Abstract
Background and aims
Citrate transporters responsible for Aluminum (Al)-induced citrate efflux have not been identified in soybean.
Method
Three soybean multi-drug and toxic compound extrusion (MATE) family genes were characterized by transcriptional expression, subcellular localization and overexpression experiments.
Results
GmMATE75, GmMATE79 and GmMATE87 are localized to plasma membrane. Their over-expression respectively resulted in more citrate efflux and less Al contents in soybean hairy roots, alleviated root elongation inhibition in Arabidopsis and partially restored root growth in atmate mutant under Al stress. Al increased their transcriptional expression at either the root apex or the base zone. Cu2+, Cd2+, La3+ increased the expression of GmMATE79 and GmMATE87. Ten day of –Fe culture increased the expression of GmMATE75 and GmMATE79. Al treatment extended β-glucuronidase (GUS) staining from central cylinder to cortical and epidermis cells for pGmMATE75::GUS or pGmMATE79::GUS soybean hairy roots transformation. But GUS staining restricted within central cylinder for pGmMATE87::GUS transformation with or without Al treatment.
Conclusion
GmMATE75, GmMATE79 and GmMATE87 are plasma-membrane-localized citrate transporters and have capabilities to increase citrate efflux. They played different role in Al-induced citrate secretion from soybean because their different tissue localization and expression patterns.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (Al)-induced organic acid excretion has been convincingly demonstrated as a major mechanism of Al resistance through Al exclusion in many plant species (Ma et al. 2001; Kochian et al. 2015). The first resistance gene in plants was cloned from wheat as TaALMT1 (aluminum-activated malate transporter) encoding an Al3+-activated malate channel (Sasaki et al. 2004). Citrate transporters responsible for Al-activated citrate efflux were later identified by positional cloning in sorghum (SbMATE, multidrug and toxic compound extrusion, Magalhaes et al. 2007) and barley (HvAACT1,aluminum-activated citrate transported 1, Furukawa et al. 2007), respectively; these transporters are the members of the multi-drug and toxic compound extrusion (MATE) family. Transgenic Arabidopsis and tobacco showed enhanced Al resistance with the overexpression of SbMATE (Magalhaes et al. 2007) and HvAACT1 (Furukawa et al. 2007), respectively. The transporter genes of the MATE and ALMT families can provide effective ways to enhance the Al resistance of plants. Unravelling additional genes encoding organic acid anion transporters will help more crops to adapt to Al-stresses under acidic soil environments.
MATE transporters are proposed to be involved in a wide range of biological events including xenobiotic efflux, accumulation of alkaloids and flavonoids, iron translocation, Al detoxification and plant hormone signalling (Takanashi et al. 2014). The identified citrate transporter genes in the MATE family have been suggested to function in Al resistance or iron uptake or both. First, SbMATE (Magalhaese et al. 2007), AtMATE (Liu et al. 2009), BoMATE (Wu et al. 2014), EcMATE1 and EcMATE3 (Sawaki et al. 2013), OsFRDL4 (ferric reductase defective like 4, Yokosho et al. 2011), OsFRDL1 (Yokosho et al. 2016a) and ScFRDL2 (Yokosho et al. 2010), VuMATE1 (Yang et al. 2011), VuMATE2 (Liu et al. 2018), ZmMATE1 (Maron et al. 2010; Maron et al. 2013), FeMATE1 (Lei et al. 2017) were reported to be responsible for Al-activated citrate efflux under Al stress. Second, members such as AtFRD3 (Durrett et al. 2007), GmFRD3b (Rogers et al. 2009), OsFRDL1 (Yokosho et al. 2009) and ScFRDL1 (Yokosho et al. 2010), LjMATE1 (Takanashi et al. 2013) were found to be localized to either the root pericycle cell or the nodules and were responsible for Fe supply by assisting the translocation of Fe by providing citrate. Thirdly, it was reported that HvAACT1 and TaMATE1b were involved in both Fe translocation and Al3+ detoxification (Fujii et al. 2012; Tovkach et al. 2013; Takanashi et al. 2014).
Various regulatory systems were found to control citrate transporter expression and thus function. The first group is represented by ZmMATE1. Its gene duplications determined their expression and thus citrate efflux (Maron et al. 2013). The second group is represented by HvAACT1 and TaMATE1b. HvAACT1, a citrate transporter in barley, provides primary transport of citrate into the root xylem for Fe translocation to the shoot as a Fe-citrate complex in the mature root zones (Fujii et al. 2012). The insertion of a 1 kb transposon in the 5′ upstream region of HvAACT1 changed its distribution and density to facilitate citrate secretion from the root apex to the rhizosphere for Al detoxification (Fujii et al. 2012). Fujii et al. (2018) also found that retrotransposon insertion and DNA methylation in the upstream genomic region are involved in regulating the expression of HvAACT1 then Al resistance in barley especially distributed in European areas with acid soil. A transposon-like element in Triticum aestivum increased citrate transporter TaMATE1B expression in root apex, where it facilitates citrate efflux and enhances Al tolerance (Tovkach et al. 2013). The third group is represented by AtMATE and VuMATE1. Their expression is regulated by Cys2His2 zinc finger-type transcription factor STOP1 (sensitive to proton rhizotoxicity1). The expression of AtMATE is regulated by the transcription factor AtSTOP1 (Liu et al. 2009). Yeast one-hybrid examination has shown that there is an interaction between VuSTOP1 and the promoter region of VuMATE1and VuMATE2 (Yang et al. 2011; Fan et al. 2015; Liu et al. 2018). A 1.2 kb insertion in the OsFRDL4 promoter region in the rice japonica subspecies increased the number of cis-acting elements of ART1 then enhanced OsFRDL4 expression level, but did not alter its spatial expression or cellular localization (Yokosho et al. 2016b).
Multiple Al resistance mechanisms have been suggested to be involved in Al resistance in soybean (Nian et al. 2004). Correspondingly, multigenetic traits have been suggested to relate to mechanisms underlying Al resistance in soybean by genetic studies (Bianci-Hall et al. 1998, 2000). Compared with the progress made in other important crops and model plant species, little was known about the molecular mechanisms of Al resistance in soybean. Al-induced citrate efflux has been well established as an Al resistance mechanism in soybean (Yang et al. 2000, 2001; Silva et al. 2001). Liu et al. (2016) identified and named a total of 117 genes encoding MATE transporters in the whole soybean genome, which are unevenly distributed on the 20 soybean chromosomes. However, the citrate transporter responsible for citrate efflux from soybean under Al stress has not been identified until now.
Rogers et al. (2009) showed that GmFRD3a and GmFRD3b were induced by iron deficiency in an iron-efficient soybean cultivar. GmFRD3b, but not GmFRD3a, was expressed at higher levels in the iron-efficient cultivar than its iron-inefficient near-isogenic line, and thus GmFRD3b is suggested to function similarly to AtFRD3 to mediate citrate efflux into the xylem. Their function has not been further characterized until now. Our previous microarray analysis found a MATE-family transcript (GmMATE75) was up-regulated by Al treatment in the Al-tolerant genotype Jiyu 70 (You et al. 2011) which was also predicated to be the candidate Al tolerance gene according to its gene expression pattern and cis-elements (Liu et al. 2016). The expression of GmALCT1 (GmMATE 79) was also highly induced in soybean root apices under Al stress (Xu et al. 2010). In the present study, we compared three genes in the MATE family, GmMATE75, GmMATE79 and GmMATE87 (GmFRD3a), to reveal their functions in the process of Al-induced citrate efflux from soybean root apices.
Material and methods
Plant materials and growth conditions
Compared with Jiyu 62, soybean cultivar Jiyu 70 exhibited higher Al resistance and higher amount of citrate efflux under Al stress (Zhou et al. 2018a). They were used as plant materials in the present study. For hydroponic culture, soybean seeds were surface sterilized in 1.0% (v/v) sodium hypochlorite for 5 min, washed 3–4 times with tap water, and germinated in peat moss for 3 days at 25 °C in the dark. After germination, uniform seedlings were cultured in 1 L plastic pots filled with nutrient solution (pH 4.5) as described by Horst et al. (1992) containing 750 μM KNO3, 250 μM Ca(NO3)2, 325 μM MgSO4, 10 μM KH2PO4, 20 μM Fe-EDTA, 8 μM H3BO3, 0.2 μM (NH4)6Mo7O24. The solution was aerated and renewed every other day. Seven-day-old seedlings were transferred to 0.5 mM CaCl2 solution overnight for the following treatments.
In the time course experiment, the seedlings were exposed to 30 μM AlCl3 in 0.5 mM CaCl2 solution. The root apices (0–1 cm) were excised at 0 h, 2 h, 4 h, 8 h, 12 h, and 24 h. For the other metal stresses, the seedlings were transferred into nutrient solution (pH 4.5) containing 25 μM CdCl2, 10 μM LaCl3, or 1 μM CuCl2. Root apices (0–1 cm) were excised after 4 h metal exposure. For the iron deficiency experiment, the germinated seedlings were cultured in nutrient solution (Horst et al. 1992) (pH 4.5) without the addition of Fe-EDTA. Root apices (0–1 cm) were excised after 10 days of culture. For the root localization experiment, the seedlings were exposed to 0 or 30 μM AlCl3 in 0.5 mM CaCl2 solution for 4 h. Root segments of 0–1 cm, 1–2 cm, and 2–3 cm were excised respectively. All the excised root segments were put into liquid nitrogen instantly and stored in a − 80 °C freezer for RNA isolation. All hydroponic experiments were carried out in a controlled growth chamber at 25 °C day/22 °C night temperatures, 60% constant relative humidity, 14 h light/10 h dark cycles, and 300 μmol m−2 s−1 of light intensity during the day.
Gene cloning of GmMATE75, GmMATE79 and GmMATE87 and their bioinformatics analysis
With Affymetrix probe sequence, the CDS of GmMATE75 was got by Blast in the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). Then, the encoding amino acid sequence of GmMATE75 was searched against Phytozome (www.phytozome.net) to find its homologues. A phylogenetic tree was generated with TreeView 5.1 with GmMATE75, its homologues and other known citrate transporter. Primers were designed according to CDS sequences of GmMATE79 (Glyma.13G339800), GmMATE87 (Glyma.15G274600), GmMATE75 (Glyma.13G203000) considering the enzyme cutting locus of BamHI in the modified vector pCAMBIA3301 (Table S1). Total RNA was isolated from 4 h Al-treated soybean root apices (Jiyu70 and Jiyu 62) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reversely transcribed with M-MLV reverse transcriptase (TaKaRa Bio Tokyo, Japan) according to the manufacturer’s protocols. With the cDNA as template, the three genes were cloned by RT-PCR. The PCR products were purified by a TransGen Biotech kit according to the manufacturer’s protocol and confirmed by sequencing. Jiyu 70 and Jiyu 62 displayed same sequences in the CDS region of GmMATE79 (Glyma.13G339800), GmMATE87 (Glyma.15G274600) or GmMATE75 (Glyma.13G203000).
Transcriptional expression of putative citrate transporter genes
Quantitative real-time PCR (qRT-PCR) was performed to determine transcriptional expression of the three putative citrate transporter genes. Gene-specific primers were designed using Primer 3.0 online (http://primer3.ut.ee/) and listed in Table S1. The housekeeping gene β-Tubulin (GenBank ID: 100811275) was used as an internal standard. qRT-PCR was conducted in an Mx3005P machine (Stratagene, USA). The reaction system (25 μl) was as follows: 2 μl cDNA template (50–100 ng), 0.5 μl 10 mM specific forward primer, 0.5 μl 10 mM specific reverse primer, 12.5 μl 2× SYBR Premix Ex Taq (TaKaRa, Bio Inc.), and 9.5 μl double-distilled H2O. The procedure was performed as follows: 1 cycle for 30 s at 95 °C, 30 cycles for 5 s at 95 °C and 20 s at 60 °C, and 1 cycle for 60 s at 95 °C, 30 s at 55 °C, and 30 s at 95 °C for the melting curve analysis. The relative expression level of each gene was computed by the 2-ΔΔCt method (Livak and Schmittgen 2001).
Subcellular localization of GmMATE75, GmMATE79 and GmMATE87
The CDSs of GmMATE75, GmMATE79 and GmMATE87 were cloned into the pENSG-N-GFP vector under the control of a CaMV 35S promoter. The transient expression of the translational in-frame fusion was achieved in Arabidopsis protoplast cells. Transformed Arabidopsis protoplast cells were stained by cell plasma membrane Marker stain(Cell Mask™ Orange plasma membrane stain,C10045,USA). The GFP fluorescence was observed by laser-scanning confocal microscopy (Leica TCS SP8X DLS, Wetzlar, Germany).
Overexpression of GmMATE75, GmMATE79, and GmMATE87 in soybean hairy roots
The CDSs of GmMATE75, GmMATE79, GmMATE87 were amplified and respectively cloned into the modified pCAMBIA3301 vector with CaMV 35S as promoter and labelled with Nanoluciferase. The transformed plasmids were sequenced then electroporated into the Agrobacterium strain K599. The transformation of soybean Jiyu 62 cotyledons was performed as described by Subramanian et al. (2005). Luciferase activity in hairy roots was measured by luminometer (Centro LB960 XS3, Bert-hold, Germany) with Coelenterazine (Prolume Ltd., Pinctop, USA) as substrate. The hairy roots with luciferase value greater than 3000 were considered as successful transformants. Hairy roots transformed by only K599 were considered untransformed type. Both transgenic and untransformed hairy roots were exposed to 0.5 mM CaCl2 solution (pH 4.5) with or without 30 μM AlCl3 within a 5-ml plastic tube. Root exudates were collected at 4 h for citrate efflux measurement. Citrate concentrations were measured by the enzymatic method according to Delhaize et al. (1993). Ten root apices (0–1 cm) were excised for Al content measurements. Al concentrations in hairy root apices were extracted by 2 M HCl and determined by an atomic adsorption spectrophotometer with a graphite furnace atomizer (PerkinElmer AAnalyst 700, USA).
Arabidopsis transformation of GmMATE75, GmMATE79 and GmMATE87
The constructs of modified pCAMBIA3301 vector prompted by CaMV 35S and labelled by Nanoluciferase were introduced into an Agrobacterium tumefaciens strain (Agl0) and subsequently transformed into Arabidopsis ecotype (Col-4) or the atmate mutant (SALK_081671) by the floral dip method (Clough and Bent 1998). The transgenic seedlings were screened by spraying with Basta herbicide and confirmed by luciferase activity and reverse transcription-polymerase chain reaction (RT-PCR) measurements. Arabidopsis leaves with luciferase activity value greater than 5000 were considered as successful transformation. Homozygous T3 lines were cultured in Al-containing mediums for measuring relative root elongation according to Sun et al. (2014). The root growth of representative plants from two independent transgenic lines grown in MS medium for 5 d was recorded; uniform seedlings were grown on solid agar medium supplied with 4.3 mM CaCl2 and 3% sucrose (with 0 or 100 μM AlCl3 at pH 4.5 for 2 d).
Tissue-level localization of GmMATE expression via histochemical staining of GUS activity
The sequences from upstream of the start condon of GmMATEs respectively were isolated from Jiyu70 genomic DNA, then constructed to modified pCAMBIA3301 vector with GUS label. The promoter sequences of GmMATE75, GmMATE79, GmMATE87 were 1499 bp, 2022 bp and 2000 bp respectively. The resulting constructs were sequenced and electroplated into the Agrobacterium strain K599. The transformation of soybean Jiyu 62 cotyledons was performed according to Subramanian et al. (2005). After 4 h exposure to 0.5mMCaCl2 solution (pH 4.5) containing 0 or 30 μM AlCl3, hairy roots were rinsed within a X-gluc solution (RTU4032 Real-times Biotechnology, Beijing, China) for staining observation. The staining in roots were observed and photographed with microscope (Zeiss 2012 Observer A1, Göttingen, Germany). For transection observation, roots were dissected at 0–9 mm and immediately fixed in solution containing ethanol/ acetic acid /formaldehyde at a 9:1: 1 ratio. After 24 h fixation, the samples were rinsed by 70% ethanol for 3 times and dehydrated gradually in ethanol (70%, 85%, 95% and 100%), and embedded in paraffin wax. Slices with (10 μm) were dissected from the root apex part by a microtome (LEICA Biosystems RM2245, Wetzlar, Germany) and placed on a glass slides. The slides were covered with glycerol and observed by microscope.
Statistical analysis
Significant difference among treatments or transgenic lines were evaluated by Data Processing System (Tang and Zhang 2012).
Results
Bioinformatics analysis
GmMATE75, GmMATE79 and GmMATE87 highly conserved in including 12 transmembrane regions and a 97 amino acids loop between 2nd and 3rd transmembrane domains (Fig. S1). Much variation was present in the N-terminal and in this long loop between TM2 and TM3 among the three putative soybean citrate transporters (Fig. S1).
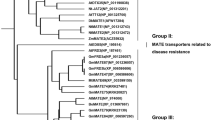
A sequence divergence of citrate transporters in the MATE family existed between monocots and dicots but was not strongly related to their functions in Al resistance or iron deficiency (Fig. S2). TaMATE1-4B, HvAACT1, ScFRDL1, OsFRDL1, SbMATE, OsFRDL4, ZmMATE1, and ZmMATE2 in monocots clustered closely and showed less similarity to EcMATE3, GmMATE87, AtMATE, BoMATE, EcMATE1, AtFRD3, GmMATE75, VuMATE1, LjMATE, GmMATE87 and GmFRD3b in dicots. GmMATE75 showed high amino acid sequence homology to VuMATE (with 78% identity) from Vigna umbellata and AtFRD3 (with 60% identity). GmMATE79 showed high similarity to AtMATE (65% identity) and BoMATE (65% identity). GmMATE87, encoding 553 amino acids, clustered closely with AtFRD3 (61%) and LjMATE (72%) (Fig. S2). The MATE proteins indicated with different colours represented different functions. Their varied distribution suggested their physiological role related less to their protein similarity.
The transcriptional patterns of GmMATE75, GmMATE79 and GmMATE87
Jiyu70 has been reported to have a larger amount of Al-induced citrate efflux thus confers to higher Al resistance compared to Jiyu 62 (Zhou et al. 2018a).GmMATE75, Gm MATE79 and GmMATE87 all constitutively expressed in soybean roots (Fig. 1a-c). In the time course experiment, both genotypes displayed high transcriptional expression level of GmMATE75 under Al stress. Nearly a 200-fold increase in GmMATE75 expression was found in both genotypes after 4 h Al treatment (Fig. 1a). GmMATE75 expression in Jiyu 70 was higher than that of Jiyu 62 starting from 8 h Al treatment (Fig. 1a). This result is consistent with our previous study, which showed that more citrate secretion from Jiyu 70 than Jiyu 62 was found after 8 h Al treatment (Zhou et al. 2018a). Almost 7-fold higher transcription expression of GmMATE79 was induced in Jiyu 70 at 2 h, then decreased to about 2-fold within the following Al treatment duration (Fig. 1b). The expression of GmMATE79 remained constant in Jiyu 62 over 24 h Al treatment (Fig. 1b). Higher expression of GmMATE87 was also induced after 2 h Al treatment but then returned to basal level (Fig. 1c). The expression of GmMATE87 sustained 2-fold increase from 4 h Al treatment duration for Jiyu62 (Fig. 1c). Spatial expression analysis showed that GmMATE75, GmMATE79 and GmMATE87 were distributed evenly in the root segments at 0–1 cm, 1–2 cm and 2–3 cm under control conditions (Fig. 1d-f). Four hours of Al treatment enhanced transcription levels of these three MATE-family genes in 0–3 cm root segments, but the increase was more profound in mature root zones especially in the root segment of 2–3 cm (Fig. 1d-f).
Temporal and spatial expression ofGmMATE75(ad),GmMATE79(be) andGmMATE87(cf) in soybean under Al stress. Seven-day-old soybean seedlings (Jiyu70 and Jiyu62) were exposed to 0.5 mM CaCl2 solution containing 30 μM AlCl3 (pH 4.5). The 0–1 cm root apices were excised after 0, 2, 4, 8, 12, and 24 h Al treatment to study temporal expression. The 0–1 cm, 1–2 cm and 2–3 cm root segments (Jiyu70) were also excised from 4 h Al-treated or control soybean roots to study spatial expression. The expression of GmMATE75, GmMATE79 and GmMATE87 were examined by qRT-PCR with β-tubulin as the reference gene. Data are represented as means±standard deviation (SD) of three biological replicates. Different letters above columns represented significantly different (p < 0.05)
The transcript levels of GmMATE79 and GmMATE87 (but not of GmMATE75) were also up-regulated under Cd2+, La3+, and Cu2+ stress (Fig. 2a-c). Ten days of -Fe treatment increased the transcript abundance about 2-fold for GmMATE79 and approximately 30-fold for GmMATE75, but decreased the transcript level of GmMATE87 by half compared with those in + Fe culture (Fig. 3a-c).
Transcriptional expression ofGmMATE75(a),GmMATE79(b) andGmMATE87(c) under Cd2+, La3+, Cu2+and Al3+stresses in soybean root apices. Seven-day-old soybean seedlings (Jiyu70) were exposed to 0.5 mM CaCl2 solutions containing 25 μM Cd2+, 10 μM La3+, 1 μM Cu2+ and 30 μM Al3+ (pH 4.5). The 0–1 cm root apices were excised after 4 h Al treatment to study gene expression. Data are represented as means ±SD of three biological replicates. Different letters above columns represented significantly different (p < 0.05)
Transcriptional expression ofGmMATE75(a),GmMATE79(b) andGmMATE87(c) under Fe deficiency conditions. Germinated soybean seedlings (Jiyu 70) were cultured in nutrient solution containing Fe-EDTA or not. After 10 days of culture, soybean root apices (0–1 cm) were excised to extract RNA to study gene expression. Data are represented as means ±SD of three biological replicates. Different letters above columns represented significantly different (p < 0.05)
Subcellular localization of GmMATE75, GmMATE79 and GmMATE87
The cellular localization was characterized by a transient expression assay with translational fusions of the separate citrate transporter gene with GFP in Arabidopsis protoplast cells. In comparison of the non-specific fluorescence distribution of cell transformed with the empty GFP vector, transient expression of GFP -GmMATE75, GFP-GmMATE79 and GFP-GmMATE87 can respectively induce fluorescence in the cell periphery of the Arabidopsis protoplasts that was overlapped with the plasma membrane marker staining (Fig. 4). Thus, all of GmMATE75, GmMATE79 and GmMATE87 are localized to the plasma membrane (Fig. 4).
Subcellular localization of proteins by transient expression of GFP-GmMATE75, GFP -GmMATE79 and GFP-GmMATE87 fusion proteins in Arabidopsis protoplast cells. The first-row images show GFP alone, and the second-, third- and fourth-row images are the fusion proteins GFP-GmMATE75, GFP -GmMATE79 and GFP-GmMATE87, respectively
Transgenic hairy roots with overexpression of GmMATE75, GmMATE79 or GmMATE87
Promoter CaMV35S driven GmMATE75, GmMATE79, and GmMATE87 were separately overexpressed in soybean hairy roots (Jiyu 62). GmMATE75-OE, GmMATE79-OE and GmMATE87-OE hairy roots increased their corresponding transcript levels (Fig. 5a-c), and citrate efflux under either –Al or + Al treatment (Fig. 5d-f). The Al-treated GmMATE79-OE, GmMATE75-OE, GmMATE87-OE hairy roots also contained less Al in their root apices than wild-type hairy roots (Fig. 5g). Consistent with its less citrate efflux, GmMATE87-OE contained more Al in their root apices compared with GmMATE79-OE or GmMATE75-OE (Fig. 5g). The above results indicated that the overexpression of GmMATE75, GmMATE79 and GmMATE87 resulted in increased citrate efflux and decreased Al content in root apices, which underscores their contribution to Al resistance.
Phenotype of Al tolerance in soybean hairy roots overexpressingGmMATE75,GmMATE79andGmMATE87. Transcriptional expression of GmMATE75 in GmMATE75-OE soybean hairy roots (a), GmMATE79 in GmMATE79-OE soybean hairy roots (b) and GmMATE87 in GmMATE87-OE soybean hairy roots (c). Citrate efflux from soybean hairy roots of GmMATE75-OE (d), GmMATE79-OE (e) and GmMATE87-OE (f). (g)Al content in the root apices of soybean hairy roots of GmMATE75-OE, GmMATE79-OE and GmMATE87-OE. The induction of soybean hairy roots and treatment procedure was described in Material and Methods. Al content in root apices of GmMATE75-OE, GmMATE79-OE and GmMATE87-OE(g). Data are represented as means ±SD of three biological replicates. Different letters above columns represented significantly different (p < 0.05)
Heterologous overexpression of GmMATE75, GmMATE79 and GmMATE87 in Arabidopsis
RT-PCR has shown that the transcript levels of GmMATE75, GmMATE79 and GmMATE87 were separately increased in their corresponding Arabidopsis overexpression lines (Fig. S3). Without Al, transgenic and Col-4 Arabidopsis showed similar root growth (Fig. 6). Root elongation was inhibited by 55% by 100 μM Al stress in Col-4 Arabidopsis (WT) or vector transgenic line (Vector). GmMATE75-OE lines showed very similar root elongation inhibition at approximately 53% (GmMATE75-OE1, GmMATE75-OE2) (Fig. 6ad. Two GmMATE79-OE lines showed 51% and 55% root elongation inhibition, respectively, under Al stress (GmMATE79-OE1, GmMATE79-OE2) (Fig. 6b, e). Root elongation inhibition was approximately 55% and 58% in two GmMATE87-OE lines under Al stress (GmMATE87-OE1, GmMATE87-OE2) (Fig. 6c, f). Increases in root growth were found in the GmMATE79-OE, GmMATE75-OE and GmMATE87-OE lines comparing with Col-4 or vector transformation line under Al stress (Fig. 6a-f). Consistently, more citrate efflux were found in the GmMATE79-OE, GmMATE75-OE and GmMATE87-OE lines compared with the wild type under Al stress (Fig. 6g).
Phenotype of Al resistance in transgenic Arabidopsis overexpressingGmMATE75,GmMATE79orGmMATE87. Phenotype analysis of Arabidopsis Col-4, GmMATE75-OE (a), GmMATE79-OE(b) and GmMATE87-OE (c). Relative root elongation of Arabidopsis Col-4, GmMATE75-OE (d), GmMATE79-OE (e), and GmMATE87-OE (f) under Al stress. Scale bar = 5 mm. Citrate efflux from Arabidopsis Col-4 and transgenic Arabidopsis overexpressing roots of GmMATE75-OE, GmMATE79-OE and GmMATE87-OE (g).The gene transformation and treatment procedure was described in Material and Methods. Data are represented as means ±SD (n = 10 for root elongation measurement, n = 3 for the citrate efflux measurement). Different letters above columns represented significantly different (p < 0.05)
RT-PCR monitor has shown that GmMATE75, GmMATE79 and GmMATE87 were separately complementally expressed in Arabidopsis atmate mutant (Fig. S3). Root elongation inhibition of 75% was found in the atmate mutant or atmate-vector line under Al stress (Fig. 7). The Al-induced root elongation inhibition was approximately 62% and 70%, respectively, in the two complementary lines of GmMATE75 (Gm MATE75-CE1, GmMATE75 -CE2) (Fig. 7a, d). Complementary expression of GmMATE79 decreased the root elongation inhibition to 62% and 69%, respectively, in two Arabidopsis lines (GmMATE79-CE1, GmMATE79-CE2) (Fig. 7b, e). Root elongation inhibition under complementary expression of GmMATE87 was 70% and 71% under Al stress (GmMATE87-CE1, GmMATE87-CE2) (Fig. 7c, f). Thus, each of GmMATE75, GmMATE79 and GmMATE87 can partially restore root elongation in the atmate mutant under Al stress (Fig. 7). The amounts of citrate efflux from GmMATE75-CE, GmMATE79-CE and GmMATE87-CE lines was also higher than those in the control (Fig.7g).
Phenotype of Al resistance of Arabidopsisatmatemutants complementarily expressingGmMATE75,GmMATE79orGmMATE87. Phenotypic analysis of WT, atmate, GmMATE75-CE(a), GmMATE79-CE (b) and GmMATE87-CE(c). Scale bar = 5 mm. Relative root elongation of atmate, WT, GmMATE75-CE(d), GmMATE79-CE(e) and GmMATE87-CE(f) under Al stress. The bars represent the means ± SE, n = 10. Citrate efflux from Arabidopsis Col-4, Arabidopsis atmate mutants and atmate mutants complementarily expressing of GmMATE75-CE, GmMATE79-CE and GmMATE87-CE (g).The gene transformation and treatment procedure was described in Material and Methods. Data are represented as means ±SD (n = 10 for root elongation measurement, n = 3 for the citrate efflux measurement). Different letters above columns represented significantly different (p < 0.05)
β-Glucuronidase (GUS) staining
Jiyu 62 exhibited different sequences in the promoter region of three GmMATEs from Jiyu 70. We successfully cloned 1499 bp, 2022 bp, and 2000 bp DNA sequences respectively from the upstream of start condon of GmMATE75, GmMATE79 and GmMATE87 from Jiyu 70. But we failed to clone those sequences from Jiyu 62. As a transcription factor, GmSTOP1a was found to regulate Al and proton resistance in soybean (Zhou et al. 2018b). Jiyu 70 and Jiyu 62 displayed same GmSTOP1a sequence and very similar expression pattern under Al stress (Data not shown). It is easy to induce strong hairy roots from cytocledon of Jiyu 62. Thus, each promoter fragment fused with a GUS reporter gene from Jiyu 70 was introduced into Jiyu 62 hairy roots with Agrobacterium transformation. In the absence of Al stress, GUS staining was restricted in the central cylinder zone of root apex for each of pGmMATE75::GUS, pGmMATE79::GUS and pGmMATE87::GUS transformation (Fig. 8a-d, i-l, q-t). After 4 h Al exposure, GUS staining extended from central cylinder to cortical and epidermis cells for pGmMATE75::GUS or pGmMATE79::GUS transformation (Fig.8e-h, m-p). But for pGmMATE87::GUS transformation, Al exposure strengthened the GUS staining, but still restricted within central cylinder region for the three root segments within 0–9 mm root apical zone (Fig. 8u-x).
Tissue-level localization ofGmMATE75,GmMATE79andGmMATE87expression via promoter-Gus fusion. Transgenic soybean hairy roots were exposed to 0.5 mM CaCl2 solution with or without 30 μM AlCl3 for 4 h. Activation of the GmMATEs promoter was observed by Gus staining in intact root (0–3 mm, 3–6 mm and 6–9 mm) or its cross section slices (within 0–9 mm). a-d showed promoter of GmMATE75 under no Al stress. e-h showed promoter of GmMATE75 under stress. i-l showed promoter of GmMATE79 under no Al stress. m-p showed promoter of GmMATE79 under Al stress. q-t showed promoter of GmMATE87under no Al stress. u-x showed promoter of GmMATE87 under Al stress
Discussion
Al-induced citrate efflux has been well-proven as one of the most important Al resistance mechanism in soybean (Yang et al. 2000, 2001; Silva et al. 2001). In this report, GmMATE75, GmMATE79 and GmMATE87 in the MATE family are proposed as plasma-membrane-localized citrate transporters (Fig. 4) conferring Al-induced citrate efflux in soybean (Figs. 5, 6, and 7).The transcript levels of GmMATEs displayed almost evenly within root apices and base regions (Fig. 1d-f). GUS staining of transgenic hairy roots indicated that three GmMATEs mainly expressed in the central cylinder under –Al stress but the expression of GmMATE75 and GmMATE79 was strengthen and expanded to the cortex and epidermal cells in root apical region under Al stress (Fig. 8). The three MATE genes exhibited distinct expression patterns under Al stress (Figs. 1, 2, and 3), which might result from various regulation methods that contribute differently to Al-induced citrate efflux in soybean.
GmMATE75, GmMATE79 and GmMATE87 are all citrate transporters conferring citrate efflux from soybean roots
The 22 reported citrate transporters in the MATE family were conserved between dicots and monocots, but loosely correlated with their known biological functions, such as Al resistance or Fe acquisition (Fig. S1, Fig. S2). GmMATE79, GmMATE75 and GmMATE87 showed the highest similarities to AtMATE (Liu et al. 2009), VuMATE1 (Liu et al. 2013) and LjMATE (Takanashi et al. 2013) respectively (Fig. S2). The former two are Al-activated citrate transporters that conferred Al-induced citrate efflux in Arabidopsis andrice bean, respectively (Liu et al. 2009; Yang et al. 2011), and the latter was a citrate transporter responsible for iron supply to the nodule infection zone of Lotus japonicus (Takanashi et al. 2013).
Consistent with the above multiple sequence alignment and phylogenetic tree, subcellular localization analysis in Arabidopsis protoplast verified that GmMATE79, GmMATE75 and GmMATE87 are all localized to the plasma membrane (Fig. 4). CaMV 35S promoter-driven overexpression of GmMATE75, GmMATE79 or GmMATE87 in soybean hairy roots, Arabidopsis or Arabidopsis mutant all resulted in increased citrate efflux under either –Al or + Al stress (Figs. 5, 6, and 7). Thus, we speculated each of GmMATE79, GmMATE75 or GmMATE87 have the capability to release citrate. The lower Al content in GmMATE75-OE, GmMATE79-OE or GmMATE87-OE soybean hairy roots (Fig. 5g), and higher relative root elongation in Arabidopsis GmMATE75-OE, GmMATE79-OE, GmMATE87-OE (Fig. 6) or GmMATE75-CE, GmMATE79-CE, GmMATE87-CE (Fig. 7) also indicated their acquirement of Al exclusion and Al resistance.
There existed an approximately 4 h lag phase preceding a large amount of citrate efflux and Al exposure in soybean (Yang et al. 2001), which was clearly classified in pattern II (Ma et al. 2001). Citrate exudation was non-detectable by HPLC when soybean was grown in the absence of Al (Silva et al. 2001; Yang et al. 2001). A small amount of citrate efflux from soybean roots can be detected after 2 h or 3 h Al treatment (Silva et al. 2001; Yang et al. 2001). However, in the present study, the more sensitive enzymatic assay detected some citrate efflux from both wild-type and transgenic hairy roots without Al treatment. GmMATE75-OE, GmMATE79-OE or GmMATE87-OE hairy roots each with higher transcript levels of GmMATEs (Fig. 5a-c), secreted more citrate without Al treatment compared to the untransformed hairy roots (Fig. 5d-f). GmMATE75, GmMATE79 and GmMATE87 constitutively contribute to citrate efflux and do not require Al activation, which is similar to AtFRD3 overexpression in barley but different from HvAACT1 or SbMATE with necessarity of Al to activate citrate efflux (Zhou et al. 2014).
CaMV 35S was used to constitutively express the MATE proteins, thus abrogating the tissue specific localization factor, which has been regarded as critical in determining their biological functions in Fe acquisition or Al detoxification (Yokosho et al. 2011; Maron et al. 2010; Fujii et al. 2012). For example, AtFRD3 functions as a citrate transporter mainly facilitating Fe acquisition (Durrett et al. 2007), but its CaMV 35S -driven overexpression can enhance Al resistance in Arabidopsis (Durrett et al. 2007) and barley (Zhou et al. 2014). VuMATE1 was proven as citrate transporter conferring Al resistance (Fan et al. 2015). But, complementation of frd3–1 with CaMV 35S mediated VuMATE1 expression could restore its Fe deficiency phenotype (Liu et al. 2016). Thus, tissue expression or Cis-regualtion of citrate transporters seemed to play more important role in determining their biological function.
The expression of GmMATE75, GmMATE79 and GmMATE87
Tissue localization of citrate transporter was pivotal for its Al detoxification capability. Typically, Fujii et al. (2012, 2018) indicated that barley accessions in East Asia and Europe have developed independent strategies including 1 kb transponson insertion or 15.3 kb multi-retrotransposon-like sequence insertion to regulate HvAACT1 spatial expression to adapt to acid soils. qRT-PCR revealed that three GmMATEs display similar spatial expression patterns with distinct level in response to Al stress (Fig. 1a-c). They were constitutively expressed in soybean roots with slightly higher expression in the more mature region (2–3 cm) (Fig. 1d-f). Their spatial expression and distribution were different from those of other MATE citrate transporter homologues, such as SbMATE (Magalhaes et al. 2007), HvAACT1 (Fuji et al. 2012) and OsFRDL4 (Yokosho et al. 2011), whose expression levels were found to be higher at the root apices than that in the mature zones. The VuMATE1 in rice bean even showed restricted expression in only the root apex zone (Yang et al. 2011). The distinct spatial expression features of the soybean MATE genes might relate to the following two factors. (1) Soybean Al accumulation characteristics: In soybean, Al accumulation was detected in all of the root segments with significantly higher Al concentrations in the 2–3 and 3–4 cm segments (Nian et al. 2004). The hematoxylin-stained region was confined to the mature root zone and extended to include the root apices with increasing Al treatment concentrations (Nian et al. 2004); (2) The three MATE genes might also be required for both characterized and still-uncharacterized functions in the absence of Al stress. Small amounts of citrate and malate efflux were detected from soybean roots under Cu2+, Cd2+, or La3+ treatment (Nian et al. 2004; Silva et al. 2001) Consistently, the expression of GmMATE79 and GmMATE87 was less specific to Al stress and responded notably to Cu2+, Cd2+ or La3+ stress (Fig. 2b, c). It has been known that homologues of citrate transporters are involved in either Fe translocation or Al tolerance depending on individual genes (Yokosho et al. 2011; Maron et al. 2010). After 10 days -Fe culture resulted in 2 folds increase of GmMATE79 (Fig. 3b) and almost 30-folds increase in GmMATE75 expression (Fig. 3a), implying their putative roles in Fe acquisition. However, the transcript abundance of GmMATE87, which clustered more closely with AtFRD3a and LjMATE, known citrate transporters functioning in Fe acquisition (Durret et al. 2007; Takanashi et al. 2013), was decreased after 10 days of Fe deficiency. GUS-staining observation indicated Al treatment strengthened and extended pGmMATE75::GUS or pGmMATE79::GUS from central cylinder to outer tissues including cortex and epidermis cells (Fig.8). Considering that citrate need to release to rhizosphere to protect Al injury, GmMATE75 and GmMATE79 might be more important in Al detoxifying for soybean. But the capabilities of GmMATEs were probably underestimated with GUS staining observation after promoter of Jiyu 70 transformation into Jiyu 62. (1) The regulatory systems of MATE family citrate transporters are complex. STOP1/ART1 type transcription was reported to regulate citrate transporter genes such as AtMATE (Liu et al. 2009), VuMATE1and VuMATE2 (Yang et al. 2011; Fan et al. 2015; Liu et al. 2018) and OsFRDL4 (Yokosho et al. 2016b). Its regulation was affected by other factors. WRKY22 was found to function together with ART1 in regulation of OsFRDL4 (Li et al. 2018). An F-box protein-encoding gene regulation of Atalmt1 expression 1 (RAE1) was reported to interact with and promote the degradation of AtSTOP1 via the ubiquitin-26S proteasome pathway (Zhang et al. 2018). Even Jiyu 70 and Jiyu 62 displayed same GmSTOP1a sequence and very similar expression pattern under Al stress (Data not shown). We can’t ensure the homologies of GmSTOP1a or other genes such as WRKY or RAE1 displayed same sequence and expression pattern between Jiyu 70 and Jiyu 62; (2) The capacity of GmMATEs regulation also depended on promoter length. Typically, very important regulatory elements including transponson insertion or DNA methylation localized at more than 5000 bp upstream of 5’ UTR of HvAACT1 (Fujii et al. 2012, 2018). More effective regulatory elements were expected to reveal in promotor regions of GmMATEs.
Fine-tuning root citrate secretion with two separate root citrate transport systems was reported in rice bean to regulate biphasic Al-induced citrate efflux (Liu et al. 2018). The constitutively expressed VuMATE2 and Al-increased expressed VuMATE1 were respectively suggested to be responsible for the early phase of minor citrate secretion and late phase of large amount citrate efflux under Al stress (Liu et al. 2018). In present study, the rapid and strong induction of GmMATE75 is similar to or even greater than that of OsFRDL4 (Yokosho et al. 2011). Its expression was greatly induced at 2 h Al treatment (near 100-fold) and reached approximately 500-fold at 8 h (Fig. 1a). GmMATE79 (Fig. 1b) was also induced after 2 h Al treatment with lower level as AtMATE (Liu et al. 2009), ZmMATE1 (Maron et al. 2010) and ScFRDL2 (Yokosho et al. 2010), but decreased its expression in the following Al treatment duration. According to their expression patterns, we propose as follows: the constitutive expression and smaller increase of GmMATE79 and GmMATE75 expression resulted in minor citrate efflux from soybean roots under no Al or less than 4 h Al treatment. However, the large amount of Al-induced citrate efflux after 4 h might result from the strong induction of GmMATE75. The higher expression of GmMATE75 in Jiyu70 (Fig. 1) could also partially explain the genotypic difference that Jiyu 70 secreted more citrate than Jiyu 62 after 8 h Al treatment (Zhou et al. 2018a).
The origin of the Al-induced citrate efflux pattern in soybean roots is complex. (1) The contribution of citrate metabolism to citrate efflux can not be neglected. Our previous study has shown that cytosolic malic enzyme (GmME1) contributed to internal citrate content then Al-induced citrate efflux from soybean (Zhou et al. 2018a). GmME1 displayed higher expression in Jiyu70 in comparison to Jiyu62 under Al stress (Zhou et al. 2018a) (2) The contribution of the other putative soybean citrate transporters has not been clarified; (3) More effective regulatory elements might exist in 5’UTR-uptream of GmMATEs to promote the high expression of GmMATEs under Al stress. Jiyu 70 was bred from Jilin province of China with less acidic soils and exhibited moderate Al resistance. Efficient regulation of GmMATEs mechanisms deserved to be revealed in soybean cultivars with higher Al resistance capability. The genetics studies have revealed the complexity of Al-induced citrate efflux mechanisms in soybean. Some quantitative trait loci analyses have indicated that Al tolerance in soybean is a multigene trait (Abdelhaleem et al. 2014; Sharma et al. 2010; Qi et al. 2008). Using recombinant inbred lines (RILs) derived from the cross of ‘Essex’ with ‘Forrest’ identified two major loci encompassing genes implicated in citrate metabolism (Sharma et al. 2010). The population originating from Young × PI 416937 identified sixteen single nucleotide polymorphisms in the citrate synthase homologue on chromosome Gm08 (Abdelhaleem et al. 2014). However, no genetic report has stated the loci or genes conferring citrate transport implicated in Al resistance in soybean until now.
In summary, GmMATE75, GmMATE79 and GmMATE87 are proposed as plasma-membrane-localized citrate transporters, having capability to release citrate efflux under –Al or + Al stress. Their additive effects might contribute to Al-induced citrate efflux from soybean. Their expression levels were also affected under iron deficiency conditions, but their functions in iron acquisition are not discussed here.
Abbreviations
- Al:
-
Aluminum
- CaMV:
-
Cauliflower mosaic virus
- GFP:
-
Green Fluorescent protein
- MATE:
-
Multidrug and toxic compound extrusion
References
Abdelhaleem H, Carter TE, Rufty TW, Boerma HR, Li Z (2014) Quantitative trait loci controlling aluminum tolerance in soybean: candidate gene and single nucleotide polymorphism marker discovery. Mol Breeding 33:851–862
Bianci-Hall CM, Carter TE, Rufty TW, Arellano C, Boerma HR, Ashley DA (1998) Heritability and resource allocation of aluminum tolerance derived from soybean PI 416937. Crop Sci 38:513–522
Bianci-Hall CM, Carter TE, Bailey MA, Mian MAR, Rufty TW, Ashley DA (2000) Aluminum tolerance associated with quantitative trait loci derived from soybean PI 416937 in hydroponics. Crop Sci 40:538–544
Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum-Aestivum L.) (II. Aluminum-stimulated excretion of malic acid from root apices). Plant Physiol 103:695–702
Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144:197–205
Fan W, Lou HQ, Gong YL, Liu MY, Cao MJ, Liu Y, Yang JL, Zheng SJ (2015) Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and Al tolerance mechanisms. New Phytol 208:456–468
Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, Sato K, Nakazono M, Ma JF (2012) Acquisition of aluminum tolerance by modification of a single gene in barley. Nat Commun 3:713
Fujii M, Yokosho K, Yamaji N, Yamane M, Saisho D, Sato K, Ma JF (2018) Retrotransposon insertion and DNA methylation regulate aluminum tolerance in European barley accessions. Plant Physiol 178(2):716–727
Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF (2007) an aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091
Horst WJ, Asher CJ, Cakmak L, Szulkiewicz P, Wissemeier AH (1992) Short-term response of soybean roots to aluminum. Plant Physiol 140:174–178
Kochian LV, Pineros MA, Liu JP, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 66:571–598
Lei GJ, Yokosho K, Yamaji N, Ma JF (2017) Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat. Plant Cell Physiol 58(12):2179–2189
Li GZ, Wang ZQ, Yokosho K, Ding B, Fan W, Gong QQ, Li GX, Wu YR, Yang JL, Ma JF, Zheng SJ (2018) Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol 219:149–162
Liu JP, Magalhaes JV, Shaff J, Kochian LV (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 57:389–399
Liu MY, Chen WW, Xu JM, Fan W, Yang JL, Zheng SJ (2013) The role of VuMATE1 expression in aluminum-inducible citrate secretion in rice bean (Vigna umbellata) roots. J Exp Bot 64:1795–1804
Liu J, Li Y, Wang W, Gai JY, Li Y (2016) Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17:223
Liu MY, Lou HQ, Chen WW, Pineros MA, Xu JM, Fan W, Kochian LV, Zheng SJ, Yang JL (2018) Two citrate transporters coordinately regulate citrate secretion from rice bean root tip under aluminum stress. Plant Cell Environ 41(4):809–822
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25(4):402–408
Ma JF, Ryan PR, Delhaize E (2001) Aluminum tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6:273–278
Magalhaes JV, Liu J, Guimaraes CT, Lana UGP, Alves VMC, Wang YH, Schaffert RE, Hoekenga OA, Pineros MA, Shaff JE, Klein PE, Carneiro NP, Coelho CM, Trick HN, Kochian LV (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39:1156–1161
Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SN, Kochian LV (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61:728–740
Maron LG, Guimarães CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, Buckler ES, Coluccio AE, Danilova TV, Kudrna D, Magalhaes JV, Pineros MA, Schatz MC, Wing RA, Kochian LV (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Pro Natl Acad Sci 110:5241–5246
Nian H, Yang ZM, Hang H, Yan X, Matsumoto H (2004) Citrate secretion induced by aluminum stress may not be a key mechanism responsible for differential aluminum tolerance of some genotypes. J Plant Nutri 27:2047–2066
Qi B, Koriir P, Zhao T, Yu DY, Chen SY, Gai JY (2008) Mapping quantitative toxin tolerance in NJRIKY recombinant inbred line population of soybean (Glycine max). J Intergr Plant Biol 50:1085–1095
Rogers EE, Wu XL, Stacey G, Nguyen HT (2009) Two MATE proteins play a role in iron efficiency in soybean. J Plant Physiol 166:1453–1459
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653
Sawaki T, Kihara-Doi T, Kobayashi Y, Nishikubo N, Kawazu T, Koyama H, Sato S (2013) Characterization of Al-responsive citrate excretion and citrate-transporting MATEs in Eucalyptus camaldulensis. Planta 237:979–989
Sharma AD, Sharma H, Lightfoot DA (2010) The genetic control of tolerance to aluminum toxicity in the ‘Essex’ by ‘Forrest’ recombinant inbred line population. Theor Appl Genet 122:687–694
Silva IR, Smyth TJ, Raper CD, Carter TE, Rufty TW (2001) Differential aluminum tolerance in soybean: an evaluation of the role of organic acids. Physiol Plant 112:200–210
Subramanian S, Graham MY, Yu O, Graham TL (2005) RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol 137:1345–1353
Sun LL, Liang CY, Chen ZJ, Liu PD, Tian J, Liu GD, Liao H (2014) Superior aluminum (Al) tolerance of Stylosanthes is achieved mainly by malate synthesis through an Al-enhanced malic enzyme. SgME1 New phyt 202:209–219
Takanashi K, Yokosho K, Saeki K, Sugiyama A, Sato S, Tabata S (2013) LjMATE1: a citrate transporter responsible for iron supply to the nodule infection zone of lotus japonicus. Plant &Cell Physiol 54:585–594
Takanashi K, Shitan N, Yazaki K (2014) The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotech 31:417–430
Tang QY, Zhang CX (2012) Data processing system (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci 20(2):254–260 (in China)
Tovkach A, Ryan PR, Richardson AE, Lewis DC, Rathjen TM, Ramesh S, Tyerman SD, Delhaize E (2013) Transposon mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root apices. Plant Physiol 161:880–892
Wu X, Li R, Shi J, Wang J, Sun Q, Zhang H, Xing Y, Zhang N, Guo YD (2014) Brassica oleracea MATE encodes a citrate transporter, and enhances aluminum tolerance in Arabidopsis thaliana. Plant Cell Physiol 55:1426–1436
Xu MY, You JF, Hou NN, Zhang HM, Chen GA, Yang ZM (2010) Mitochondrial enzymes and citrate transporter contribute to the aluminum-induced citrate secretion from soybean (Glycine max) roots. Funct Plant Biol 37:886–892
Yang ZM, Sivaguru M, Horst WJ, Matsumoto H (2000) Aluminum tolerance is achieved by exudation of citric acid from roots of soybean (Glycine max). Physiol Plant 110:72–77
Yang ZM, Nian H, Sivagur M, Tanakamaru S, Matsumoto H (2001) Characterizaton of aluminum-induced citrate secretion in aluminum-tolerant soybean (Glycine max) plants. Physiol Plant 113:64–71
Yang XY, Yang JL, Zhou Y, Pineros MA, Kochian LV, Li GX, Zheng SJ (2011) A De nove synthesist citrate transporter, multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant Cell Environ 34:2138–2148
Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol 149:297–305
Yokosho K, Yamaji N, Ma JF (2010) Isolation and characterisation of two MATE genes in rye. Funct Plant Biol 37:296–303
Yokosho K, Yamaji N, Ma JF (2011) An Al-induced MATE gene is involved in external detoxification of Al in rice. Plant J 68:1061–1069
Yokosho K, Yamaji N, Ma JF (2016a) OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. J Exp Bot 67:5485–5494
Yokosho K, Yamaji N, Fujii-Kashino M, Ma JF (2016b) Retrotransposon-mediated aluminum tolerance through enhanced expression of the citrate transporter OsFRDL4. Plant Physiol 172:2327–2336
You JF, Zhang HM, Liu N, Gao LL, Kong LN, Yang ZM (2011) Transcriptomic responses to aluminium stress in soybean roots. Genome 54:923–933
Zhang Y, Zhang J, Guo JL, Zhou FL, Singh S, Xu X, Xie Q, Yang ZB, Huang CF (2018) F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Aarbidopsis. PNAS 116:319–327. https://doi.org/10.1073/pnas.1814426116
Zhou GF, Pereira JF, Delhaize E, Zhou M, Jurandir V, Magalhaes JV, Peter PR (2014) Enhancing the aluminum tolerance of barley by expressing the citrate transporter genes SbMATE and FRD3. J Exp Bot 65:2381–2390
Zhou Y, Yang ZM, Sun HR, Sun ZT, Lin B, Sun WJ, You JF (2018a) Soybean NADP-malic enzyme functions in malate and citrate metabolism and contributes to their efflux under Al stress. Frontiers Plant Sci 8:2246
Zhou Y, Yang ZM, Gong L, Liu RK, Sun HR, You JF (2018b) Molecular characterization of GmSTOP1 homologs in soybean under Al and proton stress. Plant Soil 427(1–2):213–270
Acknowledgements
Financial support for this research was provided by the National Natural Science Foundation of China (No. 31372124) and Jilin Natural Science Foundation of China (20130101084JC).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Jian Feng Ma.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 8058 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Wang, Z., Gong, L. et al. Functional characterization of three MATE genes in relation to aluminum-induced citrate efflux from soybean root. Plant Soil 443, 121–138 (2019). https://doi.org/10.1007/s11104-019-04192-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04192-w