 4(10):1610-1613. Available online at http://www.recentscientific.com
Masson-Boivin C, Giraud E, Perret X, Batut J (2009) Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol 17(10):458–466. https://doi.org/10.1016/j.tim.2009.07.004
Mateus RG, Barrios SC, Figueiredo UJ, Do Valle CB (2013) Agronomic evaluation of 324 intraspecific hybrids of Brachiaria decumbens in Brazil. Tropical Grasslands 1:99–100
Mehdipour-Moghaddam MJ, Emtiazi G, Bouzari M, Mostajeran A, Salehi Z (2010) Novel phytase and cellulase activities in endophytic Azospirilla. W Appl Sci J 10(10):1129–1135
Miethke M, Marahiel MA (2007) Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71(3):413–451. https://doi.org/10.1128/MMBR.00012-07
Miranda CHB, Urquiaga S, Boddey RM (1990) Selection of ecotypes of Panicum maximum for associated biological nitrogen fixation using the 15N isotope dilution technique. Soil Biol Biochem 22:657–663
Mommer L, Hinsinger P, Prigent-Combaret C, Visser EJ (2016) Advances in the rhizosphere: stretching the interface of life. Plant Soil 407(2016):1–8. https://doi.org/10.1007/s11104-016-3040-9
Mostajeran A, Amooaghaie R, Emtiazi G (2007) The participation of the cell wall hydrolytic enzymes in the initial colonization of Azospirillum brasilense on wheat roots. Plant Soil 291(1–2):239–248. https://doi.org/10.1007/s11104-006-9189-x
Mutai C, Njuguna J, Ghimire S (2017) Brachiaria grasses (Brachiaria spp.) harbor a diverse bacterial community with multiple attributes beneficial to plant growth and development. MicrobiologyOpen 6(5): e00497. https://doi.org/10.1002/mbo3.497
Nandal V, Solanki M (2017) Isolation and molecular characterization of a potential zinc solubilizing bacteria for sustainable agriculture. International journal of basic and applied biology 4(1):18-21. Available online at http://www.krishisanskriti.org/Publication.html
Nautiyal C (1999) An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett 170(1):265–270. https://doi.org/10.1111/j.1574-6968.1999.tb13383.x
Oksanen, Jari, Blanchet F G, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P R, O'Hara R B, Simpson G L, Solymos P, Henry M, Stevens H, Szoecs E and Wagner H (2019). Vegan: community ecology package. R package version 2.5–4. https://CRAN.R-project.org/package=vegan
Oliveira JTC, Silva GT, da Diniz WPS, Figueredo EF, dos Santos IB, de Lima DRM, Quecine MC, Júlia KuklinskySobral JK, Freire FJ (2018) Diazotrophic bacteria isolated from Brachiaria spp.: genetic and physiological diversity. Ciencia e Investigación Agraria 45(3):277–289. https://doi.org/10.7764/rcia.v45i3.1949
Pedraza RO, Ramírez-Mata A, Xiqui ML, Baca BE (2004) Aromatic amino acid aminotransferase activity and indole-3-acetic production by associative nitrogenfixing bacteria. FEMS Microbiol Lett 233(1):15–21. https://doi.org/10.1016/j.femsle.2004.01.047
Peng G, Wang H, Zhang G, Hou W, Liu Y, Wang ET, Tan Z (2006) Azospirillum melinis sp. nov., a group of diazotrophs isolated from tropical molasses grass. Int J Syst Evol Microbiol 56(6):1263–1271. https://doi.org/10.1099/ijs.0.64025-0
Peng G, Yuan Q, Li H, Zhang W, Tan Z (2008) Rhizobium oryzae sp. nov., isolated from the wild rice Oryza alta. Int J Syst Evol Microbiol 58(9):2158–2163. https://doi.org/10.1099/ijs.0.65632-0
Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. https://doi.org/10.1016/S0923-2508(00)01172-4
Prakamhang J, Minamisawa K, Teamtaisong K, Boonkerd N, Teaumroong N (2009) The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.). Appl Soil Ecol 42(2):141–149. https://doi.org/10.1016/j.apsoil.2009.02.008
Reetha S, Selvakumar G, Bhuvaneswari G, Thamizhiniyan P, Ravimycin T (2014) Screening of cellulase and pectinase by using Pseudomonas fluorescence and Bacillus subtilis. International Letters of Natural Sciences 13:75–80. https://doi.org/10.18052/www.scipress.com/ilns.13.75
Reis Junior FB, Reis VM, Teixeira KRDS (2006) Restriction of 16S-23S intergenic rDNA for diversity evaluation of Azospirillum amazonense isolated from different Brachiaria spp. Pesq Agropec Bras 41(3):431–438. https://doi.org/10.1590/S0100-204X2006000300009
Reis Junior FB, Silva MF, Teixeira KRS, Urquiaga S, Reis VM (2004) Identificação de isolados de Azospirillum amazonense associados a Brachiaria spp., em diferentes épocas e condições de cultivo e produção de fitormônio pela bactéria. R Bras Ci Solo 28(1):103–113
Reis VM, Baldani VLD, Baldani JI (2015). Isolation, identification and biochemical characterization of Azospirillum spp. and other nitrogen-fixing bacteria. In: Cassán F, Okon Y, Creus C (eds) springer, Cham, pp. 3-26
Reis VM, Reis Junior FB, Quesada DM, de Oliveira OC, Alves BJ, Urquiaga S, Boddey RM (2001) Biological nitrogen fixation associated with tropical pasture grasses. Austral J Plant Physiol 28(9):837–844. https://doi.org/10.1071/pp01079
Richardson AE, Barea JM, Mcneill AM, Prigent-Combaret C (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321:305–339. https://doi.org/10.1007/s11104-009-9895-2
Rodrigues EP, Rodrigues LS, de Oliveira ALM, Baldani VLD, Teixeira KRdosS, Urquiaga S, Reis VM (2008) Azospirillum amazonense inoculation: effects on growth, yield and N 2 fixation of rice (Oryza sativa L.). Plant Soil 302(1–2):249–261. https://doi.org/10.1007/s11104-007-9476-1
Rodriguez H, Gonzalez T, Goire I, Bashan Y (2004) Gluconic acid production and phosphate solubilization by the plant growth-promoting bacterium Azospirillum spp. Naturwissenschaften 91(11):552–555. https://doi.org/10.1007/s00114-004-0566-0
Roesch LFW, de Quadros PD, Camargo FA, Triplett EW (2007) Screening of diazotrophic bacteria Azopirillum spp. for nitrogen fixation and auxin production in multiple field sites in southern Brazil. World J Microbiol Biotechnol 23(10):1377–1383. https://doi.org/10.1007/s11274-007-9376-9
R Core Team (2018) R: a language and environment for statistical computing. In: R Foundation for statistical computing. Austria. URL, Vienna https://www.R-project.org/
Santos JDS, Viana TDO, Jesus CMD, Baldani VLD, Ferreira JS (2015) Inoculation and isolation of plant growth-promoting bacteria in maize grown in Vitória da Conquista, Bahia, Brazil. R Bras Ci Solo 39(1):78–85. https://doi.org/10.1590/01000683rbcs20150725
Santos MCM, dos Santos DR, Bakke OA, Bakke IA (2013). Ocorrência e atividade de bactérias diazotróficas em forrageiras cultivadas na Região semiárida no Brasil. Revista Caatinga 26(1):27-34. Available online at http://www.redalyc.org/articulo.oa?id=23712 7564005
Saranraj P, Sivasakthivelan P, Sakthi SS (2013) Prevalence and production of plant growth promoting substance by Pseudomonas fluorescens isolated from paddy rhizosphere soil of Cuddalore district, Tamil Nadu. Afr J Basic Appl Sci 5(2):95–101. https://doi.org/10.5829/idosi.ajbas.2013.5.2.2934
Saravanan VS, Madhaiyan M, Thangaraju M (2007) Solubilization of zinc compounds by the diazotrophic, plant growth promoting bacterium Gluconacetobacter diazotrophicus. Chemosphere 66:1794–1798. https://doi.org/10.1016/j.chemosphere.2006.07.067
Sarwar M, Kremer RJ (1995) Enhanced suppression of plant growth through production of L-tryptophan-derived compounds by deleterious rhizobacteria. Plant Soil 172(2):261–269. https://doi.org/10.1007/BF00011328
Saxena B, Modi M, Modi VV (1986) Isolation and characterization of siderophores from Azospirillum lipoferum D-2. Microbiol 132(8):2219–2224. https://doi.org/10.1099/00221287-132-8-2219
Sayyed RZ, Chincholkar SB (2009) Siderophore producing A. feacalis more biocontrol potential Vis-a’-Vis chemical fungicide. Curr Microbiol 58(1):47–51. https://doi.org/10.1007/s00284-008-9264-z
Schwab S, Terra LA, Baldani JI (2018) Genomic characterization of Nitrospirillum amazonense strain CBAmC, a nitrogen-fixing bacterium isolated from surface-sterilized sugarcane stems. Molecular Genetics and Genomics 293(4):997–1016. https://doi.org/10.1007/s00438-018-1439-0
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160(1):47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Shigenaga AM, Argueso CT (2016) No hormone to rule them all: interactions of plant hormones during the responses of plants to pathogens. Semin Cell Dev Biol 56:174–189. https://doi.org/10.1016/j.semcdb.2016.06.005
Silva MCP, Figueiredo AF, Andreote FD, Cardoso EJBN (2013) Plant growth promoting bacteria in Brachiaria brizantha. World J Microbiol Biotechnol 29(1):163–171. https://doi.org/10.1007/s11274-012-1169-0
Singh RK, Mishra RP, Jaiswal HK, Kumar V, Pandey SP, Rao SB, Annapurna K (2006) Isolation and identification of natural endophytic rhizobia from rice (Oryza sativa L.) through rDNA PCR-RFLP and sequence analysis. Curr Microbiol 52(5):345–349. https://doi.org/10.1007/s00284-005-0138-3
Souza MS, de Baura VA, Santos SA, Fernandes-Júnior PI, Junior FBR, Marques MR, Paggi GM, Brasil MS (2017) Azospirillum spp. from native forage grasses in Brazilian Pantanal floodplain: biodiversity and plant growth promotion potential. World J Microbiol Biotechnol 33(4):1–13. https://doi.org/10.1007/s11274-017-2251-4
Sturz AV, Christie BR, Nowak J (2000) Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit Rev Plant Sci 19(1):1–30. https://doi.org/10.1080/07352680091139169
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tian B, Zhang C, Ye Y, Wen J, Wu Y, Wang H, Li H, Cai S, Cai W, Cheng Z, Lei S, Ma R, Lu C, Cao Y, Xu X, Zhang K (2017) Beneficial traits of bacterial endophytes belonging to the core communities of the tomato root microbiome. Agric Ecosyst Environ 247:14–156. https://doi.org/10.1016/j.agee.2017.06.041
Tortora M, Díaz-Ricci J, Pedraza R (2011) Azospirillum brasilense siderophores with antifungal activity against Colletotrichum acutatum. Arch Microbiol 193:275–286. https://doi.org/10.1007/s00203-010-0672-7
Tuimala, J (2004) Using ClustalX for multiple sequence alignment. Methods Enzymol 226:383–402
USDA 2018- United States Department of Agriculture. Foreign Agriculture Service. Available in: http://www.usdabrazil.org.br/pt-br/. Accessed 27 Nov 2018
Valle CB, Macedo MCM, Euclides VPB, Jank L, Resende RMS (2010) Gênero Brachiaria. In: da Fonseca DM, Martuscello JA (eds) Plantas forrageiras. Viçosa, UFV, pp 30–77
Videira SS, de Oliveira DM, de Morais RF, Borges WL, Baldani VLD, Baldani JI (2012) Genetic diversity and plant growth promoting traits of diazotrophic bacteria isolated from two Pennisetum purpureum Schum. Genotypes grown in the field. Plant Soil 356(1–2):51–66. https://doi.org/10.1007/s11104-011-1082-6
Verma SC, Ladha J, Tripathi AK (2001) Evaluation of plant growth promotion and colonization ability of endophytic diazotrophs from deep water rice. J Biotechnol 91(2–3):127–141. https://doi.org/10.1016/S0168-1656(01)00333-9
Wang RF, Cao WW, Cerniglia CE (1996) Phylogenetic analysis of Fusobacterium prausnitzii based upon the 16S rRNA gene sequence and PCR confirmation. Int J Syst Bacteriol 46(1):341–343. https://doi.org/10.1099/00207713-46-1-341
Acknowledgments
This study was funded by the project Embrapa MP2 (n° 02.13.08.004.00.02.003). The first author was supported by a fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. José Ivo Baldani was supported by fellowship from CNPq (process n° 306011/2017-4) and a bench fee from FAPERJ–CNE (E26/203.011/2016). The authors thank Dr. Ederson da Conceição Jesus for the multivariate statistical analysis and PCA analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Anton Hartmann.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
Venn diagram representation of the diazotrophic strains isolated from the different plant compartments (non and desinfected roots and rhizosphere soil) with respect to bacterial genera from the Brachiaria species: (A) B. brizantha, (B) B. humidicola, (C) B. decumbens and (D) Brachiaria spp. plus B. arrecta. Signals: no *: belongs to Brachiaria spp., *: belongs to B. arrecta and **: belongs to both Brachiaria spp. and B. arrecta. (PNG 2066 kb)
Figure S2
Examples of in vitro halo formation characteristic of solubilization of inorganic phosphate (A) and zinc oxide (B) by strains NRB081 and NRB082 grown in NBRIP medium containing insoluble β-tricalcium phosphate (Ca3(PO4)2) or zinc oxide (ZnO). (JPG 79 kb)
Figure S3
. Production of siderophore in vitro of catecholate type by strains isolated from Brachiaria genotypes and evaluated in solid NFBb medium with 0.1% (NH4)2SO4 of CAS. The blue/yellow color change was observed 72 h after incubation at 30 °C. (A) strains with no production capacity; (B) strain with low production capacity; (C) strain with medium capacity; and (D) strain with high capacity of siderophore production. (JPG 47 kb)
Figure S4
Production of siderophore in vitro of hydroxamate type by strains isolated from Brachiaria genotypes and analysed in solid LGI medium, with 0.1% (NH4)2SO4 of CAS. The change of color blue/pink was observed 72 h after incubation at 30 °C. (A) strain with no production capacity; (B) strain with low production capacity; (C) strain with medium capacity; and (D) strain with high capacity of siderophore production. (JPG 40 kb)
Figure S5
Examples of in vitro cellulolytic activity of strains isolated from Brachiaria genotypes. Bacteria were inoculated onto CMC plate medium containing 0.2% of carboxymethylcellulose. (A) strain showing no activity; (B) strain with low activity; (C) strain with medium capacity; and (D) strain with high cellulose degradation capacity. (JPG 68 kb)
Figure S6
Detailed cluster of strains closely related to Azospirillum lipoferum from the phylogenetic tree presented in Fig. 2. (PNG 208 kb)
Figure S7
Detailed cluster of strains closely related to Azospirillum brasilense from the phylogenetic tree presented in Fig. 2. (PNG 154 kb)
Figure S8
Detailed cluster of strains closely related to Azospirillum formosense from the phylogenetic tree presented in Fig. 2. (PNG 150 kb)
Figure S9
Detailed cluster of strains closely related to Azospirillum melinis from the phylogenetic tree presented in Fig. 2. (PNG 178 kb)
Figure S10
Detailed cluster of strains closely related to Nitrospirillum amazonense from the phylogenetic tree presented in Fig. 2. (PNG 367 kb)
Table S1
(PDF 86 kb)
Rights and permissions
About this article
Cite this article
Ribeiro, N.V.d.S., Vidal, M.S., Barrios, S.C.L. et al. Genetic diversity and growth promoting characteristics of diazotrophic bacteria isolated from 20 genotypes of Brachiaria spp.. Plant Soil 451, 187–205 (2020). https://doi.org/10.1007/s11104-019-04263-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04263-y