Abstract
Key message
OscWRKY1 from Ocimum sanctum positively regulates phenylpropanoid pathway genes and rosmarinic acid content. OscWRKY1 overexpression promotes resistance against bacterial pathogen in Arabidopsis.
Abstract
WRKY transcription factor (TF) family regulates various developmental and physiological functions in plants. PAL genes encode enzymes which are involved in plant defense responses, but the direct regulation of PAL genes and phenylpropanoid pathway through WRKY TF’s is not well characterized. In the present study, we have characterized an OscWRKY1 gene from Ocimum sanctum which shows induced expression by methyl jasmonate (MeJA), salicylic acid (SA), and wounding. The recombinant OscWRKY1 protein binds to the DIG-labeled (Digoxigenin) W-box cis-element TTGAC[C/T] and activates the LacZ reporter gene in yeast. Overexpression of OscWRKY1 enhances Arabidopsis resistance towards Pseudomonas syringae pv. tomato Pst DC3000. Upstream activator sequences of PAL and C4H have been identified to contain the conserved W-box cis-element (TTGACC) in both O. sanctum and Arabidopsis. OscWRKY1 was found to interact with W-box cis-element present in the PAL and C4H promoters. Silencing of OscWRKY1 using VIGS resulted in reduced expression of PAL, C4H, COMT, F5H and 4CL transcripts. OscWRKY1 silenced plants exhibit reduced PAL activity, whereas, the overexpression lines of OscWRKY1 in Arabidopsis exhibit increased PAL activity. Furthermore, the metabolite analysis of OscWRKY1 silenced plants showed reduced rosmarinic acid content. These results revealed that OscWRKY1 positively regulates the phenylpropanoid pathway genes leading to the alteration of rosmarinic acid content and enhances the resistance against bacterial pathogen in Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
O. sanctum, commonly referred to as "holy basil," is a member of the Lamiaceae family. It has been significantly used worldwide due to its immense pharmacological properties and economically important aromatic oils related to terpenoid biosynthesis (Rastogi et al. 2014). O. sanctum leaves contain many biologically active compounds like triterpenoids, flavonoids, saponins and tannins (Jaggi et al. 2003). O. sanctum has antihistaminic, larvicidal, antibacterial, radioprotective, cardio-protective, anti-genotoxic, neuro-protective, anti-anaphylactic, wound healing, and antidiabetic activity (Rahman et al. 2011).
Transcription factors (TFs) usually interacts with a cis-element in the promoter of downstream genes, hence providing the regulation of the signaling cascade. (Agarwal and Jha 2010). Generally, a plant-specific WRKY TF family is characterized by the presence of a highly conserved WRKYGQK peptide sequence and a C2H2 or C2HC zinc finger motif (Eulgem et al. 2000). These regulatory TFs preferentially bind with the W box cis-elements (T/C-TGAC-T/C) present in their promoter, thereby differentially regulating the expression of target genes. Activation or repression of target genes is mainly regulated at transcriptional, translational, and domain levels through W-box consensus sequences (Phukan et al. 2016).
Based on the number of WRKY domains present, WRKY proteins are categorised into three major types. Group-I WRKY proteins contain two WRKY domains, whereas Group-II and -III WRKY proteins include a single WRKY domain. A zinc finger motif of the C2H2 type (C-X4-5-C-X22-23-H-X1-H) is found in Group-I and Group-II WRKY proteins, whereas the C2-HC type (C-X7-C-X23-H-X1-C) is found in Group-III WRKY proteins. Based on their primary amino acid sequence, Group-II WRKY proteins are classified into different subgroups IIa, IIb, IIc, IId, and IIe (Rushton et al. 2010). Group-I WRKY proteins feature functionally distinct WRKY domains, and it was discovered that the C-terminal domain of proteins binds to target DNA in a sequence-specific manner (Eulgem et al. 2000). The WRKY TF family is well known for its important function in controlling plant biotic and abiotic stress tolerance. WRKY members regulate the genes related to metabolite synthesis, hence regulating the production of valuable natural products. They are found to regulate the three major classes of plant secondary metabolite biosynthetic pathways i.e., phenylpropanoids, alkaloids, and terpenoidal biosynthesis (Schluttenhofer and Yuan, 2014; Mishra et al. 2013). It has been reported that several WRKY genes were induced during pathogen infection, elicitor treatment, or hormonal challenges in several plant species (Dong et al. 2003). AtWRKY70 in Arabidopsis is a key player in the defense signaling pathways mediated by jasmonic acid and SA, which leads to the enhanced pathogen resistance against specific bacteria and fungi (Li et al. 2004). Similarly, AtWRKY33 confers increased pathogen resistance against necrotrophic Botrytis cinerea via activation of tryptophan-derived camalexin biosynthesis (Mao et al. 2011). A WRKY TF from potato, StWRKY1 was involved in the regulation of phenylpropanoid biosynthesis against infection of Phytophthora infestans (Yogendra et al. 2015).
Phenylalanine gives rise to cinnamic acid, which acts as an intermediate in the formation of phenylpropanoids. The non-oxidative deamination of phenylalanine results in the formation of trans-cinnamate, which is catalyzed by PAL (Phenylalanine ammonia-lyase) enzyme. This is an important regulatory step between primary and secondary metabolism (Huang et al. 2010; Vogt T 2010). PAL plays a crucial role in plant defense mediated via the biosynthesis of SA, which acts as an essential signaling molecule in plant systemic resistance (Chaman et al. 2003; Nugroho et al. 2002). Expression studies of PAL reveal its corresponding response to various abiotic and biotic stresses like UV irradiation, nutrient depletion, wounding, extreme temperatures, and pathogen infection (Jin et al. 2013; Payyavula et al. 2012; Huang et al. 2010; Shine et al. 2016). Another important enzyme of the phenylpropanoid biosynthetic pathway is C4H, which is involved in synthesizing the building blocks of the lignin polymer. C4H mainly converts the trans-cinnamic acid into p-coumaric acid, which is the first hydroxylation step in the biosynthesis of lignin, hydroxycinnamic acid esters, and flavonoids (Blount et al. 2000; Blee et al. 2001).
In the present study, we elucidated the functional role of OscWRKY1 TF from O. sanctum in regulating the phenylpropanoid pathway. The interaction of OscWRKY1 with PAL and C4H promoters in both O. sanctum and Arabidopsis suggest the possible mechanism of OscWRKY1 in regulating the phenylpropanoid pathway leading to the enhanced Pst DC3000 resistance in the over-expression (OE) lines of Arabidopsis. We have utilized the VIGS for the first time in O. sanctum to functionally characterize the OscWRKY1. OscWRKY1 silencing provides a novel molecular insight into its role in directly regulating the phenylpropanoid pathway leading to the altered rosmarinic content in O. sanctum.
Materials and methods
Identification of some putative WRKY transcripts from O. sanctum
The various WRKY protein sequences were identified using O. sanctum transcriptome data (SRA Study accession number SRP039008). Full-length WRKY protein sequences in Arabidopsis and Oryza sativa (http://www.arabidopsis.org/) were used as reference sequences. The BLASTX search tool was used to manually validate selected WRKY proteins. All WRKY sequences, whether complete or partial, were included in this quest. From all these WRKY sequences full-length WRKY protein sequences were selected and grouped into WRKY subgroup I, II, and III, according to the classification data available from TAIR (Arabidopsis) WRKY TF database.
Phylogenetic analysis based on conserved WRKY domains
CLUSTALW performed multiple sequence alignment of the amino acid sequences of both full-length and partial WRKY sequences. MEGA11 with the maximum likelihood method was used to evaluate the phylogenetic and evolutionary relationships between various WRKYs. Only protein sequences having the highest homology with OscWRKY1 sequence were used in the multiple sequence alignment and phylogenetic study.
Plant growth and treatments
O. Sanctum two-month-old plants (var: CIM Ayu) were grown and collected from CSIR-CIMAP, hariti growth chamber. Plants were sprayed with a solution of 200 µM MeJA and SA in dimethyl sulfoxide (DMSO) and Triton-X. In control plants, only DMSO and Triton-X-containing solutions were sprayed. To maintain the proper transpiration rate, perforated plastic bags were used to cover the samples. At different time intervals, samples treated with MeJA/SA were collected and thoroughly washed with autoclaved water to eliminate contaminants. To avoid substantial injury, the whole plant was perforated with sterile needles to create wounds in the O. sanctum. For further analysis, samples were taken from treated and control plants at 1, 3, and 5 h time intervals. Arabidopsis (WT Col-0) and transgenic OscWRKY1 seeds were sown in a soilrite mixture and maintained in the growth chamber at 22 °C, 16:8 light: dark, and 65% relative humidity.
Total RNA isolation, cDNA synthesis, and qRT-PCR
The leaf and root samples were rinsed twice with DEPC treated water before being crushed in liquid N2. The NucleoSpin RNA kit (Macherey Nagel) was utilized to isolate RNA from plant samples. The quality of total RNA was monitored by spectrophotometry and running on 2% agarose gel. First-strand cDNA was synthesized from 1 µg of total RNA utilizing a high-efficiency cDNA reverse transcription kit (Thermo Scientific). Total cDNA products were quantified and checked for purity. The qRT-PCR reactions were set up for 20 μL, each containing 100 ng of total cDNA, 0.4 μM of forward and reverse primers, 10 μL of SYBR Premix Ex Taq™ II (Takara), and 0.4 μL of reference dye. The amplification conditions for all the primer pairs were: 95 °C initial denaturation for 10 min; 95 °C denaturation for 15 s, 60 °C primer annealing/elongation for 1 min (40 cycles). The fluorescence was detected and measured during the last step of each cycle.
After each PCR loop, a melting curve analysis was performed by progressively rising the fluorescence temperature from 60 to 95 °C. The qRT-PCR experiments used the 7500 FAST Real-Time PCR system (Applied Biosystems, USA) (Fig. S11). Actin and ubiquitin were used to normalize gene expression as endogenous controls. To evaluate relative transcript expression, the 2−ΔΔCt method was used. Both tests were carried out with three biological and experimental replicates, and the data were then statistically analyzed. Real-time PCR primers were produced using the unique region of the transcript in Table S1.
Electrophoretic mobility shift assay (EMSA) and transactivation β‐galactosidase assay
The O. sanctum transcriptome data set was used to select OscWRKY1, which was then cloned into the bacterial expression vector pGEX4T2 using EcoRI and XhoI restriction sites. The construct was confirmed using digestion and sequencing. The reading frame was retained in addition to GST. After that, the OscWRKY1-GST cloned plasmid was transformed into BL21 (CodonPlus, DE3 strain), and then induced with IPTG (0.4 mM). The recombinant OscWRKY1 was affinity purified using GST beads (Amersham). The W-box sequence TTGAC[C/T] cis-element probes were developed, as were their mutated counterparts (TTGACG, TTCACT) (Table S1). The EMSA was carried out utilizing the manufacturer's instructions (DIG-gel shift kit 2nd generation kit, Roche).
OscWRKY1 was cloned into the Saccharomyces cerevisiae expression vector pGBKT7 and fused to the GAL4-DNA-binding domain. Positive clones were confirmed using sequencing and restriction enzyme digestion (NdeI and BamHI). The positive cloned plasmid was transformed into yeast Y187 (S. cerevisiae), containing LacZ gene with modified GAL4 promoter, and the resulting colonies were selected in the dropout medium (SD-Trp-Ura). The selected positive colonies were further streaked on an SD-Trp-Ura replica plate for the positive transactivation assay. The blue color intensity was determined using X-gal as a substrate and a colony lift assay with galactosidase (Phukan et al. 2018).
Yeast one-hybrid (Y1H) assay
We identified the upstream promoter sequences of PAL and C4H from the genome sequence of both O. sanctum and Arabidopsis. Promoters were amplified using specific forward and reverse primers from their respective genomic DNA. OscWRKY1 was cloned into the yeast expression vector pGADT7 for the Y1H assay, and the promoters of O. sanctum and Arabidopsis (PAL and C4H) were cloned into the pHIS-2.0 vector. In the yeast S. cerevisiae Y187, both cloned plasmid DNAs were co-transformed was checked by lithium acetate-mediated (LiAc) yeast transformation. Desired cloned plasmid DNA and excess carrier DNA were transformed into the yeast competent cells (Gietz et al. 1992). The positive interaction was checked by plating it on SD-His-Leu medium. Positive colonies were streaked on YPD and SD-His-Leu selection dropout media. All yeast experiments were carried out using the Yeast Protocols Handbook (Clontech).
PAL activity assay
By using 100 mM phosphate buffer (pH 6.0) with 4 mM dithiothreitol, 2 mM EDTA, and 2% polyvinylpyrrolidone, total protein was isolated from the leaf samples of Arabidopsis. In the leaf extract, PAL activity was determined as described in the earlier protocol (Song and Wang 2011). The protein extract was incubated for 50 min at 30 °C containing 2 mL borate buffer (0.01 M, pH 8.7) and L-phenylalanine (0.02 M, 1 ml). The absorbance of the sample was taken at 290 nm before and after the incubation. For blank control, a reaction mixture without substrate was used and the experiment was performed in biological replicates, and for each extract, triplicate assays were performed. PAL activity was expressed in nkatal per mg of protein.
Bacterial resistance assay
Fully developed rosette leaves of WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 Arabidopsis plants were syringe-infiltrated with Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) resuspended in 10 mM MgCl2. Disease phenotype was observed and photographed at regular time points. For the quantification of bacterial growth, the leaf disc area of 1 cm2 from each genotype were collected at 0- and 2 days post-infection (DPI) and ground in 1000 µL MilliQ water. The homogenate was then serial diluted and plated on LA medium containing 50 µg mL−1 rifampicin. The bacterial colony-forming unit (CFU) was counted and the bacterial population [Log (CFU/cm2)] are presented. Two independent experiments were conducted and three biological replicates were taken in each experiment.
In planta pathogen colonization using confocal microscopy
For the in planta pathogen colonization assay, WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 Arabidopsis plants were syringe-infiltrated with GFP-tagged Pst DC3000 strain. At 2 DPI, images were collected from the infected leaf discs using confocal laser scanning microscopy. GFP fluorescence (488 nm excitation/493–598 nm emission) and red chlorophyll fluorescence (633 nm excitation/647–721 nm emission) was acquired on a Zeiss LSM 880 confocal microscope. Image acquisition was performed using ZEN blue software (Carl Zeiss, Oberkochen, Germany).
Electrolyte leakage measurement
Four leaf discs each of diameter 4 mm were excised from each WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 plants infiltrated with Pst DC3000 at 2 DPI. The leaf discs (each tube containing four discs per genotype) were then gently agitated in deionized water for 3 h at 28 °C. The initial conductivity was determined with the help of a conductivity meter (HORIBA Scientific, F74BW). Then, these tubes were boiled at 121 °C for 20 min to release the total electrolytes and the conductivity was again measured. The values of the initial electrical conductivity, and conductivity obtained after autoclaving were used to calculate the percentage of electrolyte leakage relative to total electrolytes in each genotype. Two independent experiments were conducted and two biological replicates were taken in each experiment.
Virus-induced gene silencing (VIGS) in O. sanctum
The pTRV1 and pTRV2 vectors based on the tobacco rattle virus (TRV) were used to conduct VIGS in O. sanctum. (Liu et al. 2002a, b; Senthil-Kumar and Mysore 2014). The cDNA sequences of OscPDS and OscWRKY1 were retrieved from the transcriptome resource of the O. sanctum. Specific primers were designed utilizing the conserved domains of OscPDS and OscWRKY1. Both fragments were amplified using PCR and cloned into the pTRV2 vector, which was validated using restriction digestion and sequencing. Cloned plasmids were then individually transformed into Agrobacterium tumefaciens strain (GV3101). Overnight grown Agrobacterium suspensions containing pTRV1 and pTRV2 (pTRV2-OscPDS and pTRV2-OscWRKY1) were centrifuged and re-suspended in infiltration buffer (10 mM MES pH5.6, 10 mM MgCl2, 200 μM acetosyringone). Vacuum-aided agro-infiltration (Misra et al. 2020; Deng et al. 2021; Wang et al. 2018; Zhang et al. 2017) was employed to perform VIGS. Seeds of O. sanctum were surface sterilized and sown in the nursery bed. The cotyledonary leaf stage plants were tested for agro-infiltration using vacuum. The Agrobacterium pTRV1 and pTRV2 (pTRV2-OscPDS and pTRV2-OscWRKY1) suspensions were mixed in similar concentrations to achieve the optimal optical density OD600. Agrobacterium suspension was generated by mixing Agrobacterium possessing pTRV1 and pTRV2 vectors, served as control whilst using the infiltration buffer for mock control. Agrobacterium density was optimized for efficient VIGS and so suspension at OD600 of 0.5, 1.0, and 1.5 were also considered. Approximately, 100 mL of the final suspension was used. Healthy cotyledonary leaf stage seedlings with uniform growth were utilized for the vacuum-aided infiltration method. The seedlings were uprooted, washed with Milli-Q water, and were then inverted into the suspension solution in a desiccator jar connected to the vacuum pump. A vacuum was generated with the help of a pump to further generate 750 mm Hg pressure. The pressure was applied for an additional 90 s (Misra et al. 2020) and was released slowly causing infiltration of the Agrobacterium suspension into the immersed plant leaves. The seedlings designated to serve as a mock were also immersed in the infiltration buffer. The seedlings were removed and the excess buffer was drained in blotting paper. The seedlings were then transplanted into plastic pots containing soilrite mix, covered with PVC cling film to maintain an elevated humidity level, and left in the dark overnight at room temperature. Plants were then held in light at 22–24 °C at 16 h: 8 h, light: dark cycle, and further after 3 days the cling film was removed. The VIGS phenotypes were observed for up to 29 days. At 24 DPI (days post infiltration), first-pair leaves of infected seedlings along with the mock seedlings were harvested and stored at − 80 °C for specific transcript analysis.
HPLC estimation of VIGS silenced plants
The solution of reference compound was prepared in methanol at different concentrations of 0.1875–3 µg/mL (Fig. S2). The first-pair leaves of infected seedlings were harvested and the methanolic extract of the sample for identification was prepared (Sarfaraz et al. 2021). The plant extracts were used for the quantification of rosmarinic acid using HPLC (Waters 2998, PDA detector) and the elution was followed according to the method described in the earlier studies (Gharibi et al. 2019; Sarfaraz et al. 2021). The stationary phase consisted of a 250 mm × 4.6 mm (5 µm) symmetry C18 column (Phenomenex, California, USA), and the mobile phase included solvent A and B with a flow rate of 1 mL min−1 with detection between 200 and 400 nm through UV detector. 0.1% of the water-formic acid was used as solvent A, while acetonitrile was applied as solvent B in the mobile phase. The gradient conditions were performed with solvent B varying from 10 to 90% in 30 min. The content in the extract was calculated comparing the peak and the retention time and reported as µg/g.
Results
Identification, phylogenetic study, and expression analysis of OscWRKY1 from O. sanctum
To identify the WRKY TF in O. sanctum, we used earlier sequence data resources available in the public domain submitted as sequence read archives (SRAs) (Rastogi et al. 2014). From the transcriptome sequence of O. sanctum, transcripts that belong to the WRKY superfamily were identified initially. Almost all identified proteins contained the highly conserved WRKYGQK motifs in their protein sequences (Fig. S1). After identifying all the WRKY transcripts in O. sanctum, we BLAST the annotated sequences to identify the full-length transcripts from the transcriptome data. From the total annotated transcripts, the full-length sequence of OscWRKY1 was amplified using gene-specific primers. It was cloned in the PTZ57R/T vector and further confirmed by sequencing. The size of OscWRKY1 transcript as per the annotation made in earlier data is 3349 bp, which includes a coding sequence (CDS) of 1023 bp encoding 340 amino acids. The full-length CDS was aligned with its encoded amino acid sequence as shown in Fig. S2. It was found that the DNA binding domain consists of 58 amino acids. Protein sequences of OscWRKY1 homologs from different plant species were retrieved from the NCBI database for sequence alignment to determine its evolutionary similarity and diversity using Clustal Omega. The phylogenetic homology of OscWRKY1 was determined using MEGAX. OscWRKY1 was showing maximum homology (75%) to the probable WRKY protein of Salvia splendens and to the hypothetical protein of Perilla frutescens (74%) (Fig. 1a). Based on the BLASTp analysis of WRKY proteins from different species, we found most of them were either putative or uncharacterized (data not shown). From the transcriptome differential gene expression data, we studied the expression of unique full-length transcripts present in O. sanctum and found that OscWRKY1, OscWRKY4, OscWRKY48, OscWRKY2, and OscWRKY15 were highly expressed (Fig. S3). To further validate the tissue-specific expression patterns of specific full-length OscWRKY1 genes in leaf and root tissues, we performed qRT-PCR. The leaf and root tissue show differential expression of WRKY genes; among them, OscWRKY1 expression is higher in leaf tissue than in root tissue (Fig. 1b).
Phylogenetic analysis and tissue specific expression of OscWRKY1 TF. a The evolutionary history was inferred by using the Maximum Likelihood method model. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test 1000 replicates are shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithm, and then selecting the topology with superior log likelihood value. Evolutionary analyses were conducted in MEGA11. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. b Heat map showing tissue-specific expression of OscWRKY transcripts. Some of the selected full-length OscWRKY transcripts from the transcriptome data were validated using the qRT-PCR analysis to study the differential expression in the leaf and root tissues. c Tissue-specific expression of OscWRKY1 after MeJA, SA treatment and wounding. The relative expression of OscWRKY1 transcript in the shoot and root tissue was studied after 1, 3, and 5 h of MeJA, SA treatment and wounding. Relative expression of transcripts was calculated taking untreated plant samples as a control. Actin and ubiquitin were used as an endogenous control for gene normalization. The color scale represents the relative fold change values
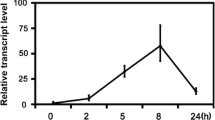
MeJA and SA are known to be important elicitors in diverse signaling cascades and are also involved in the regulation of various enzymes related to plant secondary metabolic pathways. To know the role of identified WRKYs as a signaling molecule in response to MeJA, SA, and wounding, we studied the differential expression of OscWRKY1 after 1, 3, and 5 h of MeJA, SA treatment, and wounding. We found that OscWRKY1 is highly induced after 3 and 5 h of MeJA treatment in the leaf as compared to the root (Fig. 1b). OscWRKY1 also showed induced expression in response to SA treatment. Wounding in O. sanctum also upregulates the early expression of OscWRKY1 in both leaf and root tissues (Fig. 1c). The significant upregulation of OscWRKY1 after MeJA, SA, and wound treatment indicates that it might have a role in the regulation of pathogen response or secondary metabolism.
OscWRKY1 binds to the promoter containing W-box cis-elements and transactivates the reporter gene in yeast
The promoter region contains regulatory components that are required for gene expression to be regulated on a temporal, geographic, and cell type-specific basis (Lescot et al. 2002). For bacterial protein purification and studying the DNA binding affinity, we cloned OscWRKY1 in the pGEX-4T2 expression vector. Recombinant glutathione S-transferase (GST)-OscWRKY1 fusion protein was induced and purified from E. coli BL21 (DE3). The size of (GST)-OscWRKY1 is 62.4 KDa. SDS-PAGE was used to check the purified fraction of recombinant OscWRKY1, which was then subjected to an EMSA. The EMSA results indicated that only when OscWRKY1 binds to the W-box cis-elements TTGACC (W-box 1) and TTGACT (W-box 5) (Fig. 2a) could DNA–protein complex signals be produced. We designed specific probes for TTGACC and TTGACT cis-elements as well as their mutated counterparts with a single nucleotide change. We observed that OscWRKY1 interacted with W-box 1 and W-box 5, but not with the mutated probes (Fig. 2b, Fig. S4). These results suggest that W-box 1 and W-box 5 are the novel cis-elements bound by OscWRKY1. OscWRKY1 interacts with W-box cis-element present within the promoter of target genes with the help of their DNA binding domain (DBD) and thus activates transcription with the help of their transactivation domain. To investigate the transactivation ability, we cloned OscWRKY1 in pGBKT7 in fusion with modified GAL4-DBD and then transformed into the yeast Y187. OscWRKY1 demonstrated positive transactivation by activating the reporter gene LacZ in yeast and producing blue color when X-gal was applied. In the absence of Trp and Ura, transformed yeast colonies were developed in the synthetic dextrose media (Fig. 2c).
Electrophoretic mobility shift and transactivation assay of OscWRKY1 protein. a Probes containing two different W-box cis-elements (TTGACC and TTGACT) were designed to study the DNA–protein interaction. The desired cis-elements are marked with red color while the binding site carrying mutations are highlighted with blue color. b EMSA of OscWRKY1 showed that it specifically interacts with both TTGACC and TTGACT cis-elements (L- DIG-labelled and M- mutated probe) M: TTGACG, TTCACT. DIG. Digoxigenin. c To study the transactivation property of OscWRKY1, it was cloned in yeast expression vector pGBKT-7 in fusion with GAL4-DBD and then transformed in yeast S. cerevisiae (Y187 strain). The positive colonies were selected and streaked on dropout SD-U-T media. Transactivation assay was performed using X-gal as a substrate
Overexpression of OscWRKY1 in Arabidopsis confers enhanced resistance to P. syringae pv. tomato DC3000
To assess the functional role of OscWRKY1 in plants, transgenic Arabidopsis lines were generated using the constitutive promoter CaMV35S. We selected two independent homozygous lines (OscWRKY1-L1 and OscWRKY1-L2) containing the 35S:OscWRKY1 construct. To validate transgenic lines of Arabidopsis, PCR amplification was performed using genomic DNA and pBI-121 nptII (KanR)/gene-specific primer set (Fig. S5). The predicted amplification product was obtained from transgenic lines but not from WT Col-0 plants, demonstrating that the transgene was successfully incorporated into the Arabidopsis genome. The overexpression of OscWRKY1 in transgenic plants was confirmed by semi-quantitative PCR using gene-specific primers. We observed that both the homozygous lines (OscWRKY1-L1 and OscWRKY1-L2) had enhanced expression of OscWRKY1, whereas no expression was detected in WT Col-0 plants (Fig. S6). To study the role of OscWRKY1 in plant defense, OscWRKY1 overexpressing transgenic plants (OscWRKY1-L1 and OscWRKY1-L2) were infected with bacterial pathogen Pst DC3000 and compared with WT Col-0 Arabidopsis plant. Importantly, estimation of bacterial pathogen Pst DC3000 population from the OscWRKY1-L1 and OscWRKY1-L2 plants, showed significantly reduced growth of Pst DC3000 when compared to WT Col-0 plant. At 2 DPI, Pst DC3000 growth enhanced by 1.48 [Log (CFU/cm2)] unit (~ 30-fold) in WT Col-0 plants, however, during the same time period, Pst DC3000 growth enhanced only by 0.58 [Log (CFU/cm2)] unit (~ 3.8-fold) and 0.22 [Log (CFU/cm2)] unit (~ 1.7-fold), in OscWRKY1-L1 and OscWRKY1-L2 plants, respectively (Fig. 3a). Additionally, the Arabidopsis transgenic plants overexpressing OscWRKY1 (OscWRKY1-L1 and OscWRKY1-L2) infiltrated with Pst DC3000 showed significantly reduced electrolyte leakage as compared to the WT Col-0 infiltrated with Pst DC3000 (Fig. 3b). Furthermore, a GFP-tagged Pst DC3000 strain was used to observe the in planta colonization of the pathogen in the leaves of WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 plants using confocal fluorescence microscopy. The GFP-tagged Pst DC3000 bacteria were observed as green fluorescent microcolonies inside the infected leaves. The overexpressing transgenic plants (OscWRKY1-L1 and OscWRKY1-L2) displayed reduced colonizing bacterial pathogen Pst DC3000 as compared to WT Col-0 plants (Fig. 3c). Taken together, these observations suggest that overexpression of OscWRKY1 enhances the Arabidopsis resistance to bacterial pathogen, Pst DC3000.
Overexpression of OscWRKY1 in Arabidopsis promotes resistance against P. syringae pv. tomato (Pst DC3000). a Estimation of bacterial population in leaves of WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 plants infiltrated with Pst DC3000 at 2 DPI. Data are from two independent experiments and biological triplicates per experiment. Error bars indicate mean ± SD. Student’s t-test: *, P < 0.05; **, P < 0.01. b Electrolyte leakage was measured in Arabidopsis leaves of WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 plants at 2 DPI with Pst DC3000. Electrolyte leakage values are presented (%) by calculating the ratio of initial electrical conductivity to the conductivity obtained after autoclaving (100% electrolytes). Data are from two independent experiments and biological duplicates per experiment. Error bars indicate mean ± SD. Student’s t-test: *, P < 0.05; **, P < 0.01. c Visualization of GFP-tagged Pst DC3000 in leaves of WT Col-0, OscWRKY1-L1 and OscWRKY1-L2 plants using confocal scanning microscopy. The GFP fluorescence was observed with 488 nm excitation and 493–598 nm emission and chlorophyll autofluorescence was observed with 633 nm excitation, and 647–721 emission. Confocal images are shown for GFP fluorescence (green), chlorophyll autofluorescence (red), and merged signals. Scale bars equals 100 µm
Differential expression study of the phenylpropanoid pathway and PR genes after MeJA treatment and wounding
Increased resistance to plant diseases is generally associated with increased transcript levels of PR genes implicated in the SA defense pathway (Dong et al. 2003; Loake and Grant 2007). Overexpression of OscWRKY1 increased pathogen resistance in plants; therefore, to further determine whether OscWRKY1 is involved in controlling the expression of O. sanctum phenylpropanoid pathway, we performed qRT-PCR after MeJA and wound treatment in both root and shoot tissues. Transcriptome-wide candidate genes were selected, which are related to the phenylpropanoid pathway and PR genes. Interestingly, the expression of OscPAL, OscC4H, and Osc4CL was induced early after MeJA and wound treatment in leaf tissue (Fig. S7a). PR1 protein II and thaumatin-like protein were also found to be highly upregulated after 1 h of MeJA and wound treatment in root tissue (Fig. S7b). These results suggest that OscWRKY1 elicited by MeJA/wounding might be regulating the expression of phenylpropanoid pathway genes. For further investigation, we analyzed all the promoters of MeJA inducible transcripts in O. sanctum and Arabidopsis for the presence of W-box. We found the similar W-box cis-elements present in the promoter of Arabidopsis (Fig. S8). PAL and C4H promoters from both Arabidopsis and O. sanctum were amplified from genomic DNA and cloned for further study.
Interaction study of OscWRKY1 with the promoter of AtPAL1 and AtC4H from Arabidopsis thaliana
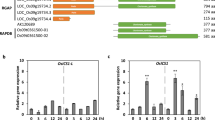
In Arabidopsis thaliana, overexpression of OscWRKY1 improved pathogen resistance by interacting with the W-box cis-element present in the promoters of AtPAL1 and AtC4H. The yeast one-hybrid assay was used to confirm the association of OscWRKY1 with the promoters of AtPAL1 and AtC4H. Reporter constructs for the AtPAL1 and AtC4H promoters were generated by cloning the promoter upstream of the HIS3 reporter gene into the pHIS2.0 vector. To investigate the yeast one-hybrid assay, effector and reporter constructs were co-transformed in S. cerevisiae strain Y187. The resulting transformants with OscWRKY1 and AtPAL1/C4H promoters could grow on SD-H–L media. Positive colonies indicated the interaction of OscWRKY1 with the promoters of AtPAL1 and AtC4H. Transformants carrying the pGAD-OscWRKY1 and pHIS2.0-AtPAL1/C4H vectors were unable to grow in the selective media (Fig. 4a, b). This indicates that pGAD-OscWRKY1 interacts with the W-box cis-element in the promoters of both AtPAL1 and AtC4H, causing transcription of the reporter Histidine (his) gene to be activated.
OscWRKY1 interacts with the promoters of AtPAL1 and AtC4H from Arabidopsis thaliana. a Yeast one-hybrid assay was performed in S. cerevisiae (Y187 strain) to study the in vivo interaction of OscWRKY1 with the AtC4H promoter. The effector construct was prepared by cloning the OscWRKY1 transcript in pGAD-T7 vector and the reporter construct was prepared by cloning the desired AtC4H promoter containing the W-box cis-elements in pHIS2.0 vector. Both the effector and reporter constructs were then co-transformed in yeast Y187. The positive colonies containing the resulting co-transformants obtained in the selection media were streaked in the YPD and dropout selection media (SD-his-leu). PAL phenylalanine lyase, C4H cinnamate-4-hydroxylase. b Similarly, the lower panel is the Y1H assay between OscWRKY1 and AtPAL1 promoter in yeast Y187. Here the reporter construct was prepared by cloning the desired AtPAL1 promoter containing the W-box cis-elements in pHIS2.0 vector
Expression study of AtPAL1 and AtC4H in transgenic Arabidopsis thaliana
To further study how OscWRKY1 regulates the transcript level of AtPAL1 and AtC4H in Arabidopsis, we performed qRT-PCR analysis in control and transgenic lines. We observed an increase in AtPAL1 expression in both overexpressing lines as compared to wild-type WT Col-0 plants. Additionally, AtC4H expression was observed to be elevated in both transgenic lines as compared to WT Col-0 plants. (Fig. 5a). These results suggest the hypothesis that OscWRKY1 might be regulating AtPAL1 and AtC4H in Arabidopsis by binding to the W-box cis-element within their promoters and involved in increased pathogen resistance in the plant.
Expression study of PAL, C4H and PAL activity assay in OscWRKY1 Arabidopsis overexpression lines. a Relative AtPAL1 and AtC4H expression levels were measured through qPCR in the leaves of OscWRKY1 overexpressing lines. The relative expression of AtPAL1 and AtC4H were positively correlated in both OscWRKY1 overexpression lines. The data represents the run from two independent biological and experimental replicates. Ubiquitin, was used as endogenous control. b PAL activity is measured in the leaves of OscWRKY1 overexpression lines and WT Col-0 plants under both control conditions and during infection with Pst DC3000. PAL enzyme activity is calculated in nKatal/mg protein. PAL activity is significantly higher in the leaves of OscWRKY1 transgenic lines. Error bars indicate mean ± SD. Student’s t-test: **, P < 0.01
Enhanced PAL activity in OscWRKY1 transgenic lines of Arabidopsis thaliana
Overexpression study of OscWRKY1 in Arabidopsis reveals that it might play a crucial role in increased pathogen resistance against Pst DC3000. Leaves of wild-type WT Col-0 plants showed severe disease symptoms as compared to transgenic OscWRKY1 plants. Our study shows the binding of OscWRKY1 with the promoter of AtPAL1, which might be involved in regulating PAL content in transgenic plants. We next analyzed PAL activity in both WT Col-0 and OscWRKY1 overexpression lines under control conditions and during infection with Pst DC3000. We observed the level of PAL activity and found that it is significantly higher in the leaves of OscWRKY1 overexpression lines as compared to WT Col-0 plants under control conditions. As expected, infection with virulent Pst DC3000 significantly induces a higher amount of PAL activities in transgenic lines of OscWRKY1 plants as compared to the WT Col-0 plants (Fig. 5b).
OscWRKY1 interacts with the promoter of OscPAL and OscC4H from O. sanctum
To validate the DNA binding ability of OscWRKY1 in yeast, the fragment of interest was fused with the GAL4 activation domain present in the pGADT7 vector encoding the leu reporter gene. Reporter constructs for the OscPAL and OscC4H promoters were made by cloning the promoter upstream of the HIS3 reporter gene into the pHIS2.0 vector. Effector and reporter constructs were co-transformed into S. cerevisiae strain Y187 to perform the yeast one-hybrid assay. The resulting transformants containing OscWRKY1 with OscPAL/C4H promoters were unable to grow on SD-H–L media. Transformants containing pGAD-OscWRKY1 and pHIS2.0-OscPAL/C4H vector could not grow on the selective media (Fig. 6a, b). This occurs as a result of the interaction of pGAD-OscWRKY1 with the W-box cis-element in the promoters of both OscPAL and OscC4H, which activates the reporter histidine (His) gene transcription.
OscWRKY1 interacts with the promoter of OscC4H and OscPAL. a To study the in vivo interaction of OscWRKY1 with the OscC4H promoter, Y1H assay was performed in yeast S. cerevisiae (Y187 strain). The effector construct was prepared by cloning the OscWRKY1 transcript in pGAD-T7 vector and the reporter construct was prepared by cloning the OscC4H promoter containing the W-box cis-elements in pHIS2.0 vector. The effector and reporter constructs were co-transformed in Y187. The resulting co-transformants obtained in the selection media were streaked in the YPD and dropout selection media (SD-his-leu). b Yeast one-hybrid assay between OscWRKY1 and the OscPAL promoter in yeast Y187
VIGS of OscWRKY1 confers reduced PAL activity in O. sanctum
To assess the efficacy of the silencing vacuum-aided agro-infiltration approach was optimized. OscPDS was chosen as a marker gene from O. sanctum to ensure the efficiency of the protocol, which leads to the photobleaching of young leaves owing to impaired chlorophyll biosynthesis. The photobleached leaf phenotype as a sign of OscPDS silencing started appearing after 13 DPI with Agrobacterium strains carrying pTRV1 and pTRV2‐OscPDS vectors (Fig. 7a). However, the symptom gradually became more prominent by 24 DPI. We also studied the transcript level of OscPDS to understand the effective gene silencing in O. sanctum plants. We found that OscPDS was downregulated as compared to the vector control (Fig. 7b). To further confirm the silencing of OscWRKY1, qRT-PCR was performed in two independent biological replicates. We found that OscWRKY1 was downregulated as compared to the vector control. The expression of some other WRKY genes (OscWRKY7 and OscWRKY15) used as a negative control does not show any downregulation in comparison to vector control, which confirms the specificity of VIGS-OscWRKY1 (Fig. 7c). Additionally, the coat protein (CP) and RNA-dependent RNA polymerase (RdRp) from the pTRV1 and pTRV2 vectors were detected in the agro-infiltrated plants but not in the mock-treated plants. (Fig. S9). This confirms the successful infiltration of pTRV1 and pTRV2 vectors in the seedlings.
Viral Induced Gene Silencing (VIGS) in O. sanctum achieved by vacuum mediated agro-infiltration. a O. sanctum plants infected with pTRV1 and pTRV2‐OscPDS/OscWRKY1 showing photobleached phenotype 24 days post-infiltration. b Relative OscPDS expression levels in the leaves of empty vector control (pTRV1-pTRV2) and OscPDS-pTRV2 infected O. sanctum plants based on qRT-PCR analysis. OscPDS was used as a marker gene. VC, Vector Control; PDS, Phytoene desaturase c Confirmation of the specificity of silencing by the estimation of the relative transcript level of OscWRKY1 in vector control and VIGS silenced plants. WRKY genes (OscWRKY7 and OscWRKY15) used as the negative control. d Relative expression levels of OscPAL1, OscPAL2 and OscPAL3 in the leaves of vector control (pTRV1-pTRV2) and OscWRKY1-pTRV2 infected O. sanctum plants using qRT-PCR analysis. The data represent two independent biological and experimental triplicates. Ubiquitin, was used as endogenous control. Error bars indicate mean ± SD. Student’s t-test: *, P < 0.05, **, P < 0.01. e PAL activity was measured from the leaves of three independent vector control and OscWRKY1-pTRV2 plants. PAL enzyme activity was calculated in nKatal/mg protein. The data represents error bars with mean ± SD. Student’s t-test: **, P < 0.01
Phenylpropanoid biosynthesis is one of the most important processes in secondary metabolism, leading to synthesis of a vast array of natural compounds. C4H (Cinnamate-4-hydroxylase), F5H (Ferulate-5-hydroxylase), CCR (cinnamoyl-CoA reductase), COMT (caffeic acid O-methyltransferase) along with the PAL (Phenylalanine ammonia lyase) are among the key genes involved in phenylpropanoid biosynthesis. OscPAL1, OscC4H, and Osc4CL was induced early after MeJA and wound treatment in leaf tissue (Fig. S7a) and OscWRKY1 was found to interact with the promoters of PAL and C4H in O. sanctum (Fig. 6a, b). These data imply OscWRKY1 may regulate phenylpropanoid pathway in O. sanctum. To test this theory, we evaluated phenylpropanoid gene expression patterns in OscWRKY1 VIGS silenced plants. OscPAL1, OscPAL2 and OscPAL3 were downregulated in OscWRKY1 silenced line when compared with the control (Fig. 7d). We next analyzed the PAL activity in both vector control and OscWRKY1-pTRV2 agro-infiltrated O. sanctum plants and found that PAL activity is reduced in OscWRKY1-pTRV2 as compared to the vector control (Fig. 7e). These results further indicate the role of OscWRKY1 in monitoring the level of PAL activity in the plant by modulating the activity of OscPAL. The relative transcript level of F5H, CCR, COMT, 4CL and C4H were also evaluated and were found to be downregulated in silenced lines (Fig. 8a).
Expression analysis of phenylpropanoid pathway genes and quantitative determination of rosmarinic acid using HPLC in silenced lines. a Phenylpropanoid pathway gene expression study in vector control and OscWRKY1 VIGS silenced plants. PAL, Phenylalanine ammonia lyase; C4H, Cinnamate-4-hydroxylase; F5H, Ferulate-5-hydroxylase; CCR, cinnamoyl-CoA reductase; COMT, caffeic acid O-methyltransferase. Ubiquitin, was used as endogenous control. The data represent two independent biological and experimental triplicates. b Determination of rosmarinic acid in the silenced lines using HPLC. Error bars indicate mean ± SD. Student’s t-test: *, P < 0.05, **, P < 0.01
Silencing of OscWRKY1 resulted in decreased rosmarinic acid content
HPLC analysis was used for quantitative determination of polyphenols. The concentration of rosmarinic acid varied the most among the polyphenols assessed in the methanolic extract of VIGS silenced samples. The concentration of rosmarinic acid was assessed to be reduced in lines infected with VIGS-OscWRKY1 when compared to pTRV1:pTRV2 infected lines (Fig. 8b). The calibration plot was used as the basis of the determination (Fig. S10).
Discussion
O. sanctum is an important medicinal plant well known for its economically important aromatic oils. It contains aromatic compounds like limonene, eugenol, camphene, α-Pinene, and camphor in their essential oil. O. sanctum is rich in phytochemicals, which include mainly terpenoids and phenylpropanoids. Plant WRKY transcription factors are of particular importance as they participate in a variety of biotic and abiotic stress responses and biological processes (Jiang et al. 2015). In this study, we elucidate the functional role of OscWRKY1 from O. sanctum in regulating phenylpropanoid pathway genes. From the earlier published transcriptome data of O. sanctum, we identified the OscWRKY1 transcripts expressing in the leaf tissue (Rastogi et al. 2014). The phylogenetic study revealed that OscWRKY1 belongs to subgroup III. MeJA and wounding function as elicitors playing key roles in varied plant metabolic processes related to secondary metabolism and defense response (Cheong et al. 2002). The expression of OscWRKY1 after MeJA, SA, and wounding was found to be higher in comparison with other WRKY transcription factors from O. sanctum. We started characterizing OscWRKY1 due to its higher comparative expression and having a blastX homology with probable WRKY and hypothetical protein from Salvia splendens and Perilla frutescens.
WRKY genes are known to play a central role in SA- and JA- dependent processes (Thaler et al. 2012). WRKY TF controls gene expression by binding to the W-box cis-element, and hence plays an important role in many biological processes. Previous studies revealed the ability of different WRKY TFs to bind with W-box cis-elements and their involvement in both positive and negative gene regulation (Wang et al. 2009; Peng et al. 2016; Hu et al. 2018). For example, WRKY12 negatively regulates cadmium tolerance in Arabidopsis by interacting with the promoter of glutathione GSH-1 and represses the expression of phytochelatin biosynthesis genes (Han et al. 2019). PsWRKY (Papaver somniferum), and WsWRKY1 (Withania somnifera) positively regulates the secondary metabolic pathway genes by binding with the consensus W-box cis-element (Mishra et al. 2013; Singh et al. 2017). The present work showed that recombinant OscWRKY1 protein binds with the W-box cis-elements (TTGAC[C/T]) and activates the reporter gene in yeast. The yeast one-hybrid experiments further proved that OscWRKY1 binds with the conserved W-box cis-elements present in the promoters of PAL and C4H, which suggests that they might be a direct target of the OscWRKY1 TF.
Plant WRKY TFs acts as both positive and negative regulators, thus exhibiting a crucial role in plant defense response (Pandey and Somssich 2009). In this work, we have studied the biological role of OscWRKY1 in regulating the phenylpropanoid pathway and plant defense response. The OscWRKY1 overexpression results in reduced bacterial population density and electrolyte leakage as compared to Col-0 WT Arabidopsis infected with the phytopathogenic bacteria, Pst DC3000. Microscopic visualization of Arabidopsis leaves infected with GF- tagged Pst DC3000 showed decreased colonization in OscWRKY1 overexpressing lines as compared to the Col-0 WT. Taken together, these results indicate that the OscWRKY1 lines are more resistant towards Pst DC3000. In a study, overexpression of jasmonic acid-inducible WRKY40 from chickpea triggers a defense response against Pst in Arabidopsis (Chakraborty et al. 2018). CaWRKY41, from Capsicum annuum coordinates the responses against pathogenic bacterium Ralstonia solanacearum and enhances plant immunity (Dang et al. 2019). Regulation of benzylisoquinoline alkaloids biosynthesis by StWRKY8 conferred enhanced resistance to late blight disease in potatoes (Yogendra et al. 2017). In a recent study, the transcriptomic changes in the leaves of rose on interaction with either biotrophic and a hemibiotrophic leaf pathogen was analayzed and it was revealed that a common set of differentially regulated genes comprised of WRKY genes involved in the activation of phenylpropanoid biosynthesis and other factors of the SA signaling pathway (Neu et al. 2019). Phenylpropanoid derivatives can protect plants from biotic infections caused by viruses, bacteria, or fungi. Phenylalanine ammonia lyase knockout mutants of Arabidopsis increased susceptibility to a virulent strain of Pseudomonas syringae (Huang et al. 2010). Caffeate O-methyltransferase mutants demonstrated reduced resistance to bacterial and fungal infections (Quentin et al. 2009). In certain circumstances, disrupting phenylpropanoid metabolism improved pathogen resistance (Gallego-Giraldo 2011). External application of a combination of three phenylpropanoids protected plants from P. humuli infection (Feiner et al. 2021). OsWRKY67 from rice directly binds with the promoter of pathogen-related genes PR1a and PR10 and provides bacteria blight resistance (Liu et al. 2018). In cotton, wound responsive GhWRKY40 plays an important role in enhancing susceptibility to bacterial infection in Ralstonia solanacearum in transgenic tobacco (Wang et al. 2014).
Due to the unavailability of functional mutant in O. sanctum, we utilized VIGS to deliver the more efficient TRV-based VIGS for O. sanctum. TRV‐based VIGS and its applications have been studied in some medicinal plants, but not in O. sanctum (Singh et al. 2015; Liscombe and O'Connor, 2011; Hileman et al. 2005). This is the first report of the VIGS-based gene silencing in O. sanctum utilizing PDS (phytoene desaturase) as the marker gene. We further confirmed the role of OscWRKY1 in regulating the PAL gene and modulating the PAL activity. Increased PAL activity has been observed during plant-pathogen interactions, however the molecular mechanism of PAL activation in response to the pathogen is not known (Li et al. 2015; Abbas et al. 2018; Song et al. 2015; Yu et al. 2016). Overexpression of CaPAL1 from Capsicum annum in Arabidopsis conferred increased pathogen resistance to Pseudomonas syringae with increased PAL activity (Kim and Hwang 2014). In rice, OsMYB30 directly regulates the expression of OsPALs in response to brown plant hopper infestation (He et al. 2020). The leaves of O. sanctum were vacuum infiltrated with Pst DC3000. Since, Pst DC3000 is a non-adapted host for this plant species; the plant leaves exhibited HR cell death during the early stage of infection suggesting non-host resistance against Pst DC3000 (Fig. S12) which could not appropriately further confirm the bacterial disease resistance mechanism in the O. sanctum. The rosmarinic acid is one of the most abundant phenolic acids present in basil leaves (Scagel et al. 2019). In contrast to other Lamiaceae species, sweet basil cell suspensions accumulated primarily the rosmarinic acid during the initial growth phase (Kintzios et al. 2003). Accumulation of rosmarinic acid was reduced in phenolic extracts of PAL suppressed lines (Song and Wang 2011). Significant reduction in the rosmarinic acid content was observed in VIGS-OscWRKY1 lines, indicating the role of OscWRKY1 in regulation of phenylpropanoid pathway.
We conclude that MeJA/SA and wound responsive TF, OscWRKY1 binds with the W-box cis-elements present within the promoter of phenylpropanoid pathway genes and regulates them in both O. sanctum and Arabidopsis. Overexpression of OscWRKY1 promotes resistance against Pst DC3000 in Arabidopsis. Further, OscWRKY1 silencing using VIGS significantly reduces the five transcripts of phenylpropanoid pathway genes leading to reduction in rosmarinic acid content suggesting its major role in the regulation of phenylpropanoid pathway.
Data availability
The transcriptome data of O. sanctum is available as SRA accession number SRP039008. The sequences of O. sanctum gene and promoter can be found using the following accession number in the NCBI data base. OscWRKY1-MN393294, OscPAL promoter- MT446024, OscC4H promoter- MT438691.
References
Abbas HMK, Xiang J, Ahmad Z, Wang L, Dong W (2018) Enhanced Nicotiana benthamiana immune responses caused by heterologous plant genes from Pinellia ternata. BMC Plant Biol 18(1):357
Agarwal PK, Jha B (2010) Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol Plant 54(2):201–212
Blee K, Choi JW, O’Connell AP, Jupe SC, Schuch W, Lewis NG (2001) Antisense and sense expression of cDNA coding for CYP73A15, a class II cinnamate 4-hydroxylase, leads to a delayed and reduced production of lignin in tobacco. Phytochemistry 57(7):1159–1166
Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122(1):107–116
Chakraborty J, Ghosh P, Sen S, Das S (2018) Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under Fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen. Plant Sci 6:250–267
Chaman E, Sylvia V, Copaja J, Victor H (2003) Relationships between salicylic acid content, phenylalanine ammonia-lyase (PAL) activity, and resistance of barley to aphid infestation. J Agr Food Chem 51(8):2227–2231
Cheong YH, Chang HS, Gupta R, Wang H, Zhu T (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129:661–677
Dang F, Lin J, Chen Y, Li GX, Guan D, Zheng SJ (2019) A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J Exp Bot 70(5):1581–1595
Deng C, Zhang F, Wang J, Li Y, Huang H, Dai S (2021) Tobacco rattle virus-induced phytoene desaturase (PDS) silencing in Centaurea cyanus. Hortic Plant J 7(2):159–166
Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51:21–37
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Feiner A, Pitra N, Matthews P, Pillen K, Wessjohann LA, Riewe D (2021) Downy mildew resistance is genetically mediated by prophylactic production of phenylpropanoids in hop. Plant Cell Environ 44(1):323–338
Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci 108:20814–20819
Gharibi S, Tabatabaei BES, Saeidi G, Talebi M, Matkowski A (2019) The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech. f. Phytochemistry 162:90–98
Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425
Han Y, Fan T, Zhu X, Wu X, Ouyang J, Jiang L (2019) WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol Biol 99(1–2):149–159
He J, Liu Y, Yuan D, Duan M, Liu Y, Shen Z (2020) An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc Natl Acad Sci USA 117(1):271–277
Hileman LC, Drea S, Martino G, Litt A, Irish VF (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44:334–434
Hu Z, Wang R, Zheng M, Liu X, Meng F, Wu H, Yao Y, Xin M, Peng H, Ni Z, Sun Q (2018) TaWRKY51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum L.). Plant J 96(2):372–388
Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153(4):1526–1538
Jaggi RK, Madaan R, Singh B (2003) Anticonvulsant potential of holy basil, Ocimum sanctum Linn, and its cultures. Indian J Exp Bot 41:1329–1333
Jiang W, Wu J, Zhang Y, Yin L, Lu J (2015) Isolation of a WRKY30 gene from Muscadinia rotundifolia (Michx) and validation of its function under biotic and abiotic stresses. Protoplasma 252:1361–1374
Jin Q, Yao Y, Cai Y, Lin Y (2013) Molecular cloning and sequence analysis of a phenylalanine ammonia-lyase gene from Dendrobium. PLoS ONE 8:e62352
Kim DS, Hwang BK (2014) An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot 65:2295–2306
Kintzios S, Makri O, Panagiotopoulos E, Scapeti M (2003) In vitro rosmarinic acid accumulation in sweet basil (Ocimum basilicum L.). Biotechnol Lett 25:405–408
Lescot M, Patrice D, Gert T, Kathleen M, Yves M, Yves VP, Pierre R, Stephane R (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Li JB, Luan YS, Liu Z (2015) Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol Plant 155(3):248–266
Liscombe DK, O’Connor SE (2011) A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry 72(16):1969–1977
Liu Y, Schiff M, Dinesh KSP (2002a) Virus induced gene silencing in tomato. Plant J 31(6):777–786
Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30:415–429
Liu Q, Li X, Yan S, Yang J, Dong J, Zhang S (2018) OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol 18(1):257
Loake G, Grant M (2007) Salicylic acid in plant defence-the players and protagonists. Curr Opin Plant Biol 10(5):466–472
Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23:1639–1653
Mishra S, Triptahi V, Singh S, Phukan UJ, Gupta MM, Shanker K, Shukla RK (2013) Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS ONE 8(1):52784
Misra RC, Sharma S, Garg A, Ghosh S (2020). Virus-induced gene silencing in sweet basil (Ocimum basilicum). In virus-induced gene silencing in plants humana. New York, NY. 123–138
Neu E, Domes HS, Menz I, Kaufmann H, Linde M, Debener T (2019) Interaction of roses with a biotrophic and a hemibiotrophic leaf pathogen leads to differences in defense transcriptome activation. Plant Mol Biol 99(4–5):299–316
Nugroho LH, Verberne MC, Verpoorte R (2002) Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants. Plant Physiol Biochem 40(9):755–760
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Payyavula RS, Navarre DA, Kuhl JC, Pantoja A, Pillai SS (2012) Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biol 12:39
Peng X, Wang H, Jang JC, Xiao T, He H, Jiang D, Tang X (2016) OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice 9(1):1–14
Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci 7:760
Phukan UJ, Jeena GS, Tripathi V, Shukla RK (2018) MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol J 16(1):221–233
Quentin M, Allasia V, Pegard A, Allais F, Ducrot PH, Favery B, Levis C, Martinet S, Masur C, Ponchet M, Roby D, Schlaich NL, Jouanin L, Keller H (2009) Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog 5(1):e1000264
Rahman S, Islam R, Kamruzzaman M, Alam K, Jamal AHM (2011) Ocimum sanctum L.: a review of phytochemical and pharmacological profile. Am J Drug Discov. Dev 2:159–162
Rastogi S, Meena S, Bhattacharya A, Ghosh S, Shukla RK, Sangwan NS, Lal RK, Gupta MM, Lavania UC, Gupta V, Nagegowda DA (2014) De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genom 15(1):1–18
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Sarfaraz D, Rahimmalek M, Saeidi G (2021) Polyphenolic and molecular variation in thymus species using HPLC and SRAP analyses. Sci Rep. 11(1):5019
Scagel CF, Lee J, Mitchell JN (2019) Salinity from NaCl changes the nutrient and polyphenolic composition of basil leaves. Ind Crops Prod 127:119–128
Schluttenhofer C, Yuan YL (2014) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol 114:251–769
Senthil-Kumar M, Mysore K (2014) Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc 9:1549–1562
Shine MB, Yang JW, El-Habbak M, Nagyabhyru P, Fu DQ, Navarre D (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol 212(3):627–636
Singh AK, Dwivedi V, Rai A, Pal S, Reddy SG, Rao DK, Shasany AK, Nagegowda DA (2015) Virus-induced gene silencing of Withania somnifera squalene synthase negatively regulates sterol and defence-related genes resulting in reduced withanolides and biotic stress tolerance. Plant Biol J 13:1287–1299
Singh AK, Kumar SR, Dwivedi V, Rai A, Pal S, Shasany AK, Nagegowda DA (2017) A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol 215(3):1115–1131
Song J, Wang Z (2011) RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosysthesis. J Plant Res 124:183–192
Song Y, Chen D, Lu K, Sun Z, Zeng R (2015) Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front Plant Sci 6:786
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plants Sci 17(5):260–270
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3(1):2–20
Wang C, Fu D (2018) Virus-induced gene silencing of the eggplant chalcone synthase gene during fruit ripening modifies epidermal cells and gravitropism. J Agric Food Chem 66(11):2623–2629
Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X (2009) A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230(6):1155–1166
Wang X, Yan Y, Li Y, Chu X, Wu C, Guo X (2014) GhWRKY40, a multiple stress-responsive cotton WRKY gene, plays an important role in the wounding response and enhances susceptibility to Ralstonia solanacearum infection in transgenic Nicotiana benthamiana. PloS ONE 9(4):e93577
Yogendra KN, Kumar A, Sarkar K, Li Y, Pushpa D, Mosa KA (2015) Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J Exp Bot 66:7377–7389
Yogendra KN, Dhokane D, Kushalapp AC, Sarmiento F, Rodriguez E, Mosquera T (2017) StWRKY8 transcription factor regulates benzylisoquinoline alkaloid pathway in potato conferring resistance to late blight. Plant Sci 256:208–216
Yu XY, Bi Y, Yan L, Liu X, Wang Y, Shen KP, Li YC, Yu XY, Bi Y, Yan L, Liu X, Wang Y, Shen KP, Li YC (2016) Activation of phenylpropanoid pathway and PR of potato tuber against Fusarium sulphureum by fungal elicitor from Trichothecium roseum. World J Microb Biot 32(9):142
Zhang J, Yu D, Zhang Y, Liu K, Xu K, Zhang F, Wang J, Tan G, Nie X, Ji Q, Zhao L, Li C (2017) Vacuum and co-cultivation agroinfiltration of (Germinated) seeds results in tobacco rattle virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize. Front Plant Sci 8:393
Acknowledgements
The authors acknowledge CSIR-Central Institute of Medicinal and Aromatic Plants (CIMAP) National gene bank for supplying O. sanctum seeds. Authors acknowledge Yogesh Kumar and Dr. Feroz Khan for helping in retrieving the genomic sequences of promoters of O. sanctum. Authors acknowledge Payal Srivastava for her help during vacuum infiltration.
Funding
AJ, GSJ and S acknowledge UGC and CSIR for fellowship. RKS acknowledges CSIR-CHEMBIO and CSIR-Aroma Mission-I for funding. S and RKS acknowledge Academy of Scientific and Innovative Research (AcSIR). AP is grateful to DST-INSPIRE Faculty Award (IFA13-LSPA-20).
Author information
Authors and Affiliations
Contributions
AJ performed the cloning experiments (Arabidopsis promoters, pTRV2, pGADT7 and pHIS2.0), semi-qPCR experiment, Y1H (O. sanctum promoter), PAL assay (O. sanctum and Arabidopsis), VIGS, in planta pathogen colonization experiments, quantification of ion leakage, HPLC determination of rosmarinic acid, wrote the manuscript and prepared the figure for the respective part. GSJ performed the cloning experiments (Ocimum promoter, pGEX4T2, pGBKT7, pBI121), qRT-PCR analysis, protein purification, GRA, transactivation assay, Y1H (Arabidopsis promoter), PAL assay (O. sanctum), maintained transgenic lines, wrote the manuscript, and prepared the figures. S and R performed expression analysis of WRKY transcripts from the transcriptome. AP supervised the pathogenicity assay, bacterial colonization experiments, electrolyte assay and confocal microscopy study. AJ, GJ, AP and RKS analyzed the result. RKS conceived the theme supervised the work, and edited the manuscript. All authors finally read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11103_2022_1297_MOESM1_ESM.pptx
Supplementary file1 (PPTX 2239 kb)— Fig. S1. Multiple sequence alignment of OscWRKY1. Almost all identified WRKY proteins from different plant species contained a highly conserved WRKYGQK motif in their protein sequences. Fig. S2. Nucleotide and amino acid alignment of OscWRKY1. OscWRKY1 contains 1023 bp of ORF, encoding a protein with 340 amino acids as shown. The amino acid sequence is shown in a single alphabet below each triplet codon. The underlined sequence represents the 58 amino acids long DBD containing the highly conserved WRKYGQK motif shown in the box. Fig. S3. Differential gene expression analysis from the transcriptome data of O. sanctum. From the transcriptome differential gene expression data, unique full-length transcripts present in O. sanctum OscWRKY1, OscWRKY4, OscWRKY2, and OscWRKY15 were found to be highly expressed in leaf and root tissue. Fig. S4. EMSA of OscWRKY1. Probes containing mutated probes (TTGACG and TTCACT) were designed to study the DNA-protein interaction. Fig. S5. Confirmation of OscWRKY1 transgenic lines in Arabidopsis. Genomic DNA PCR of transgenic lines was confirmed by using NptII (KanR) primers, along with an untransformed WT control plant. The expected size of the PCR amplification is 786 bp. Fig. S6. Expression analysis of OscWRKY1 in Arabidopsis transgenic lines. The overexpression of OscWRKY1 in transgenic plants was studied by performing semi-quantitative PCR using gene-specific primers. Fig. S7. Differential expression study of the phenylpropanoid pathway and PR genes after MeJA treatment and wounding. Relative expression of phenylpropanoid pathway genes (a) and PR genes (b) after 1, 3, and 5 hours of MeJA treatment and wounding in both leaf and root tissues was monitored and analyzed. All the samples were carried out in three independent biological (n=3) and experimental replicates. Actin was used as an endogenous control to normalize gene expression. The color scale representing normalized fold induction, as shown in the Figure. Fig. S8. Sequence study of PAL and C4H promoters. PAL and C4H promoter sequences showing the conserved W-box cis-element in both O. sanctum and Arabidopsis. W-box cis-elements are highlighted in purple color, and the primer sequences for cloning are highlighted in yellow. The sequence shown with blue color is used for cloning. Fig. S9. Determination of efficiency of vacuum mediated agro-infiltration. Detection of TRV2 coat protein (CP) and TRV1 RNA-dependent RNA polymerase (RdRp) in mock-infiltrated (10 mM MES pH 5.6, 10 mM MgCl2, 200 μM acetosyringone), empty vector control (pTRV1-pTRV2) and pTRV2‐OscPDS/OscWRKY1 -infected plants of O. sanctum by RT-PCR at 24 days post-infiltration. Fig. S10. Calibration plot of rosmarinic acid. A linear dependence of the peak area on Rosmarinic acid concentration was found with a correlation coefficient of r = 0.9934. Fig. S11. Melting curve analysis. After each PCR loop, a melting curve analysis was performed by progressively rising the fluorescence temperature from 60 to 95°C. The qRT-PCR experiments used the 7500 FAST Real-Time PCR system (Applied Biosystems, USA). Actin and ubiquitin were used to normalize gene expression as endogenous controls. The following figure represents the melting curve for ubiquitin in mock infiltrated samples. Fig. S12. Vacuum mediated infiltration of O. sanctum with the bacterial pathogen Pst DC3000. Representative images of O. sanctum leaves infected with the bacterial pathogen Pst DC3000. The leaves infiltrated with 10 mM MgCl2 were used as mock control. The red circle denotes the necrosis associated with the HR response in the leaves of vacuum-infiltrated seedlings treated with Pst DC3000. Photographs were taken at 3 DPI. The experiment was performed in two independent biological replicates in triplicates with similar results.
Rights and permissions
About this article
Cite this article
Joshi, A., Jeena, G.S., Shikha et al. Ocimum sanctum, OscWRKY1, regulates phenylpropanoid pathway genes and promotes resistance to pathogen infection in Arabidopsis. Plant Mol Biol 110, 235–251 (2022). https://doi.org/10.1007/s11103-022-01297-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-022-01297-2