Abstract
Key message
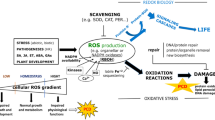
Chloroplast singlet oxygen initiates multiple pathways to control chloroplast degradation, cell death, and nuclear gene expression.
Abstract
Chloroplasts can respond to stress and changes in the environment by producing reactive oxygen species (ROS). Aside from being cytotoxic, ROS also have signaling capabilities. For example, the ROS singlet oxygen (1O2) can initiate nuclear gene expression, chloroplast degradation, and cell death. To unveil the signaling mechanisms involved, researchers have used several 1O2-producing Arabidopsis thaliana mutants as genetic model systems, including plastid ferrochelatase two (fc2), fluorescent in blue light (flu), chlorina 1 (ch1), and accelerated cell death 2 (acd2). Here, we compare these 1O2-producing mutants to elucidate if they utilize one or more signaling pathways to control cell death and nuclear gene expression. Using publicly available transcriptomic data, we demonstrate fc2, flu, and ch1 share a core response to 1O2 accumulation, but maintain unique responses, potentially tailored to respond to their specific stresses. Subsequently, we used a genetic approach to determine if these mutants share 1O2 signaling pathways by testing the ability of genetic suppressors of one 1O2 producing mutant to suppress signaling in a different 1O2 producing mutant. Our genetic analyses revealed at least two different chloroplast 1O2 signaling pathways control cellular degradation: one specific to the flu mutant and one shared by fc2, ch1, and acd2 mutants, but with life-stage-specific (seedling vs. adult) features. Overall, this work reveals chloroplast stress signaling involving 1O2 is complex and may allow cells to finely tune their physiology to environmental inputs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants experience a variety of cellular stresses, such as reactive oxygen species (ROS) produced within their energy-producing organelles (i.e., chloroplasts and mitochondria). Within chloroplasts during photosynthesis, harnessed light energy can lead to ROS production, causing damage to nearby macromolecules (e.g., lipids, proteins, DNA). Plants detoxify ROS through several enzymatic and non-enzymatic mechanisms (e.g., ROS scavenging enzymes, antioxidant production) (Apel et al. 2004; Noctor et al. 2018). However, these safety measures can be overwhelmed, especially under various environmental stresses including excess light (EL) (Triantaphylides et al. 2008), drought (Chan et al. 2016), salinity (Suo et al. 2017), and pathogen attack (Lu et al. 2018).
Photosynthesis produces the ROS hydrogen peroxide (H2O2) and superoxide (O2−) primarily at Photosystem I (PSI) and singlet oxygen (1O2) primarily at Photosystem II (PSII) (Asada 2006; Triantaphylides et al. 2008). These molecules can inhibit photosynthesis by causing photo-oxidative damage to photosynthetic machinery leading to photoinhibition (Asada 2006). Although ROS are cytotoxic molecules, they can report on a plant’s current environment (Chan et al. 2015; Foyer 2018; Mittler 2017). For instance, 1O2 can lead to signals that initiate cellular degradation (chloroplast degradation and cell death) and nuclear gene reprogramming via retrograde (chloroplast-to-nucleus) signaling (D'Alessandro et al. 2020; Dogra et al. 2019; Woodson 2022). As 1O2 has an extremely short half-life (~ 0.5–1.0 µsec (Ogilby 2010)), the bulk of chloroplast 1O2 likely remains within the organelle of origin. Thus, secondary messengers are expected to be involved in propagating the chloroplast 1O2 signal(s) to affect nuclear gene expression and cellular degradation (Dogra et al. 2019; Woodson 2022). Researchers have discovered several signaling factors, but their mechanisms still remain mostly unclear.
A major challenge in deciphering ROS signaling in plant cells is that natural stresses can lead to the production of multiple types of ROS (Noctor et al. 2014; Pospíšil 2016) in multiple sub-cellular compartments (Choudhury et al. 2017; Rosenwasser et al. 2013). To specifically understand how 1O2 signals, researchers use several Arabidopsis thaliana mutants that conditionally accumulate chloroplast 1O2 under specific growth conditions to dissect the genetic and biochemical attributes of 1O2 signaling pathways. The fluorescent in blue light (flu-1, referred to as flu henceforth) mutant was one of the first 1O2-producing mutants described (Meskauskiene et al. 2001). The flu mutant over-accumulates the tetrapyrrole (e.g., chlorophyll and heme) intermediate protochlorophyllide (Pchlide) when grown in the dark. When the mutant is exposed to light, the energized Pchlide (like other free tetrapyrroles) reacts with nearby ground-state oxygen (3O2) to produce 1O2 within the thylakoid membranes (Wang et al. 2016). This burst of 1O2 leads to the reprogramming of hundreds of genes in the nucleus through a retrograde signal, followed by initiation of bleaching and cell death (op den Camp et al. 2003; Wagner et al. 2004). A smaller set of 168 genes, called Early Singlet Oxygen Response Genes (ESORGs), may represent the initial response (within 60 min) to chloroplast 1O2 signals in flu mutants (Dogra et al. 2017). A forward genetic screen for signaling components in this pathway identified EXECUTER1 (EX1) as playing an important role (Lee et al. 2007; Wagner et al. 2004). When the ex1-2 loss-of-function mutation (referred to as ex1 henceforth) is introduced into the flu background, induction of nuclear gene expression and cell death (but not the accumulation of Pchlide or 1O2) is blocked (Lee et al. 2007). This breakthrough discovery was among the first evidence that 1O2-induced cell death and cellular degradation is due to a genetically encoded response rather than 1O2 toxicity.
Recent studies reveal EX1 may physically sense 1O2 in the grana margins (site of tetrapyrrole synthesis and photosystem II repair) through oxidation of tryptophan 643 in a domain of unknown function (DUF) (Dogra et al. 2019). The chloroplast metalloprotease FtsH2 degrades this oxidized EX1 protein, which is necessary for EX1 signaling (Dogra et al. 2017; Wang et al. 2016). These studies hypothesize an EX1 degradation peptide may act as a signaling factor. A conserved homolog of EX1, EX2 plays a role in 1O2 signaling (Lee et al. 2007; Page et al. 2017). Like EX1, EX2 undergoes oxidation by 1O2 at a conserved tryptophan residue and is subsequently degraded by FtsH2. However, degraded EX2 does not initiate retrograde signaling and cell death like EX1 (Dogra et al. 2022). Thus, EX2 may act as a decoy to protect EX1 and attenuate 1O2 signals to prevent excessive responses. Researchers also demonstrated, using flu protoplasts, the blue light photoreceptor CRYPTOCHROME 1 (CRY1) is involved in transducing the 1O2 cell death signal, leading to the possibility that blue light is involved in chloroplast stress signaling (Danon et al. 2006). However, such a signal only represents part of the 1O2 response as the impact of cry1 on 1O2–induced nuclear gene expression is limited.
A second 1O2 over-producing mutant is chlorina 1 (ch1-1, referred to as ch1 henceforth). This mutant lacks chlorophyll b and does not have a properly functioning/assembled photosystem II antennae complex that could protect the reaction center and quench 1O2 (Ramel et al. 2013). When ch1 is grown under EL conditions (≥ 1100 μmol photons m−2 s−1), PSII accumulates 1O2 in its reaction center located in the grana core. As in the flu mutant, the 1O2 initiates retrograde signaling to the nucleus and causes cell death. When ch1 mutants experience a mild level of photo-oxidative stress (≥ 450 μmol photons m−2 s−1) prior to EL treatments, they are more tolerant to subsequent EL stress, suggesting low levels of 1O2 can lead to stress acclimation (Ramel et al. 2013; Shumbe et al. 2017). In the case of the ch1 mutant, EX1 and EX2 appear to be dispensable for 1O2 signaling (a ch1 ex1 ex2 mutant still experiences cell death under EL stress) (Ramel et al. 2013). Instead, 1O2-triggered nuclear gene expression and cell death depends on oxidative signal inducible 1 (OXI1), a nuclear-localized serine/threonine kinase originally identified for its role in pathogen defense (Shumbe et al. 2016). Furthermore, accumulation of volatile carotenoid oxidation products [e.g., β-cyclocitral (β-cc)] produced by 1O2 accumulation at PSII are another part of this response (Ramel et al. 2012; Shumbe et al. 2014). Interestingly, signals induced by β-cc trigger nuclear gene expression and acclimation, but do not cause cellular degradation (Ramel et al. 2012). As such, we hypothesize that 1O2 signaling is a complex network controlling multiple physiological responses in the cell.
A third 1O2-producing mutant is accelerated cell death 2 (acd2-2, referred to as acd2 henceforth). This mutant experiences 1O2 bursts when grown under standard growth light conditions and produces seemingly random cell death lesions that spread across leaves (Mach et al. 2001). The acd2 mutant accumulates the chlorophyll breakdown intermediate, red chlorophyll catabolite (RCC) (Pruzinská et al. 2007). Similar to Pchlide accumulated in flu mutants, photosensitive RCC can absorb light energy and produce 1O2 in the cell (Pattanayak et al. 2012; Pruzinská et al. 2007). While the bulk (if not all) 1O2 in flu and chl mutants likely accumulates in chloroplasts (the grana margins and the grana cores, respectively), the acd2 mutants produce at least some 1O2 in the mitochondria. Previous work did not reveal how this 1O2 may signal or lead to cell death, but this pathway acts independently of EX signaling (acd2 ex1 ex2 mutants have similar lesion formation as acd2) (Pattanayak et al. 2012).
In addition to cell death and retrograde signaling, 1O2 can lead to chloroplast degradation. plastid ferrochelatase two (fc2) mutants accumulate the tetrapyrrole intermediate protoporphyrin-IX (Proto) immediately after dawn (Papenbrock et al. 2001; Woodson et al. 2015). Like Pchlide, Proto absorbs light energy and produces 1O2. The 1O2 leads to chloroplast degradation, retrograde signaling, and eventually cell death (Woodson et al. 2015). Even under permissive constant light conditions, a subset of chloroplasts (up to 35%) are selectively degraded in otherwise healthy cells, likely due to moderately high levels of Proto and 1O2 (Fisher et al. 2022). To understand the molecular signal in the fc2 mutant, we previously conducted a forward genetic screen to identify suppressors of 1O2–triggered cell death and identified 24 ferrochelatase two suppressor (fts) mutations that allow fc2-1 (hereafter referred to as fc2) seedlings to survive under diurnal cycling light conditions (Woodson et al. 2015).
When we cloned these fts mutants, we identified an important role for plastid gene expression in initiating the 1O2 signal in fc2 chloroplasts. Mutations affecting PENTATRICOPEPTIDE REPEAT CONTAINING PROTEIN 30 (PPR30) or “MITOCHONDRIAL” TRANSCRIPTIONAL TERMINATION FACTOR 9 (mTERF9) block cell death and the induction of nuclear stress genes when 1O2 accumulates in chloroplasts (Alamdari et al. 2020). These mutations lead to a broad reduction of plastid-encoded RNA-polymerase (PEP)-encoded transcripts, which is likely due to the predicted functions of PPR and mTERF proteins in post-transcriptional gene expression within plastids (Barkan et al. 2014; Wobbe 2020). In addition, we identified a third gene, CYTIDINE TRIPHOSPHATE SYNTHASE 2 (CTPS2), that is necessary for 1O2 signaling in the fc2 mutant (Alamdari et al. 2021). ctps2 mutants are deficient in chloroplast dCTP, leading to reduced chloroplast DNA content and plastid transcripts. Based on these mutations decreasing plastid gene expression, we hypothesized that a chloroplast-encoded protein (or RNA) is essential for the fc2 1O2 signaling pathway (Woodson 2022).
The same genetic screen revealed the cellular ubiquitination machinery is involved with 1O2 signaling in fc2 mutants. FTS29 encodes the cytoplasmic plant U-box E3 ubiquitin ligase (PUB4) protein, which is necessary to induce 1O2-dependent cell death (Woodson et al. 2015). As an E3 ligase, PUB4 is likely responsible for controlling the placement of ubiquitination modifications on a group of proteins in the cell (Callis 2014). Although its targets are unknown, 1O2-stressed chloroplasts do accumulate ubiquitin-tagged proteins. Thus, PUB4 may lead (directly or indirectly) to the ubiquitination of proteins associated with the chloroplast envelope during 1O2 and photo-oxidative stress (Jeran et al. 2021; Woodson et al. 2015). Together, these conclusions suggest posttranslational modifications are a possible mechanism a cell could use to identify damaged chloroplasts for turnover or repair (Woodson 2019).
Although researchers have identified several signaling 1O2 factors, they primarily study these components in the 1O2 accumulating genetic backgrounds in which they were identified. Presently, some evidence suggests these pathways are independent; ex1 does not suppress cell death in the fc2 (Woodson et al. 2015), ch1 (Ramel et al. 2013), or acd2 mutants (Pattanayak et al. 2012). As such, we hypothesize that multiple chloroplast 1O2 signaling pathways exist to control cellular degradation and nuclear gene expression. Here, we compare the fc2, flu, ch1, and acd2 mutants to test if they elicit separate chloroplasts signals. A meta-analysis of transcriptional responses in these mutants suggests a core response with unique signatures exists. However, a genetic analysis of these mutants and their suppressors revealed two major 1O2 signaling pathways. The flu mutant likely uses one distinct EX-dependent signal, while fc2, ch1, and acd2 share a second 1O2 signaling pathway with life-stage-specific (seedling vs. adult) features. Together these results demonstrate chloroplast 1O2-signaling is complex and may depend on the exact sites of 1O2 production, even within a single chloroplast.
Methods
Biological material, growth conditions, and treatments
The Arabidopsis ecotype Columbia (Col-0) was used as wt and the genetic background for all lines. Mutant lines used in this study are listed in Table S1. The fc2-1 T-DNA insertion line (GABI_766H08) (Woodson et al. 2011) and the oxi1 T-DNA insertion line (GABI_355H08) (Camehl et al. 2011) used were from the GABI (Kleinboelting et al. 2012) T-DNA collections and were described previously. The ex1-2 (SALK_002088), ex2-2 (SALK_021694), and ex2-3 (SALK_121009) T-DNA insertion lines used were from the SALK T-DNA collections (Alonso et al. 2003). SALK_002088 (Lee et al. 2007) and SALK_021694 (Uberegui et al. 2015) were previously described. The cry1-304 (Bruggemann et al. 1996), pub4-6 (Woodson et al. 2015), acd2-2 (Mach et al. 2001), flu-1 (Meskauskiene et al. 2001), ppr30-1 (Alamdari et al. 2020), and ch1-1 (Havaux et al. 2007) mutants were described previously. Higher order mutant combinations were generated by crossing and confirmed by PCR genotyping where applicable (primer sequences listed in Table S2).
Seeds were surface sterilized and plated using one of two methods; (1) a previously described liquid bleach washing protocol (Alamdari et al. 2020). Briefly, seeds were washed in 30% bleach with 0.04% Triton X-100 (v/) and then rinsed with sterile water three times by pelleting seeds at 3500×g for 1 min. (2) Chloride gas sterilization. For chloride gas surface sterilization, approximately 25–100 µl of seed was placed in 2 mL microcentrifuge tubes and placed in an airtight chamber with their lids open. Five mL of concentrated HCl were added to 150 mL of bleach (3.33% v/v) and the lid to the chamber was put on immediately. Seeds were removed 24 h later and allowed to air out for 15 min before plating. Sterilization Method 1 was used for plants monitored or assayed in the seedling stage. Sterilization Method 2 was used for growing plants for bulking seed, genotyping, and adult-stage experiments. Seeds were plated on Linsmaier and Skoog medium pH 5.7 (Caisson Laboratories North Logan, UT) with 0.6% micropropagation type-1 agar powder. Seeds were stratified for 4 to 5 days in the dark at 4 °C and were germinated in control conditions: constant white light (~ 120 μmol photons m−2 s−1) at 22 °C. To initiate stress signaling in fc2 mutant seedlings, plates were germinated under 6 h light/18 h dark diurnal light cycling conditions. To initiate cell signaling in flu seedlings, plates were germinated under control conditions, shifted to the dark after 5 days for up to 24 h (by wrapping in aluminum foil), and re-exposed to light.
To test adult phenotypes, seedlings were grown under seedling control conditions, transferred to soil, and grown under adult control conditions (100 μmol photons m−2 s−1 of constant light at 22 °C). To initiate stress signaling in fc2 and flu adult mutants, plants were shifted to 16 h light/8 h dark diurnal light cycling conditions when 21 days old. Seeds used for experiments were harvested from plants of a similar age. Photosynthetically active radiation was measured using a LI-250A light meter with a LI-190R-BNC-2 Quantum Sensor (LiCOR). All above experiments were performed in chambers with cool white fluorescent bulbs.
To initiate and monitor EL signaling in seedlings, plants were grown as described above, but in a Percival LED-30L1 with white LED lights at 120 μmol photons m−2 s−1. When 7 days old, the seedlings were then transferred to an EL chamber (a Percival LED 41L1 chamber with SB4X All-White SciBrite LED tiles) and exposed to 1200 μmol photons m−2 s−1 white light at 10 °C for 24 h. Maximum PSII quantum yield (Fv/Fm) was monitored in a FluorCam chamber (Closed FluorCam FC 800-C/1010-S, Photon Systems Instruments) as previously described (Lemke et al. 2021). For adult plants, seven-day-old seedlings were transferred to soil and grown under 70 μmol m−2 s−1 white light from LED panels until plants were 18 days old. The plants were then exposed to 1300 μmol photons m−2 s−1 white light at 10 °C in the EL chamber. Fv/Fm was monitored the same as for the seedlings.
Transcriptome data analysis
Previously published microarray expression data was gathered from studies describing fc2 (Woodson et al. 2015), flu (op den Camp et al. 2003), ch1 (Ramel et al. 2013), and ß-cc treated wt plants (Ramel et al. 2012). RNAseq data of flu mutant seedlings to identify ESORGs was from (Dogra et al. 2017). As the Affymetrix GeneChip Arabidopsis ATH1 Genome Array (fc2 and flu datasets, Table S3) and the CATv5 microarray (ch1 and ß-cc datasets, Table S4) have different gene coverage, only genes contained in both were used in the analysis. Gene groups recognized by a single probe were also removed as expression values could not be assigned to one specific gene. Finally, organellar gene transcript levels were removed from the analyses and only nuclear-encoded transcripts were considered. This left a total 19,895 genes for comparative analyses (Table S5).
Differentially expressed genes (DEGs) (induced or repressed) were identified from each dataset. For the fc2 dataset, 4-day-old etiolated wt and fc2 seedlings were compared 2 h after light exposure (Table S6). For the flu dataset, wt and flu adult plants were compared after 8 h dark and 1 h light re-exposure (Table S7). For the ch1 dataset, ch1 adult plants were treated with 2 days of EL and compared to untreated ch1 (Table S8). For the ß-cc treatment dataset, wt plants treated with ß-cc for 4 h were compared to water-treated controls (Table S9). For these datasets, we applied cutoff values of ± ≥ twofold mean expression and adjusted (Bonferroni) p-value ≤ 0.05. However, for the adult flu mutant dataset (op den Camp et al. 2003), an unreported significance cutoff was already applied by the authors. For the ESORG dataset, flu seedlings were placed in the dark for 4 h and then exposed to 30 or 60 min of re-illumination and compared to flu seedlings without re-illumination. A list of 168 ESORGs were identified (≥ twofold induction, FDR ≤ 0.05 cutoffs) that overlapped with an earlier analysis of induced transcripts in the flu mutant (Chen et al. 2015) (Table S10). These gene lists were then compared using the program Venny 2.1 by Juan Carlos Oliveros (https://bioinfogp.cnb.csic.es/tools/venny/index.html) [Oliveros (2007–2015)]. Genes overlapping between mutants are listed in Tables S11 (up-regulated) and S12 (down-regulated), between mutants and ß-cc treatment are listed in Tables S13 (up-regulated) and S14 (down-regulated), and between mutants and ESORGs are listed in Table S15. Table S16 displays additional details regarding plant growth, and RNA extraction/processing to generate the published datasets.
Gene ontology enrichment analyses
Using gene lists from Tables S11–15, gene ontology (GO) terms were identified using GO::TermFinder (https://go.princeton.edu/cgi-bin/GOTermFinder) (Boyle et al. 2004) and GO terms were selected based on a p-value ≤ 0.01. Qualifying GO terms were exported to REVIGO for visualization (http://revigo.irb.hr) (Supek et al. 2011).
Polymerase chain reactions and genotyping
Approximately 100 mg of fresh tissue was flash-frozen in liquid nitrogen for 5 min and crushed using 2 silica beads in a Mini-BeadBeater (Biospec Products) for 1 min in 2 mL microcentrifuge tubes. DNA was extracted using 750 μL of 2xCTAB (2% w/v) solution (1.4 M NaCl, 100 mM Tris–Cl pH 8.0, 20 mM EDTA) with 0.3% v/v beta-mercaptoethanol. Samples were incubated for 20–24 h at 65 °C. Debris was pelleted for 5 min at 10,000×g at room temperature. 700 uL of the supernatant was moved to a clean tube and a 1:1 chloroform extraction was performed. Tubes were mixed for 2 min and left to rest for 5 min. Next, tubes were centrifuged for 10 min at 10,000×g to separate the aqueous and organic phases. 600 μL of the aqueous layer was moved to a new tube containing 240 μL 5 M NaCl and 840 μL of 100% isopropyl alcohol. Tubes were mixed for 2 min, incubated at room temperature for 10 min, and incubated at 4 °C for 24 h. DNA samples were pelleted for 30 min at 4 °C at 21,000×g. Two 1 mL 75% ethanol washes were performed, mixing the tubes by hand and pelleting for 2 min at 4 °C at 21,000×g, pouring the supernatant off each time. The tubes were spun at 21,000×g for 30 s and the supernatant was removed. DNA pellets were dried for 2 h in a laminar flow hood and resuspended in 75 μL of DNAse-free water, incubating for 24 h at 4 °C.
PCR samples were amplified using GoTaq Green Master Mix (Promega) according to the manufacturer’s instructions. 20 μL reactions were performed, using 10 μL of GoTaq Green Master Mix, 1 μL of 10 μM primer A, 1 μL of 10 μM primer B, 6 μL of sterile water, and 2 μL of genomic DNA sample. For PCR samples not requiring restriction enzyme digestion (see below), DNA fragments were separated in a 1% (w/w) agarose gel containing 0.625 mg/mL ethidium bromide for 30 min at 120 V. Gels were imaged using a UV box. For unknown reasons, we were unable to amplify the left border of the oxi1 T-DNA (GABI_355H08) using primers specific to the left T-DNA border and the OXI1 sequence (JP1291/JP285). Instead, this mutation was confirmed by the inability to amplify wt OXI1 using the primer set JP1291/JP1292 and 100% resistance (no segregation) to 5 µg/ml sulfadiazine (the antibiotic marker cassette in GABI T-DNA sequences).
For genotyping requiring a restriction enzyme digestion (dCAPs), 10 μL digestions were performed. In a new tube, 5 μL of PCR product, 4.4 μL of nuclease-free water, 0.5 μL of the appropriate 10 × buffer, and 0.1 μL of the appropriate enzyme were combined and mixed gently by hand. Samples were incubated at 37 °C overnight. DNA fragments were separated in a 3% agarose gel containing 0.625 mg/mL ethidium bromide until the dye front was at the end of the gel. The gel was imaged using a UV box. Table S2 lists enzymes and expected fragment sizes.
RNA extraction and real-time quantitative PCR
Total RNA extraction, cDNA synthesis, and RT-qPCR was performed as previously described (Alamdari et al. 2020), using the RNeasy Plant Mini Kit (Qiagen), Maxima first strand cDNA synthesis kit for RT-qPCR with DNase (Thermo Scientific), and the SYBR Green Master Mix (BioRad), respectively, according to the manufacturers’ instructions. RT-qPCR experiments were all performed using a CFX Connect Real Time PCR Detection System (Bio-Rad). For expression analyses, all genes were normalized using ACTIN2 as a standard. The primers used for RT-qPCR are presented in Table S2.
Chlorophyll measurements
Chlorophyll was measured as previously described (Alamdari et al. 2021). Briefly, seeds were stratified for 5 days and counted prior to germination. Seedlings were collected 7 days after germination. Approximately 30–60 seedlings were used per seed line, and at least 3 biological replicates were collected for both constant light and diurnal cycling light conditions. Seedlings were flash-frozen in liquid nitrogen for 5 min and crushed using a Mini-BeadBeater (Biospec Products) for 1 min. Constant light samples were extracted in 1.2 mL of 100% ethanol, and diurnal light samples were extracted in 0.150 mL of 100% ethanol. Cell debris was pelleted and removed at 12,000×g for 30 min at 4 °C. The debris removal process was repeated twice before readings were taken. Chlorophyll was measured spectrophotometrically at 652 nm and 665 nm with a Varioskan LUX spectrophotometer with optically clear 96 well plates. Path corrections were calculated and chlorophyll concentrations were determined based on a previously described protocol (Warren 2008). Each biological replicate is a mean of 3 technical replicates. Total chlorophyll content was normalized to the number of seedlings collected.
Protochlorophyllide measurements
Protochlorophyllide (Pchlide) was measured as previously described (Shin et al. 2009). Briefly, seeds were stratified for 4 days and counted prior to germination, which was initiated with 1 h of white light in control conditions. Seedlings were grown in the dark for 4 days at 22 °C. Tissue (10 seedlings per replicate) was collected in dim green light and stored in amber 1.5 mL tubes containing 2 silica beads after flash-freezing with liquid nitrogen. Seedlings were crushed using a Mini-BeadBeater (Biospec Products) for 1 min. Pchlide was extracted using 1 ml of 80% acetone (v/v). In a black plastic 96-well plate (Grenier Bio-One), 200 µl of sample was loaded, with 3 biological replicates per genotype. The fluorescence of the samples was measured (excitation 440 nm/emission at 638 nm) with a Varioskan LUX spectrophotometer.
Singlet oxygen measurements
Singlet oxygen was measured as previously described (Alamdari et al. 2020). Briefly, seedlings were grown on plates in 6 h light/18 h dark diurnal cycling light conditions. As day three concluded, seedlings were moved to 1.5 ml microcentrifuge tubes containing 250 µl of ½-strength Linsmaier liquid media, wrapped in foil, and incubated at 22 °C for 18 h in the dark. An hour prior to subjective dawn on day four, 50 µM of 1.5 mM Singlet Oxygen Sensor Green (SOSG, Molecular Probes) and 0.1% Tween 20 (v/v) was added to the medium under dim, green light (final concentration of 250 µM). Seedlings were vacuum infiltrated for 30 min in the dark. After 30 additional min, seedlings were exposed to light for 3 h. Seedlings were washed once with 1 ml of ½-strength Linsmaier and Skoog medium pH 5.7 prior to imaging with a Zeiss Axiozoom 16 fluorescent stereo microscope equipped with a Hamamatsu Flash 4.0 camera and a GFP fluorescence filter. At least 12 seedlings from each genotype were monitored and average fluorescence per mm2 was quantified using ImageJ, choosing the brightest cotyledon per seedling.
Assessment of cell death
Cell death was measured in plant tissue as previously described (Woodson et al. 2015). Briefly, tissue was stained with a trypan blue solution [10 ml phenol, 10 ml glycerol, 10 ml lactic acid, 10 ml H2O, and 0.02 mg trypan blue (Sigma)] diluted with 2 volumes of 100% ethanol. The tissue in the staining solution was boiled for 2 min at 100 °C and incubated at room temperature overnight. Non-specific stain was removed using 2 overnight chloral hydrate (25 g/10 mL water) incubations. Tissue was moved to 30% glycerol for imaging. The intensity of the trypan blue stain was measured with ImageJ using at least 6 seedlings from each genotype and was normalized to the area of the cotyledon and then wt. The darkest cotyledon per seedling was chosen for measurements.
Lesion counting
To assess leaf lesion formation in adult plants, plants were grown under 16 h light/8 h dark diurnal cycling light conditions until lesions became apparent in some plants (day 18). Lesions were counted for each plant for an additional 18 days.
Results
Singlet oxygen accumulation in the fc2, flu, and ch1 backgrounds leads to overlapping nuclear transcriptomic responses
The Arabidopsis fc2 (Woodson et al. 2015), flu (op den Camp et al. 2003), and ch1 (Ramel et al. 2013) mutants produce excess 1O2 in the chloroplast, which leads to the induction of nuclear genes and cell death. However, we do not know if these three mutants utilize the same 1O2 signaling pathways to promote these outcomes. To test if these mutants share 1O2 pathways, we assessed the similarity of the nuclear responses to chloroplast 1O2 accumulation. We mined publicly available gene expression datasets to identify targets of the 1O2 signal in each genetic background [i.e., differentially expressed genes (DEGs)]. For the fc2 dataset, 4-day-old etiolated (dark-grown) seedlings were exposed to light for 2 h (Woodson et al. 2015). The flu dataset was generated using plants grown to the rosette life stage, incubated in the dark for 8 h, and then collected 1 h post light re-exposure (op den Camp et al. 2003). The ch1 dataset was from plants grown for 5–8 weeks and exposed to 8 h of EL for 2 days (Ramel et al. 2013). We also analyzed datasets from β-cc-treated wt plants. These plants were grown for 4 weeks and exposed to β-cc for 4 h before sample collection (Ramel et al. 2012). Finally, we compared the list of ESORGs identified in flu seedlings grown for 5 days under constant light, dark incubated for 4 h, and re-exposed to light for 30 or 60 min before sample collection (Dogra et al. 2017). Unfortunately, there is not a publically available transcriptome dataset for the acd2 mutant. Additional information on the datasets is listed in Table S16.
Next, we filtered the datasets to identify DEGs using cutoffs of ≥ twofold difference and an adjusted p-value ≤ 0.05 (Tables S6–10). Finally, we compared these lists of genes for each background/treatment group to identify DEGs shared between groups (Tables S11–15). Of the identified 1633 DEGs from the mutants, the majority of upregulated and downregulated DEGs were unique (Fig. 1 A and B, Table S17). At the same time, a subset of DEGs were shared between the mutants (between 28.8 and 40.9% of total DEGs from one mutant overlapped with another). While the overlap appeared similar between the three mutants (8.5–31.0% DEGs within a set), the overlap between the flu and ch1 DEGs was slightly larger (31.0% of flu DEGs and 23.1% of ch1 DEGs) than with fc2 (13.8% and 8.5% of flu and ch1 DEGs, respectively). In general, we observed more overlap between up-regulated DEGs than down-regulated DEGs among the three genetic backgrounds.
Meta-analysis of transcriptome expression data from three chloroplast 1O2-producing mutant backgrounds. Transcriptome datasets from three Arabidopsis thaliana 1O2-producing mutants were analyzed; fc2 (Woodson et al. 2015), flu (op den Camp et al. 2003), and ch1 (Ramel et al. 2013). Shown are Venn diagrams of differentially expressed genes (DEGs) A up-regulated or B down-regulated during chloroplast 1O2 stress in fc2, flu, and ch1. A ≥ twofold change and ≤ 0.05 adjusted p-value cutoffs were applied for all datasets where applicable. Additional information on the experiments are provided in Table S16. Tables S11 and 12 list the DEGs. Within each circle are the number of shared DEGs and the percentage of total DEGs (1019 up-regulated and 614 down-regulated) from the analysis
Furthermore, we compared these DEGs to those identified in β-cc–treated wt plants (Fig S1A, B and Table S18). While we observed overlap with each mutant (8.0–18.7%), the largest overlap was observed between β-cc–treated wt plants and EL treated ch1 mutants (18.7%). ESORGs identified in flu seedlings also showed a degree of overlap with each mutant (4.8–11.7%), but the largest overlap was observed with adult flu mutants (11.7%) (Fig. S1C and Table S19). Together, these results suggest that a core transcriptional response to chloroplast 1O2 occurs regardless of stress type, life stage, or stress duration.
To uncover roles of the overlapping gene expression, we conducted Gene Ontology (GO) term enrichment analyses. Only 25 and 11 genes were up-regulated or down-regulated, respectively, within all three mutant datasets (Table S20). The small number of genes limited our ability to perform robust GO-term analyses. However, within the up-regulated genes we did observe an enrichment for three terms (adjusted p-values ≤ 0.05): “cellular response to hypoxia,” “response to chitin,” and “transcription.” The latter is due to the presence of eight transcription factors including three ethylene response factors (ERFs), two WRKY transcription factors, and two zinc-finger proteins. Of the 11 down-regulated genes, the only significantly enriched GO term was “chloroplast” and four of these genes encode chloroplast-localized proteins. These included two involved in tetrapyrrole synthesis; CH1 (discussed herein) and GENOMES UNCOUPLED 4 (GUN4). The latter encodes a positive regulator of chlorophyll synthesis that binds Proto (Larkin et al. 2003) and affects photo-oxidative stress in chloroplasts (Li et al. 2021).
To increase the number of genes in our analyses, we next focused on genes upregulated in at least two genetic backgrounds (fc2, flu, or ch1) and found a diverse array of GO terms (Fig. S2A). However, the majority clustered around “response to stress,” “regulation of cellular processes,” and “aromatic compound biosynthetic process.” For genes down-regulated in two or more genetic backgrounds (Fig. S2B), we identified fewer GO terms and they had lower significance scores. Nonetheless, we observed four terms associated with photosynthesis: “pigment metabolic process,” “porphyrin-containing compound metabolic process,” “tetrapyrrole metabolic process,” and “photosynthesis, light harvesting in photosystem I.” Our result is consistent with earlier studies indicating 1O2 signals reduce the expression of photosynthesis protein-encoding genes to minimize photo-oxidative damage in the light (Page et al. 2017).
Continuing our GO term enrichment analysis, we tested the DEGs in common with β-cc-treated wt and at least one other genetic background (fc2, flu, or ch1). For up-regulated genes, we observed an enrichment for many GO terms found in common between the genetic backgrounds (Fig. S3A), with one large cluster around “response to chemical.” Some small differences include GO terms related to “sulfur compound metabolic process” and “plant organ senescence.” Despite these differences, the overall similarity suggests β-cc induces a similar response to the genetic backgrounds under photo-oxidative stress, further implicating this secondary metabolite in photo-oxidative stress signaling. As before, we found fewer GO terms with less significance with the down-regulated genes, yet we identified three terms associated with cell wall modifications (Fig. S3B). These results indicate plants may change their cell walls during 1O2 stress.
Finally, we performed a GO term enrichment analysis of ESORGs (Dogra et al. 2017) up-regulated in at least one genetic background (Fig. S4). We observed a striking similarity with GO terms identified through comparing mutants (Fig. S2A), having clusters around “response to stress,” “regulation of response to stress,” and “aromatic compound biosynthetic process.” We partly expected this result as Dogra et al. (2017) identified these ESORGs from flu seedlings. However, we also observed an enrichment for the GO terms “cellular response to hypoxia” and “cellular ketone metabolic process,” the latter suggesting a role for secondary metabolite synthesis or signaling.
Overall, the similarity between the GO term analyses of up-regulated genes within the datasets suggest plants have a core transcriptional response to 1O2 stress to induce the expression of genes broadly involved with stress, signaling, and secondary metabolites and to down-regulate genes involved in chloroplast function and tetrapyrrole synthesis. However, our analysis shows a large number of unique DEGs attributed to each mutant and condition suggesting that plants use different pathways depending on the specific source and site of chloroplast 1O2 stress. To delve deeper into the uniqueness of each genotype’s response to chloroplast stress, we identified the top 28 significant GO terms associated with DEGs unique to each background totaling 61 different GO terms (Table S21). We did not include down-regulated genes in this analysis as the flu dataset contained 41 genes, too few for a robust enrichment analysis. Each mutant had GO terms unique to itself (64%, 50%, and 43% of the GO terms for fc2, flu, and ch1, respectively). For the fc2 mutant, GO terms involving heat and hypoxia were unique including “response to heat” “response to hypoxia” and “response to decreased oxygen levels.” For the flu mutant, GO terms involving defense were unique including “response to bacterium,” “defense response to bacterium,” and “regulation of defense response.” The ch1 mutant had the fewest unique GO terms, but they included “response to hormone,” “response to abscisic acid,” and “response to jasmonic acid.” Despite these differences, we found a 28% overlap of the GO terms present in at least two mutants and a 10% overlap among all three mutants (notable GO terms include “response to stimulus,” “response to stress,” and “response to chemical.”) These results illustrate these mutant backgrounds activate similar responses (as indicated by shared and related GO terms), but utilize unique gene sets.
Testing genetic interactions with the fc2 signaling pathway in seedlings
Because the fc2, flu, and ch1 1O2-producing backgrounds all conditionally trigger cell death and have overlapping nuclear responses (Fig. 1), we tested if they employ the same mechanisms to transmit chloroplast stress signals. Therefore, we tested if genetic suppressors identified for one 1O2-producing mutant would suppress the others. First, we introduced the ppr30, cry1, oxi1, pub4, ex1, and ex2 mutant alleles into the fc2 background. We previously demonstrated that ex1 alone could not suppress cell death or nuclear signaling in the fc2 mutant (Woodson et al. 2015) and repeated those results here (Fig. S5A–C). Because EX1 and EX2 may have partially redundant functions (Page et al. 2017), we also introduced two alleles of ex2 (ex2-2; SALK_021694 and ex2-3; SALK_121009) with T-DNA insertions in the eighth exon and tenth intron, respectively) (Fig. S6A). Researchers previously showed ex2-2 is a null allele (Uberegui et al. 2015), and our analysis confirmed this conclusion. A semi-quantitative analysis of EX2 transcripts did not detect full-length transcript in ex2-2 (Fig. S6B). On the other hand, ex2-3 produced normal length transcripts and a sequencing analysis of the amplified ex2-3 cDNA revealed normal splicing across the tenth intron. Furthermore, a RT-qPCR analysis showed wt levels of EX2 transcript in the ex2-3 mutant (Fig. S6C). As such, we continued our analysis with the ex2-2 null allele.
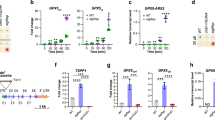
When grown under constant 24 h light, fc2 mutant seedlings appear pale, but healthy (Fig. 2A). However, when they grow under 6 h light/18 h dark diurnal cycling light conditions, the seedlings bleach and die, whereas wt is unaffected. As reported, the ppr30-1 and pub4-6 mutations suppress the bleaching phenotype and keep the seedlings green and alive. However, we did not observe any suppression of bleaching by cry1-304, oxi1, or the ex1 ex2-2 allele combination. To confirm these phenotypes, we stained the seedlings with trypan blue to assess cell death in cotyledons. As expected, fc2 mutants stained dark blue after growing under 6 h light/18 h dark diurnal cycling light conditions, confirming cell death (Fig. 2B and C). In line with their visual phenotypes, ppr30-1 and pub4-6 significantly reduced cell death in fc2, while cry1-304 and oxi1 did not. Surprisingly, the fc2 ex1 ex2-2 mutant did not suffer significant levels of cell death despite having a bleached appearance.
Genetic analysis of singlet oxygen signaling in fc2 mutant seedlings. Genetic suppressors of 1O2-producing mutants were tested for their ability to suppress the stress phenotypes of fc2 seedlings. A Shown are six-day-old seedlings grown under constant light (24 h) or diurnal cycling light (6 h light/18 h dark) conditions. B Shown are representative trypan blue stains of these seedlings. The dark blue color is indicative of cell death. C Shown are mean intensities of trypan blue (± SE, n ≥ 4 seedlings) from (B). D RT-qPCR analysis of stress gene markers of four-day-old seedlings grown under 6 h light/18 h dark conditions harvested 1 h after dawn. Shown are mean expression values (± SE, n = 3 biological replicates). Statistical analyses in (C and D) were performed using a one-way ANOVA followed by a Tukey HSD test. Statistical significance in respect to wt is indicated as follows: n.s. = p-value ≥ 0.05, * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001. E Mean levels of total chlorophyll (per seedling) of seven-day-old seedlings grown in 24 h light (± SE, n = 3 replicates). F Mean levels of protochlorophyllide (Pchlide) of four-day-old dark grown (etiolated) seedlings (± SE, n = 3 replicates). G Shown are representative images of four-day-old seedlings stained with Singlet Oxygen Sensor Green (SOSG). Seedlings were grown for 3 days in 6 h light/18 h dark diurnal light cycling conditions, dark incubated at the end of day three, and re-exposed to light on day four. Pictures were acquired 3 h post-dawn. H Shown are mean SOSG intensities (± SE, n ≥ 12 seedlings) of these seedlings. Statistical analyses in (E, F, and H) were performed using a one-way ANOVA followed by a Tukey HSD test. Different letters indicate statistical differences (p ≤ 0.05). In bar graphs, closed circles represent individual data points
Next, we tested if these mutations affect retrograde signaling to the nucleus and alter the transcriptional response. We measured steady state transcript levels in four-day-old seedlings grown under 6 h light/18 h dark diurnal cycling light conditions one hour post subjective dawn using RT-qPCR, probing for six previously identified chloroplast stress marker genes [SIB1 and HSP26.5 identified in 1O2-stressed fc2 seedlings (Woodson et al. 2015), BAP1 and ATPase identified in 1O2-stressed flu seedlings (op den Camp et al. 2003), and general oxidative stress markers ZAT12 and GST (Baruah et al. 2009)]. As shown in Fig. 2D, photo-oxidative stress significantly induces expression of five of the six stress marker genes (excluding the flu marker BAP1) in fc2 compared to wt. As expected of suppressors, both ppr30-1 and pub4-6 reduce induction of these marker genes. In line with their bleached phenotypes, cry1-304 and oxi1 did not hugely impact of expression of the marker genes. Compared to wt, fc2 cry1-304 and fc2 oxi1 experienced significant induction of all marker genes (except for ZAT12 in fc2 cry1-304). Despite its pale appearance, the fc2 ex1 ex2-2 mutant transcriptionally resembled the suppressors (fc2 ppr30-1 and fc2 pub4-6) with no significant induction of stress marker genes compared to wt. Together, these results suggest neither CRY1 nor OXI1 play a major role in 1O2-triggered cell death or retrograde signaling in fc2 mutant seedlings. However, the results reveal a potential genetic interaction between fc2 and the ex1 ex2-2 combination.
We did not expect the ex1 ex2-2 combination to suppress cell death and transcriptomic responses in fc2 as ex1 does not partially suppress these fc2 phenotypes alone [(Woodson et al. 2015) and Fig. S5A–C]. To distinguish if ex1 and ex2-2 additively suppress cell death or if ex2-2 alone is sufficient, we generated an fc2 ex2-2 mutant. Under 6 h light/18 h dark diurnal cycling light conditions, the fc2 ex2-2 mutant was visually similar to the fc2 mutant (Fig. S5A). Furthermore, trypan blue stains confirmed the ex2-2 mutation alone did not suppress cell death in the fc2 background (Fig. S5B and C).
One possible mechanism to suppress cell death in fc2 is through reducing tetrapyrrole biosynthesis (either directly or by reducing general chloroplast development). Second site mutations can accomplish this reduction by decreasing flux through the tetrapyrrole pathway and avoiding 1O2 accumulation (e.g. plastid protein import and tetrapyrrole biosynthesis mutants) (Woodson et al. 2015). Indeed, the fc2 ex1 ex2-2 mutant appeared very pale even under permissive 24 h constant light conditions (Fig. 2A). As expected, these mutant seedlings had significantly reduced levels of total chlorophyll compared to fc2 (Fig. 2E). ex2-2 alone reduced chlorophyll levels in fc2, but the ex1 ex2-2 combination reduced chlorophyll levels further. This additive effect was independent of fc2 as ex1 ex2-2 mutants accumulated less chlorophyll than wt. To determine if light-induced degradation or decreased tetrapyrrole synthesis caused a reduction in chlorophyll levels, we measured steady-state protochlorophyllide (Pchlide) levels in etiolated (dark grown) seedling to gauge the flux through the tetrapyrrole pathway. As previously shown, fc2 mutants accumulate two-to-three-fold excess Pchlide compared to wt [(Woodson et al. 2015) and Fig. 2F]. ex1 alone reduced Pchlide levels in fc2, but the ex1 ex2-2 combination further lowered Pchlide to wt levels. Next, we measured bulk 1O2 levels in four-day-old seedlings grown under 6 h light/18 h dark diurnal cycling light conditions using Singlet Oxygen Sensor Green (SOSG) (Fig. 2G and H). Two hours after subjective dawn, fc2 mutants accumulated excess 1O2 compared to wt. However, the fc2 ex1 ex2-2 mutant had wt 1O2 levels. Together, these results suggest the ex1 and ex2-2 mutations additively block fc2 phenotypes by reducing tetrapyrrole synthesis and 1O2 production rather than by directly affecting a signaling mechanism as shown in the flu mutant.
Testing genetic interactions with the fc2 signaling pathway in adult plants
As life stage could affect the ability of these mutations to suppress the fc2 cell death phenotype, we tested for suppression of cell death in adult plants. We grew plants for 21 days under 24 h constant light conditions and shifted them to 16 h light/8 h dark diurnal cycling light conditions for 6 days. As a control, we kept another set of plants in 24 h constant light for the full 27 days. Under constant 24 h light, fc2 plants appeared relatively healthy and do not present any indications of obvious cell death lesions (Fig. 3A). However, after shifting to 16 h light/8 h dark diurnal cycling light conditions, fc2 mutants developed leaf lesions. A trypan blue stain confirmed these lesions are areas of cell death (Fig. 3B and C). If a mutation causes suppression of 1O2 signaling in fc2, we expect a reduction in the appearance of leaf lesions under these conditions. We found, as expected, that the ppr30 and pub4-6 mutations suppressed the fc2 cell death phenotype, having fewer observable lesions and less trypan blue staining than the fc2 single mutant (Fig. 3A–C). Surprisingly, we found the oxi1 mutation significantly suppressed lesion formation in the fc2 mutant, suggesting OXI1 may play a role in 1O2 signaling in fc2 adult plants. The ex1 ex2-2 combination did not suppress cell death in adult leaves, consistent with these mutations leading to developmental (rather than signaling) defects. As in seedlings, cry1-304 did not suppress cell death, further suggesting CRY1 does not play a strong role in 1O2 signaling in the fc2 mutant.
Genetic analysis of singlet oxygen signaling in fc2 mutant adult plants. Genetic suppressors of 1O2-signaling were tested for their ability to suppress the cell death phenotype of fc2 adult plants. A Shown are representative 27-day-old plants grown under constant light (24 h) or under 24 h light for 21 days and then shifted to diurnal cycling light conditions (16 h light/8 h dark) for 6 additional days. White arrows indicate lesions. B Shown are representative trypan blue cell death stains for both sets of plants. The dark blue color is indicative of cell death. C Shown are mean intensities of trypan blue (± SE, n ≥ 3 leaves) from (B). Statistical analyses within each light treatment were performed using a one-way ANOVA followed by a Tukey HSD test. Statistical significance in respect to wt is indicated as follows: ** = p-value ≤ 0.01, *** = p-value ≤ 0.001, not significant (ns) = p-value ≥ 0.05. Closed circles represent individual data points
Testing genetic interactions in flu mutant seedlings
To continue our assessment of potential genetic interactions of known chloroplast 1O2 suppressors in other 1O2-generating backgrounds, we monitored phenotypes of 1O2 signaling mutations in the flu mutant background. Previously, researchers determined flu mutants accumulate chloroplast 1O2 proportionally to increasing lengths of time in the dark (Wang et al. 2020). To experimentally determine the length of time in the dark needed to induce cell death, we treated 5-day-old wt and flu seedlings to various lengths of time in the dark (0, 4, 8, 12, and 24 h) and re-exposed the seedlings to light for 36 h. Based on the outcomes shown in Fig. S7A, we decided 12 h of dark was adequate to completely bleach most flu seedlings within 36 h of light exposure. Therefore, we crossed in 1O2 signaling mutations (ex1, pub4-6, cry1-304, and oxi1) into the flu background to test which mutations may block 1O2 signaling phenotypes.
As expected, the ex1 mutation suppressed bleaching in the flu seedlings after a 12 h dark treatment (Fig. 4A), while the other mutations did not obviously appear to affect bleaching. To confirm our visual assessment, we performed a trypan blue stain using the 12 h time point to confirm flu mutants experience extensive cell death in their cotyledons, and the ex1 mutation significantly reduces this effect to near wt levels (Fig. 4B and C). Consistent with their bleached phenotypes, flu pub4-6 and flu oxi stained similarly to flu. However, we found flu cry1-304 stained significantly lower than flu (p value ≤ 0.001), confirming CRY1 plays at least a minor role in 1O2 signaling in flu mutants (Danon et al. 2006).
Genetic analysis of singlet oxygen signaling in flu seedlings. Genetic suppressors of chloroplast 1O2 signaling were tested for their ability to suppress flu phenotypes in seedlings. A Shown are (top) 6-day-old seedlings grown under constant light (24 h) and (bottom) 5-day-old seedlings grown under 24 h light, incubated in the dark for 12 h, and re-exposed to light for 36 h (7 days total). B Shown are representative images of these seedlings stained with trypan blue. The dark blue color is indicative of cell death. C Mean intensities of trypan blue signal (± SE, n ≥ 9 seedlings) from panel (B). D RT-qPCR of flu-specific stress gene markers of five-day-old seedlings grown under 24 h constant light then dark-incubated for 12 h, harvested 1 h after re-exposure to light. Shown are mean expression values (± SE, n ≥ 3 biological replicates). Statistical analyses were performed using a one-way ANOVA followed by a Tukey HSD test. Statistical significance in respect to wt is indicated as follows: n.s. = p-value ≥ 0.05, * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001. Closed circles represent individual data points
Next, we tested if our double mutants activated 1O2–triggered retrograde signaling by measuring the expression of stress marker genes [as for fc2 (Fig. 2D)]. Here, we also included NOD1, another gene induced in flu mutants (op den Camp et al. 2003). We placed 5-day-old seedlings in the dark for 12 h and exposed them to 1 h light prior to tissue collection for RNA extraction. In the flu mutant, we observed significant induction of all three flu-specific stress marker gene transcripts (Fig. 4D). However, this effect was reduced by ex1. pub4-6 and cry1-304 did not significantly reduce any one of these transcripts. However, oxi1 lowered levels of two transcripts (ATPase and NOD1). We tested the other four stress marker transcript levels in the flu mutant. We observed higher transcript levels compared to wt, but they were not significant (Fig. S7B). Furthermore, none of the suppressor mutations significantly reduced expression of these marker genes. Together, these results suggest PUB4 does not play a significant role in facilitating the 1O2 signal in flu mutants, while CRY1 plays a minor role in regulating cell death in flu seedlings. OXI1 does not play a major role in triggering cell death in the flu mutant, yet it may play a minor role in transmitting the retrograde signal to the nucleus.
Testing genetic interactions in flu adult plants
As with the fc2 mutant, we assessed if life stage affected 1O2-signaling in the flu mutant. We grew wt, flu, and the double mutant plants under 24 h constant light conditions to avoid 1O2 stress. We then shifted them to 16 h light/8 h dark diurnal cycling light conditions for 3 days to accumulate Pchlide and 1O2. As in seedlings, the flu plants developed lesions under these conditions, whereas wt appeared normal (Fig. 5A). flu ex1 plants exposed to 16 h light/8 h dark diurnal cycling light conditions for 5 days did not develop leaf lesions, consistent with EX1 playing a role in 1O2-triggered cell death regardless of life stage (Wagner et al. 2004) (Fig. 5A). As in seedlings, we did not observe obvious suppression of the cell death phenotype by cry1-304, pub4-6, or oxi1 in adult plants. We confirmed these cell death phenotypes with a trypan blue cell death stain (Fig. 5B and C). Together, we conclude 1O2 signaling in the flu mutant utilizes the EX1-dependent pathway rather than the PUB4 (and possibly OXI1)-dependent chloroplast quality control pathway implemented by fc2. However, OXI1 may contribute to the retrograde signaling in seedlings to control some nuclear gene expression in the flu mutant.
Genetic analysis of 1O2 signaling in adult flu plants. Genetic suppressors of 1O2-signaling were tested for their ability to suppress the flu cell death phenotype in adult plants. A Shown are representative 24-day-old plants grown under constant light (24 h) and 21-day-old plants grown in 24 h light and then shifted to diurnal cycling light (16 h light/8 h dark) conditions for 3 days (24 days old total). White arrows indicate lesions. B Shown are representative images of leaves from these plants stained with trypan blue. The dark blue color is indicative of cell death. C Shown are mean intensities of trypan blue (± SE, n ≥ 3 leaves) from (B). Statistical analyses within each light treatment were performed using a one-way ANOVA followed by a Tukey HSD test. Statistical significance in respect to flu is indicated as follows: ** = p-value ≤ 0.01, *** = p-value ≤ 0.001, not significant (ns) = p-value ≥ 0.05. Closed circles represent individual data points
Testing genetic interactions with the chlorina1 signaling pathway
Previously, researchers demonstrated that growing the ch1 mutant under EL stress (≥ 1,100 µmol photons m−2 s−1) induces 1O2 signaling (Ramel et al. 2013). The generated 1O2 initiates cell death, which the oxi1 mutation blocks in adult plants (Shumbe et al. 2016). Furthermore, researchers demonstrated EX1 and EX2 are not involved in ch1’s 1O2 signaling since ch1 ex1 ex2 mutants suffer from a comparable level of EL-triggered cell death to ch1 (Ramel et al. 2013). Here, we tested the involvement of PUB4 in transmitting this 1O2 signal. We grew seedlings under permissive light conditions (120 µmol photons m−2 s−1) for 7 days and shifted them to 1200 µmol photons m−2 s−1 for 24 h. We lowered the ambient temperature to 10 °C to avoid any incidental heat stress caused by the increased radiation. Within 2 h, all seedlings experienced a decrease in maximum photosystem II quantum efficiency (Fv/Fm), which continued to decrease for 24 h of treatment (Fig. 6A). Furthermore, we observed photo-bleaching of cotyledons after 12 h of EL. Photo-bleaching worsened after 24 h (Fig. 6B). At the same time points, we observed increased susceptibility (lowered Fv/Fm values and increased bleaching) of the ch1 mutant to EL stress, consistent with its chloroplasts experiencing increased photo-damage (Ramel et al. 2013). The pub4-6 mutation partially reversed these effects (increased Fv/Fm values at 2 and 6 h EL and delayed bleaching at 12 h EL), while the oxi1 mutation did not reverse them. Additionally, we observed increased tolerance of the pub4-6 single mutant to EL compared to wt, having higher Fv/Fm values at 2 and 6 h EL and delayed bleaching at 12 h EL (Fig. 6A and B). Together, these results suggest PUB4 may be involved in EL-triggered 1O2 signaling in the seedling stage.
Effect of singlet oxygen signaling mutations on excess light-induced phenotypes. The effect of the oxi1 and pub4-6 mutations were tested in excess light (EL) conditions. A Time course analysis of maximum PSII quantum efficiency (Fv/Fm) in seven-day-old seedlings during 24 h of EL (1200 µmol photons sec−1 m−2) at 10 °C (± SE, n ≥ 3 groups of seedlings). B Shown are representative seedlings immediately after the indicated length of EL treatment. C Time course analysis of Fv/Fm in 18-day-old adult plants during 48 h of EL (1300 µmol photons sec−1 m−2 (± SE, n ≥ 6 plants). Statistical analyses within each time point (for wt or ch1) were performed using a one-way ANOVA followed by a Tukey HSD test. Statistical significance in respect to wt (for oxi1 and pub4-6) or ch1 (for ch1 oxi1 and ch1 pub4-6) is indicated as follows: * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001, not significant (ns) = p-value ≥ 0.05
As researchers previously studied the ch1 EL-treated phenotype in adult plants and leaves (Ramel et al. 2013; Shumbe et al. 2016), we tested EL sensitivity in 18-day-old plants. We grew plants under permissive light conditions (70 µmol photons m−2 s−1) for 18 days. Next, we shifted them to 1300 µmol photons m−2 s−1 at 10 °C. Similar to seedlings, all plants experienced an immediate decrease in Fv/Fm values indicating photo-damage (Fig. S8A). Again, the ch1 mutant was particularly susceptible. oxi1 and pub4-6 did not significantly affect Fv/Fm values in the ch1 background (although pub4-6 single mutants had significantly higher Fv/Fm values compared to wt at 6 and 12 h EL). However, oxi1 and pub4-6 attenuated photo-bleaching in both wt and ch1 backgrounds (Figs. 6C and S8B). Together, our results suggest both OXI1 and PUB4 play a role in transmitting EL-triggered stress signals, but OXI1 may play a stage-specific role in adult leaves.
Onset of lesions in the acd2 mutant was slowed by pub4-6
As pub4-6 mitigated 1O2-induced cell death in the fc2 and ch1 mutants, we investigated if it can affect lesion formation caused by other sources of ROS. Therefore, we examined the effect of the pub4-6 mutation in the ROS and lesion accumulating acd2 mutant. Prior work determined that a cry1 mutation or an ex1 ex2 combination did not significantly alter the accumulation of lesions, suggesting the flu signaling pathway is not being used in acd2 to induce cell death (Pattanayak et al. 2012). As PUB4 appears to represent another separate 1O2 pathway, we tested if PUB4 plays a role in lesion formation in acd2 mutants by generating acd2 pub4-6 double mutants and growing them alongside wt and the corresponding single mutants under 16 h light/8 h dark diurnal light cycling conditions. Initially, acd2 mutant plants appeared healthy and comparable to wt. However, after 18 days, acd2 mutants began to randomly develop lesions of cell death on their leaves, which accumulated until senescence (Figs. 7A and B and S9). Conversely, we observed the acd2 pub4-6 double mutant appeared healthier than the acd2 single mutant, and developed fewer leaves with lesions over time. At 36 days, we stopped the experiment as we could not reliably distinguish between acd2-specific and normal leaf senescence lesions. This early senescence was particularly apparent in pub4-6 as previously reported (Woodson et al. 2015). Together, these data suggest PUB4 is involved in regulating ROS-induced lesion formation in acd2 plants.
The pub4-6 mutation slows the progression of spontaneous cell death in the acd2 mutant. The acd2 spontaneous cell death and lesion phenotypes were assessed. A Shown are representative 30-day-old adult plants grown under 16 h light/8 h dark diurnal cycling light conditions. The inflorescences were removed for the picture. White arrows indicate lesions. B Mean number of leaves with lesions per plant (± SE, n ≥ 18 plants). Statistical analyses were performed for each time point using a one-way ANOVA followed by a Tukey HSD test. Statistical significance as follows in respect to wt: ** = p-value ≤ 0.01, *** = p-value ≤ 0.001
Discussion
Despite knowing chloroplast 1O2 has signaling capabilities, natural stresses complicate its study by causing complex ROS signatures in plant cells (Choudhury et al. 2017; Rosenwasser et al. 2013). Thus, to dissect 1O2 signals, researchers use several Arabidopsis mutants that conditionally and specifically produce this ROS, including fc2 (Woodson et al. 2015), flu (Meskauskiene et al. 2001), ch1 (Ramel et al. 2013), and acd2 (Pruzinská et al. 2007). To assess how similar these signaling pathways are, we compared publicly available transcriptomic data for the fc2, flu, and ch1 mutants (the acd2 mutant did not have an available dataset for comparison). We found each mutant shared a proportion of their DEGs with the other two mutants (Fig. 1). Between all three data sets, 25 and 11 were shared among the up-regulated and down-regulated genes, respectively, which may indicate a core response to 1O2. The small number of genes limited our analysis, but it was noteworthy that many of the up-regulated genes encoded well-studied proteins involved in stress and defense, which indicates 1O2 signals converge into cellular stress networks (Table S20). These included MAP KINASE KINASE 9 (MKK9) [encodes a kinase involved in inducing leaf senescence via MPK6 (Zhou et al. 2009)], ORTHOLOG OF SUGAR BEET HS1 PRO-1 2 (HSPRO2) [encodes a protein involved in basal defense against bacterial pathogens (Murray et al. 2007)], IDA-LIKE7 (IDL7) [encodes a negative regulator of stress-induced ROS (Vie et al. 2017)], STRESS-ASSOCIATED PROTEIN 12 (SAP12) [encodes a protein involved in stress responses and is a target of the nematode effector protein MiPDI1 (Zhao et al. 2020)], DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2 (DREB2) [encodes a transcription factor involved in heat and dehydration stress (Sakuma et al. 2006)], ZAT12 [encodes a zinc finger transcription factor and is a commonly used marker gene for 1O2 and photo-oxidative stress (Baruah et al. 2009)], WRKY22, and WRKY18 [encode transcription factors involved in defense responses to aphids and microbial pathogens, respectively (Kloth et al. 2016; Xu et al. 2006)]. Of the down-regulated genes, the only GO-term enriched was “chloroplast.” Notably, two of these genes encode enzymes in tetrapyrrole synthesis (CH1 and GUN4) suggesting that turning down tetrapyrrole synthesis may be a common response to chloroplast 1O2 stress, possibly to avoid further 1O2 accumulation.
Many more genes were shared by only two mutants (Fig. 1), which were enriched with GO-terms related to stress (up-regulated genes) or photosynthesis and tetrapyrroles (down-regulated genes). We were surprised by this level of overlap since the datasets represent samples collected from different ages (seedling vs. adult) and grown in different conditions to elicit 1O2 stress in the chloroplasts. Therefore, we hypothesize these genotypes share a core transcriptomic response and that the different sources of 1O2 converge on some transcriptomic responses. To further support our hypothesis, we found ß-cc responsive genes and ESORGs in the mutant datasets (Fig. S1A-C). Notably, as we found ß-cc responsive genes in the fc2 and flu datasets, we can hypothesize these mutants produce ß-cc. Such measurements, however, have not yet been reported.
Notwithstanding the similarities found in the transcriptomic meta-analysis, we identified DEGs specific to each background (Fig. 1A). Although these DEGs were unique, we found they produced similar GO-terms between mutants for up-regulated genes (Table S21). These GO terms include “response to stimulus,” “response to stress,” and “response to chemical.” These results may indicate these mutants employ different sets of genes with similar functions for the cell to respond or acclimate appropriately to a specific stress. As each of these mutants produce 1O2 differently, we hypothesize such ROS signatures may mimic those caused by different types of environmental stress (Rosenwasser et al. 2013).
Our meta-analysis revealed the potential for the existence of multiple 1O2 signaling pathways in these mutants. Therefore, we took a genetic approach to identify potential converging points of these pathways. We hypothesized that if a secondary suppressor mutation for one mutant background has a genetic interaction within a different mutant background, these two mutant backgrounds may share a portion of their signaling cascades. First, with fc2 mutant seedlings, we observed the ppr30-1 and pub4-6 mutations suppressed cell death and retrograde signaling as previously described (Alamdari et al. 2020; Woodson et al. 2015). Initially, we did not notice obvious suppression of the fc2 cell death phenotype by suppressors of the flu or ch1 backgrounds (Fig. 2A). Nonetheless, with further analysis, we found fc2 ex1 ex2-2 seedlings had blocked cell death and retrograde signaling even though they were bleached white (Fig. 2A-C).
These results surprised us as we previously demonstrated ex1 alone does not block 1O2 controlled retrograde signaling or cell death in fc2 seedlings or adults (Woodson et al. 2015). Indeed, both the fc2 ex1 and fc2 ex2-2 double mutants had similar levels of cell death to fc2, suggesting the suppression in the triple mutant was additive (Fig. S5A–C). We hypothesize the mechanism of suppression is indirect, as the fc2 ex1 ex2-2 accumulated less bulk 1O2 compared to fc2 (Fig. 2G and H). The observed suppression is likely due to a delay in chloroplast development and a decrease in tetrapyrrole synthesis: ex1 ex2-2 led to a decrease in chlorophyll and Pchlide levels (Fig. 2E and F). Furthermore, the ex1 ex2-2 combination failed to block cell death in adult fc2 mutants, which suggests that once chloroplasts become fully developed, the ex1 ex2-2 combination does not affect the fc2 phenotype. How the ex1 ex2 combination affects chloroplast development is not clear as these proteins are primarily implicated in ROS signaling. Interestingly, we also observed that ex1 and ex2 affect chloroplasts differently and each lead to reduced Pchlide and chlorophyll levels in fc2, respectively (Fig. 2E and F). This may mean EX1 has a stronger effect on tetrapyrrole synthesis while EX2 is more generally involved in chloroplast development. Alternatively, the proteins may play different roles in etiolated and de-etiolated seedlings. Adding to the complexity, recent work suggests these two proteins have different functions. EX2 may act as a decoy to protect EX1 from oxidation and prevent premature signaling (Dogra et al. 2022). Thus, a double mutant may have a more complicated phenotype than previously assumed. Even so, we conclude the EX proteins do not play a direct role in 1O2 signaling in the fc2 mutant.
To our surprise, the oxi1 mutation suppressed the cell death phenotype in fc2 adult plants (similarly to the ppr30-1 and pub4-6 mutations). Due to the signaling role of OXI1 in ch1 mutants, we expect the oxi1 mutation is blocking a similar 1O2 signal in fc2. However, it is also possible OXI1 may be indirectly affecting local Proto or 1O2 accumulation by an unknown mechanism. Interestingly, oxi1 did not affect cell death or retrograde signaling in seedlings. To date, researchers have primarily studied the role of OXI1 in 1O2 signaling in adult plants and leaves (Shumbe et al. 2016). However, some research indicates OXI1 plays a role in basal defense against oomycete pathogens in seven-day-old seedling cotyledons (Rentel et al. 2004) indicating that OXI1 is present in this tissue. This may mean that OXI1 only participates in chloroplast 1O2 signaling in true leaf mesophyll cells, which is consistent with our EL experiments where the oxi1 mutation only mitigated cell death in adult leaves (Fig. 6A–C). Prior work indicates chloroplast physiology differs between the embryonic cotyledons in seedlings and the mesophyll cells in true leaves (Albrecht et al. 2008). Whether a different serine/threonine kinase is used in cotyledons, or there is another mechanism altogether, will require further study.
Next, we performed similar experiments in the flu mutant. In seedlings, we observed suppression of cell death by the ex1 and cry1-304 mutations, and ex1 reduced expression of all three nuclear marker genes (Fig. 4A–D). Neither pub4-6 or oxi1 reduced cell death, although oxi1 did have a minor effect on retrograde signaling. While EX1 is necessary for 1O2 signaling in flu mutants (Dogra et al. 2019), researchers have previously demonstrated CRY1’s involvement only in protoplasts (Danon et al. 2006). In protoplasts, the effect of the cry1 mutation was equally strong compared to ex1 in terms of cell death. In our study, the effect of the cry1 mutation was noticeably weaker than ex1 in seedlings, and cry1 did not significantly suppress cell death in the adult phase (Fig. 5B and C). These results may be due to a difference between systems (in vitro protoplasts vs in planta). The protoplast study also used the Landsberg erecta ecotype with different flu and cry1 alleles (this study was performed in the Columbia background), which may also account for some of the differences. When researchers performed a microarray analysis of 1O2-responsive transcripts in flu protoplasts, they found the cry1 mutation affected only ~ 3% of the DEGs in flu (Danon et al. 2006). When we pair this finding with our work here, we hypothesize CRY1 plays a minor role in the 1O2 retrograde signal, which can be uncoupled from cellular degradation. Overall, however, our results suggest the flu mutant emits a functionally unique signal that does not involve PUB4 or OXI1 to control cellular degradation.
To continue our investigation of 1O2 signaling, we next tested the ch1 mutant that produces 1O2 due to unprotected PSII reaction centers (Ramel et al. 2013) and signals for cell death with OXI1 (Shumbe et al. 2016). Previously, researchers showed cellular degradation in this background is independent of EX1 and EX2; ch1 ex1 ex2 mutants still suffer EL-induced lesions and PSII photo-inhibition (Ramel et al. 2013). Accordingly, we assessed if the fc2 pathway is active in ch1 mutants by testing the role of PUB4 in EL stress. In EL-stressed seedlings, pub4-6 attenuated cotyledon bleaching and the reduction in Fv/Fm values in the wt and ch1 backgrounds (Fig. 6A and B). Somewhat surprisingly, the oxi1 mutation did not affect these phenotypes, further suggesting OXI1 is not involved in 1O2 signaling in seedlings. In the adult phase, both oxi1 and pub4-6 attenuated EL-induced lesions in the wt and ch1 backgrounds (Fig. 6C), confirming earlier reports on OXI1 (Shumbe et al. 2016) and suggesting fc2 and ch1 mutants share a 1O2 signaling pathway induced by natural EL stress.
Finally, as pub4-6 can block 1O2-induced cell death in fc2 and ch1 mutants, we tested if we would observe a similar trend in the acd2 mutant, which produces leaf lesions due to the accumulation of chlorophyll breakdown products (such as RCC) that can produce 1O2 (Pruzinská et al. 2007). Remarkably, the acd2 pub4-6 double mutant had delayed onset of lesions compared to the acd2 single mutant, suggesting acd2 mutants activate the same pathway as fc2 and ch1 mutants (Fig. 7A and B). Previously, researchers demonstrated neither cry1 or an ex1 ex2 combination could reduce lesion formation in the acd2 background (Pattanayak et al. 2012), further indicating the 1O2-signaling pathway used by flu mutants is distinct. This result also led the authors to conclude chloroplast 1O2 and ROS were not responsible for cell death. As 1O2 was detected in acd2 mitochondria, these researchers hypothesized these organelles may trigger a cell death signal instead. Nevertheless, the observation that pub4-6 reduces lesion formation in acd2 opens the possibility for the involvement of chloroplast 1O2 during lesion formation in acd2 mutants. On the other hand, we cannot rule out the possibility that PUB4 plays a second role in mitochondrial ROS signaling. Plant chloroplasts may export the heme precursor protoporphyrinogen IX (Jacobs et al. 1993), which can then be imported into mitochondria and converted to Proto by the mitochondrial-localized protoporphyrinogen IX oxidase (Lermontova et al. 1997; Watanabe et al. 2001). Thus, mitochondria may also be able to accumulate Proto and 1O2, leading to the induction of a PUB4-dependent stress pathway. The measurement of Proto from separately isolated chloroplasts and mitochondria from fc2, acd2, ch1 mutants may help to address this possibility.
Overall, our work suggests chloroplasts use at least two 1O2 signaling pathways to control cellular degradation. One pathway depends on the PUB4 protein to control cellular degradation and is employed by fc2, ch1, and acd2 mutants along with EL-stressed wt plants. At least in fc2 and ch1 mutants, the OXI1 protein participates in this pathway (its role in acd2 was not tested), but is restricted to true leaves in adult tissue. flu mutants utilize an alternative 1O2 signaling pathway. Instead, their 1O2 signal requires EX1 and (at least in seedlings) CRY1 to initiate cell death. Retrograde signaling to the nucleus to alter the transcriptome follows a similar, yet more complex, pattern. For instance, the cry1 and oxi1 mutations partially reduce the expression of some stress marker genes in the fc2 and flu backgrounds, respectively, despite no effect on cell death. As these effects on transcript levels were relatively mild, we hypothesize some crosstalk may exist between these two 1O2 pathways.
As the fc2, flu, and ch1 mutants produce 1O2 within chloroplasts that leads to retrograde signaling (to control similar sets of genes), cell death, and (at least in flu and fc2) chloroplast degradation, we were initially surprised multiple pathways can be activated by this specific ROS. One possibility is that the exact location of 1O2 production determines which signal is activated. In flu mutants, this likely occurs in the thylakoid grana margins where the EX proteins localize (Dogra et al. 2022; Wang et al. 2016). These grana margins are the site of PSII repair and tetrapyrrole synthesis, both potential sources of 1O2 in the light. On the other hand, ch1 mutants produce 1O2 within the grana core, the site of active PSII (Ramel et al. 2013). Researchers have not yet determined the exact site of 1O2 production in fc2 and acd2 mutants. However, some work suggests Proto accumulates in the chloroplast envelope and stromal fractions of pea and beet, respectively (Mohapatra et al. 2002, 2007). Thus, 1O2 in fc2 may represent a more advanced stage of photo-oxidative stress where damage has spread throughout the chloroplast. Another possibility for multiple pathways could be that the kinetics of 1O2 accumulation affects signaling. 1O2 is produced almost instantly after light exposure in flu mutants due to the accumulation of Pchlide in the dark (Meskauskiene et al. 2001). On the other hand, 1O2 production in the other mutants is slower and accumulates over time (Ramel et al. 2013; Woodson et al. 2015). The identification of additional signaling components should help resolve these possibilities.
Ultimately, we do not yet know why chloroplasts require multiple 1O2 signaling pathways to respond to stress or what the roles of these pathways are under natural environmental stresses. However, some experiments have offered clues. Under severe photo-inhibitory conditions that lead to bleaching and cell death, the fc2/ch1 pathway may prevail. pub4-6 (shown here) and oxi1 (shown here and (Shumbe et al. 2016)) mitigate cell death under EL stress. We have demonstrated other suppressors of fc2 cell death slow light-induced photo-bleaching in leaves (i.e., ppr30, mterf9 (Alamdari et al. 2020) and cotyledons (i.e. ctps2) (Alamdari et al. 2021)). Under milder, non-photo-inhibitory light stress, plants may utilize the flu pathway. ex1 blocks the formation of microlesions under moderate light stress that did not associate with non-enzymatic lipid peroxidation (Kim et al. 2012). Furthermore, these pathways could integrate different types of stress signals. Prior work links PUB4 and OXI1 to basal defense pathways (Rentel et al. 2004; Wang et al. 2022), and PUB4 plays a role in nitrogen and carbon starvation (Kikuchi et al. 2020). These findings suggest the fc2/ch1 pathway might integrate with defense and senescence pathways. On the other hand, researchers have shown EX1 has a role in systemic acquired acclimation responses to EL stress (Carmody et al. 2016), suggesting plants use the flu pathway for distal signaling. These results along with our current findings demonstrate that chloroplast 1O2 signaling is a complex process requiring additional investigation. However, the existence of two (or more) chloroplast pathways may allow plants to better respond to their surroundings and to thrive in stressful and dynamic environments.
References
Alamdari K, Fisher KE, Sinson AB, Chory J, Woodson JD (2020) Roles for the chloroplast-localized PPR protein 30 and the “Mitochondrial” transcription termination factor 9 in chloroplast quality control. Plant J 103:735–751
Alamdari K, Fisher KE, Tano DW, Rai S, Palos KR, Nelson ADL, Woodson JD (2021) Chloroplast quality control pathways are dependent on plastid DNA synthesis and nucleotides provided by cytidine triphosphate synthase two. New Phytol 231:1431–1448
Albrecht V, Ingenfeld A, Apel K (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66:599–608
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141:391–396
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442
Baruah A, Simkova K, Apel K, Laloi C (2009) Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol Biol 70:547–563
Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO::Termfinder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 20:3710–3715
Bruggemann E, Handwerger K, Essex C, Storz G (1996) Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J 10:755–760
Callis J (2014) The ubiquitination machinery of the ubiquitin system. Arabidopsis Book/am Soc Plant Biol 12:e0174
Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, Munnik T, Hirt H, Oelmüller R (2011) The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog 7:e1002051
Carmody M, Crisp PA, d’Alessandro S, Ganguly D, Gordon M, Havaux M, Albrecht-Borth V, Pogson BJ (2016) Uncoupling high light responses from singlet oxygen retrograde signaling and spatial-temporal systemic acquired acclimation. Plant Physiol 171:1734–1749
Chan KX, Mabbitt PD, Phua SY, Mueller JW, Nisar N, Gigolashvili T, Stroeher E, Grassl J, Arlt W, Estavillo GM, Jackson CJ, Pogson BJ (2016) Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proc Natl Acad Sci USA 113:E4567–E4576
Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2015) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67:25–53
Chen S, Kim C, Lee JM, Lee HA, Fei Z, Wang L, Apel K (2015) Blocking the QB-binding site of photosystem II by tenuazonic acid, a non-host-specific toxin of Alternaria alternata, activates singlet oxygen-mediated and EXECUTER-dependent signalling in Arabidopsis. Plant Cell Environ 38:1069–1080
Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90:856–867
D’Alessandro S, Beaugelin I, Havaux M (2020) Tanned or sunburned: how excessive light triggers plant cell death. Mol Plant 13:1545–1555
Danon A, Coll NS, Apel K (2006) Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana (vol 103, pg 17036, 2006). Proc Natl Acad Sci USA 103:18875–18875
Dogra V, Duan J, Lee KP, Lv S, Liu R, Kim C (2017) FtsH2-dependent proteolysis of executer1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front Plant Sci 8:1145
Dogra V, Kim C (2019) Singlet oxygen metabolism: from genesis to signaling. Front Plant Sci 10:1640
Dogra V, Li M, Singh S, Li M, Kim C (2019) Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat Commun 10:2834
Dogra V, Singh RM, Li M, Li M, Singh S, Kim C (2022) EXECUTER2 modulates the EXECUTER1 signalosome through its singlet oxygen-dependent oxidation. Mol Plant 15:438–453
Fisher KE, Krishnamoorthy P, Joens MS, Chory J, Fitzpatrick JAJ, Woodson JD (2022) Singlet oxygen leads to structural changes to chloroplasts during their degradation in the Arabidopsis thaliana plastid ferrochelatase two mutant. Plant Cell Physiol 63:248–264
Foyer CH (2018) Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ Exp Bot 154:134–142
Havaux M, Dall’osto L, Bassi R (2007) Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol 145:1506–1520
Jacobs JM, Jacobs NJ (1993) Porphyrin accumulation and export by isolated barley (Hordeum vulgare) plastids (effect of diphenyl ether herbicides). Plant Physiol 101:1181–1187
Jeran N, Rotasperti L, Frabetti G, Calabritto A, Pesaresi P, Tadini L (2021) The PUB4 E3 ubiquitin ligase is responsible for the variegated phenotype observed upon alteration of chloroplast protein homeostasis in arabidopsis cotyledons. Genes (Basel). https://doi.org/10.3390/genes12091387
Kikuchi Y, Nakamura S, Woodson JD, Ishida H, Ling Q, Hidema J, Jarvis RP, Hagihara S, Izumi M (2020) Chloroplast autophagy and ubiquitination combine to manage oxidative damage and starvation responses. Plant Physiol 183:1531–1544
Kim C, Meskauskiene R, Zhang S, Lee KP, Lakshmanan Ashok M, Blajecka K, Herrfurth C, Feussner I, Apel K (2012) Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24:3026–3039
Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat simplesearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40:D1211–D1215
Kloth KJ, Wiegers GL, Busscher-Lange J, van Haarst JC, Kruijer W, Bouwmeester HJ, Dicke M, Jongsma MA (2016) AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J Exp Bot 67:3383–3396
Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299:902–906
Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104:10270–10275
Lemke MD, Fisher EM, Kozlowska MA, Tano DW, Woodson JD (2021) The core autophagy machinery is not required for chloroplast singlet oxygen-mediated cell death in the Arabidopsis plastid ferrochelatase two mutant. BMC Plant Biol 21:342
Lermontova I, Kruse E, Mock HP, Grimm B (1997) Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc Natl Acad Sci USA 94:8895–8900
Li RQ, Jiang M, Huang JZ, Møller IM, Shu QY (2021) Mutations of the genomes uncoupled 4 gene cause ros accumulation and repress expression of peroxidase genes in rice. Front Plant Sci 12:682453
Lu Y, Yao J (2018) Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int J Mol Sci 19:3900
Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98:771–776
Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98:12826–12831
Mittler R (2017) ROS are good. Trends Plant Sci 22:11–19
Mohapatra A, Tripathy BC (2002) Detection of protoporphyrin IX in envelope membranes of pea chloroplasts. Biochem Biophys Res Commun 299:751–754
Mohapatra A, Tripathy BC (2007) Differential distribution of chlorophyll biosynthetic intermediates in stroma, envelope and thylakoid membranes in Beta vulgaris. Photosynth Res 94:401–410
Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20:1431–1438
Noctor G, Mhamdi A, Foyer CH (2014) The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol 164:1636–1648
Noctor G, Reichheld JP, Foyer CH (2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80:3–12
Ogilby PR (2010) Singlet oxygen: there is indeed something new under the sun. Chem Soc Rev 39:3181–3209
Oliveros JC (2007–2015) Venny An interactive tool for comparing lists with Venn's diagrams https://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed 1 August 2022
op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332
Page MT, McCormac AC, Smith AG, Terry MJ (2017) Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol 213:1168–1180
Papenbrock J, Mishra S, Mock HP, Kruse E, Schmidt EK, Petersmann A, Braun HP, Grimm B (2001) Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J 28:41–50
Pattanayak GK, Venkataramani S, Hortensteiner S, Kunz L, Christ B, Moulin M, Smith AG, Okamoto Y, Tamiaki H, Sugishima M, Greenberg JT (2012) Accelerated cell death 2 suppresses mitochondrial oxidative bursts and modulates cell death in Arabidopsis. Plant J 69:589–600
Pospíšil P (2016) Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01950
Pruzinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19:369–387
Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylides C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109:5535–5540
Ramel F, Ksas B, Akkari E, Mialoundama AS, Monnet F, Krieger-Liszkay A, Ravanat JL, Mueller MJ, Bouvier F, Havaux M (2013) Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25:1445–1462
Rentel MC, Lecourieux D, Ouaked F, Usher SL, Petersen L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, Knight MR (2004) OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427:858–861
Rosenwasser S, Fluhr R, Joshi JR, Leviatan N, Sela N, Hetzroni A, Friedman H (2013) ROSMETER: a bioinformatic tool for the identification of transcriptomic imprints related to reactive oxygen species type and origin provides new insights into stress responses. Plant Physiol 163:1071–1083
Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103:18822–18827
Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106:7660–7665
Shumbe L, Bott R, Havaux M (2014) Dihydroactinidiolide, a high light-induced beta-carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis. Mol Plant 7:1248–1251
Shumbe L, Chevalier A, Legeret B, Taconnat L, Monnet F, Havaux M (2016) Singlet oxygen-induced cell death in Arabidopsis under high-light stress is controlled by oxi1 kinase. Plant Physiol 170:1757–1771
Shumbe L, D’Alessandro S, Shao N, Chevalier A, Ksas B, Bock R, Havaux M (2017) Methylene Blue Sensitivity 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of beta-cyclocitral. Plant Cell Environ 40:216–226
Suo J, Zhao Q, David L, Chen S, Dai S (2017) Salinity response in chloroplasts: insights from gene characterization. Int J Mol Sci 18:1011
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800
Triantaphylides C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148:960–968
Uberegui E, Hall M, Lorenzo O, Schroder WP, Balsera M (2015) An Arabidopsis soluble chloroplast proteomic analysis reveals the participation of the executer pathway in response to increased light conditions. J Exp Bot 66:2067–2077
Vie AK, Najafi J, Winge P, Cattan E, Wrzaczek M, Kangasjärvi J, Miller G, Brembu T, Bones AM (2017) The IDA-LIKE peptides IDL6 and IDL7 are negative modulators of stress responses in Arabidopsis thaliana. J Exp Bot 68:3557–3571
Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Wursch M, Laloi C, Nater M, Hideg E, Apel K (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306:1183–1185
Wang L, Kim C, Xu X, Piskurewicz U, Dogra V, Singh S, Mahler H, Apel K (2016) Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc Natl Acad Sci USA 113:E3792–E3800
Wang LS, Leister D, Guan L, Zheng Y, Schneider K, Lehmann M, Apel K, Kleine T (2020) The Arabidopsis SAFEGUARD1 suppresses singlet oxygen-induced stress responses by protecting grana margins. Proc Natl Acad Sci USA 117:6918–6927
Wang Y, Wu Y, Zhong H, Chen S, Wong KB, Xia Y (2022) Arabidopsis PUB2 and PUB4 connect signaling components of pattern-triggered immunity. New Phytol 233:2249–2265
Warren CR (2008) Rapid measurement of chlorophylls with a microplate reader. J Plant Nutr 31:1231–1332
Watanabe N, Che FS, Iwano M, Takayama S, Yoshida S, Isogai A (2001) Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J Biol Chem 276:20474–20481
Wobbe L (2020) The molecular function of plant mTERFs as key regulators of organellar gene expression. Plant Cell Physiol 61:2004–2017
Woodson JD (2019) Chloroplast stress signals: regulation of cellular degradation and chloroplast turnover. Curr Opin Plant Biol 52:30–37
Woodson JD (2022) Control of chloroplast degradation and cell death in response to stress. Trends Biochem Sci. https://doi.org/10.1016/j.tibs.2022.03.010
Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salome PA, Weigel D, Fitzpatrick JA, Chory J (2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350:450–454
Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21:897–903
Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18:1310–1326
Zhao J, Mejias J, Quentin M, Chen Y, de Almeida-Engler J, Mao Z, Sun Q, Liu Q, Xie B, Abad P, Favery B, Jian H (2020) The root-knot nematode effector MiPDI1 targets a stress-associated protein (SAP) to establish disease in Solanaceae and Arabidopsis. New Phytol 228:1417–1430
Zhou C, Cai Z, Guo Y, Gan S (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150:167–177
Acknowledgements
The authors wish to thank Matthew Lemke (University of Arizona) for technical assistance with trypan blue staining, Emmanuel Gonzalez (University of Arizona) for technical assistance with genotyping, and Kamran Alamdari (University of Arizona) for technical assistance propagating mutant lines.
Funding
The authors acknowledge the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy grant DE-SC0019573 awarded to JDW. DWT was supported by the University of Arizona University Fellows Award, the John Boynton Fellowship (School of Plant Sciences), and the Graduate College Completion Fellowship. MAK was supported by the University of Arizona University Fellows Award.
Author information
Authors and Affiliations
Contributions
DWT, MAK, and JDW: planned and designed the research. DWT: performed all genetic analyses, metabolite measurements, meta-data analyses, and physiological growth experiments. DWT and RAE: performed all genotyping and gene expression experiments. MAK: performed all EL growth experiments and photosynthetic measurements. DWT and JDW: performed the SOSG assays. JDW: conceived the original scope of the project and managed the project. DWT and JDW: wrote the manuscript. All authors contributed to data analysis, collection, and interpretation.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tano, D.W., Kozlowska, M.A., Easter, R.A. et al. Multiple pathways mediate chloroplast singlet oxygen stress signaling. Plant Mol Biol 111, 167–187 (2023). https://doi.org/10.1007/s11103-022-01319-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-022-01319-z