Abstract
Key message
The transcriptomes of wild and cultivated grapes consists of similar expressed genes but distinct wiring of co-expressed modules associated with environmental conditions.
Abstract
Grapevine is an important fruit crop worldwide, with high economic value and widespread distribution. Commercial production is based on Vitis vinifera, and, to a lesser extent, on hybrids with American grapes, such as V. labrusca. Wild grape relatives are important sources of resistance against biotic and abiotic factors; however, their global gene expression patterns remain poorly characterized. We associated genome-wide transcript profiling to phenotypic analyses to investigate the responses of cultivated and wild vines to vineyard conditions. The expressed genes in the Vitis reference transcriptome are largely shared by wild grapes, V. labrusca hybrids and vinifera cultivars. In contrast, significant differential regulation between wild and vinifera genotypes represents 80% of gene expression variation, regardless of the environment. In wild grapes, genes associated to regulatory processes are downregulated, whereas those involved in metabolic pathways are upregulated, in comparison to vinifera. Photosynthesis-related ontologies are overrepresented in the induced genes, in agreement with higher contents of chlorophyll in wild grapes. Co-regulated gene network analyses provide evidence of more complex transcriptome organization in vinifera. In wild grapes, genes involved in signaling pathways of stress-related hormones are overrepresented in modules associated with the environment. Consensus network analyses revealed high preservation within co-regulated gene modules between cultivated and wild grapes, but divergent relationships among the expression clusters. In conclusion, the distinct phenotypes of wild and cultivated grapes are underlain by differences in gene expression, but also by distinct higher-order organization of the transcriptome and contrasting association of co-expressed gene clusters with the environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grapevine is one of the most important fruit crops worldwide due to its economic value and widespread distribution. Most cultivated grapes belong to the Eurasian species Vitis vinifera L., originated from its wild form V. vinifera subsp sylvestris, and considered the ancestor of modern cultivars (Marrano et al. 2018; Liang et al. 2019). Environmental factors, including temperature, light, and water availability, influence growth and the principal developmental stages of the grapevine life cycle; namely bud burst, flowering, and fruit ripening (véraison) (Bigard et al. 2018). Genotypes of cultivated grapes retain a degree of phenotypic plasticity in phenology control (Dal Santo et al. 2013; Mallet 2015), which allows the successful establishment of vineyards in distinct climatic regions in temperate, subtropical, and tropical regions (Maia et al. 2015; Mallet 2015).
Wild Vitaceae species are considered valuable sources of resistance genes against biotic and abiotic stresses (Carvalho and Amâncio 2019; Daldoul et al. 2020) and have been used in interspecific crosses with V. vinifera in scion and rootstock breeding programs (Maia et al. 2015). However, a broader characterization of the mechanisms underlying the resistance responses of wild grapes has only recently began to emerge (Marrano et al. 2018; Wang et al. 2019; Daldoul et al. 2020). Genetic diversity and differential regulation of gene expression are largely responsible for the phenotypic plasticity and resistance mechanisms in grapes (Dal Santo et al. 2013; Cramer et al. 2014; Fennell et al. 2015; Ghan et al. 2017; Zhou et al. 2019), although higher order regulatory processes, such as epigenetic modifications and genome-environment interactions, also have been demonstrated to modulate grapevine responses to the environment (Dal Bosco et al. 2018; Magris et al. 2019).
Connectivity relationships, in the form of networks, help to assess the organization of complex biological systems, often described as ‘functionally modular’ (Hartwell et al. 1999). In plants, comparative analyses of large-scale transcriptome sequence data allowed the identification of consistent networks of co-expressed genes controlling growth and development in several species (Jones and Vandepoele 2020), including grapevine (Wong 2020). The intra- and inter-relations among gene networks are influenced by specific biological and environmental factors to control a wide range of physiological and developmental responses, brought about by coordinated differential gene expression (Jones and Vandepoele 2020; Wong 2020). Complex traits, such as the tolerance to water stress, were demonstrated to occur via gene network rewiring (VanBuren et al. 2018). Gene network rewiring is an effective mechanism to transform a limited number of genes in a wide range of responses to the ever changing endogenous developmental and exogenous environmental conditions.
In the current study, we combined phenological analyses and large-scale transcriptomic data to investigate cultivated and wild grapevine responses to environmental vineyard conditions. We performed genome-wide expression analyses in ten cultivated grapevine (Vitis vinifera L.) genotypes, three V. labrusca hybrids, and four wild vine species; V. flexuosa Thunb. (creeping grape), V. gigas Fennel (Florida blue grape), Parthenocissus heterophylla (Maxim.) Trautv. (Amur peppervine) and Vitis tiliifolia Humb. & Bonpl. ex Schult (Caribbean grape), growing in experimental vineyards in tropical and temperate conditions. The association between local environmental conditions with gene expression patterns was investigated to gain further insight into the mechanisms controlling growth and development of the cultivated and wild grapes. We also performed consensus analyses to study the mechanisms shared by V. vinifera and wild species in response to local abiotic conditions.
Materials and methods
Phenological characterization and climate data
Phenological stages were considered as described (Parker et al. 2013) and were evaluated for at least 10 years, under standard vineyard agricultural practices. The plants were trained in double arched guyot and submitted to late winter trimming. Data used for correlation analyses represent the average for the evaluations and is shown in Table S1. The phenology data is available at https://www.embrapa.br/uva-e-vinho/banco-ativo-de-germoplasma-de-uva/pesquisa.
Climate information was obtained from local meteorological stations and include vineyard altitude (m), temperature (°C), average rainfall (mm/month), solar irradiation (W m−2 h−1), photosynthetically active radiation (PAR, W m−2) and ultraviolet (UV) index.
Plant material
Mature leaves and stems from ten cultivated grapevine (Vitis vinifera L.) genotypes, three V. labrusca hybrids and four wild vine species; V. flexuosa Thunb. (creeping grape), V. gigas Fennel (Florida blue grape), Parthenocissus heterophylla (Maxim.) Trautv. (Amur peppervine) and Vitis tiliifolia Humb. & Bonpl. ex Schult (Caribbean grape), at developmental stage E-L 41 (Coombe 1995), were collected between May and August, from 2:00 to 4:00 ZT. Vitis gigas J.L. Fennel is an unresolved taxon, considered to be synonymous to Vitis aestivalis Michaux (FNA Editorial Committee 2016) and was excluded from further analyses. ‘Syrah’ and ‘Tempranillo’ were grafted onto ‘IAC 766 Campinas’ (Riparia do Traviù × V. caribeae) and the remaining V. vinifera cultivars and labrusca hybrids onto ‘Paulsen P1103’ (V. berlandieri × V. rupestris). Wild species were conducted as self-rooted plants.
Plant location and environmental conditions are shown in Fig. S1 and Table S2. Pedigree for vinifera cultivars and labrusca hybrids was obtained from the Vitis International Variety Catalogue (VIVC) (Maul et al. 2019), and is schematically represented in Fig. S2.
Expression validation by RT-qPCR and chlorophyll content analyses employed unrelated wild and cultivated vines, V. riparia and V. vinifera cv. Weisser Riesling, respectively. Aerial parts were harvested from 4-week old plants, grown in vitro, in 25 × 150 mm test tubes, containing 12 mL of Woody Plant medium (Lloyd and McCown 1980), at 23 °C and 37 °C under 16-h photoperiod provided by white LED sources at 75 µmol m−2 s−1.
RNA extraction, library preparation and sequencing
Approximately 30 g of plant material from three to five genetically identical biological replicates was ground to fine powder in liquid nitrogen and used for total RNA extraction employing 90 mL of STE 2 X buffer (0.2 M NaCl, 0.1 M Tris, 0.002 M EDTA, pH 7.5), 35 mL SDS (10% w/v) supplemented with bentonite (45 mg/mL) and 1.6% (v/v) β-mercaptoethanol. The mixture was vigorously shaken for 5 min at room temperature and added 0.3 X the volume of buffer-saturated phenol, pH 7.5 and 0.3 X the volume of chloroform:isoamyl alcohol (24:1 v/v), followed by shaking for 45 min and centrifugation at 16,000g for 10 min at 4 °C. The aqueous phase containing cellular nucleic acids was submitted to Whatman CF- 11 cellulose and eluted with STE buffer (0.1 M NaCl, 0.05 M Tris, 0.001 M Na. EDTA, pH 7.0) (Morris and Dodds 1979).
The quality of isolated RNA was confirmed by Agilent 2100 Bioanalyzer RNA Nano assay (Agilent Technologies, USA) (RNA Integrity Number RIN > 7.0) and two micrograms were submitted to library preparation using the TruSeq mRNA Sample Prep kit v.2 (Illumina, San Diego, CA, USA) according the manufacturers’ instructions. Libraries were quantified employing the Qubit 2.0 Fluorometer (Invitrogen, USA), quality checked by Agilent 2100 Bioanalyzer High Sensitivity or DNA 1000 assay (Agilent Technologies) and submitted to Illumina pair-end sequencing using HiSeq2000 at 4 Gb depth. Data is deposited at Short Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) under accession numbers SAMN14331022 to SAMN14331038 (BioProject PRJNA611044).
Chlorophyll quantification
Chlorophyll contents were determined as described by Hiscox and Israelstam (1979). Total chlorophyll, chlorophyll a and b contents were calculated according to Wellburn (1994).
Phenotypic and biochemical data analyses
Statistical analyses were performed in R v.4.0.0 (R Core Team 2019). Data are presented as mean ± standard error, and treatment comparisons were performed by using F statistics and Waller-Duncan K-ratio t test at P < 0.05 and 0.01. For clustering analyses, missing values were imputed by multivariate chained equations, and the dataset was scaled. Clusters were determined by hierarchical agglomeration employing Pearson’s distance as similarity measure and maximum linkage. multiple factor analysis (MFA) was performed using the packages FactoMineR and factoextra (Lê et al. 2008; Kassambara and Mundt 2019). In MF analyses, phenotype data was grouped in phenology (bud bursting, flowering, and berry ripening) and disease response (fungus and virus). Genomic background was analyzed as categorical variable, with three levels (vinifera, labrusca hybrid, and wild species), consisting of 10, three and four replicates, respectively.
Transcriptome data analyses
Raw reads were initially processed using shortRead (Morgan et al. 2009) to remove adapter sequences, low quality bases, primers, short reads, and other contaminants. Trimmed quality reads were mapped to grapevine transcriptome and quantified at the transcript-level using Salmon v 1.1.0 (Patro et al. 2017), against a transcriptome index constructed based on V. vinifera 12X v.2 assembly and its structural annotation (VCost.v3), available at https://urgi.versailles.inra.fr/Species/Vitis/Annotations (Canaguier et al. 2017). Transcript quantification data was aggregated to the gene level with the package tximport (Soneson et al. 2015) and used for differential expression analyses employing DESeq2 (Love et al. 2014). Differential gene expression analyses were conducted by fitting two-factorial generalized linear models of the negative binomial family (NB) to the read counts Kij for gene i in sample j with a logarithmic link (Love et al. 2014). The normalization constant sj and the dispersion parameter αi were calculated as described (Love et al. 2014), where qij is proportional to the expectation value of the true concentration of fragments from gene i in sample j, xjr denotes the elements of the design matrix X, and βir denote the coefficients for gene i and parameters corresponding to columns of the design matrix r. Genes with significant differences (fold change > 2.0 and false discovery rate [FDR] = 0.05) in transcript abundance between three replicates of ‘vinifera’ (cultivars ‘Syrah’, ‘Tempranillo’ and ‘Trajadura’) and ‘wild’ species (Pathernocissus heterophylla, V. flexuosa and V. tiliifolia), were considered as differentially expressed (DEGs).
Gene networks and consensus eigengene networks between wild species and vinifera cultivars were constructed using the package WGCNA v. 1.69 (Langfelder and Horvath 2008) in R, using normalized transcription data. Adjacency matrices were used to summarize co-expression modules using a soft-threshold power selected based on the lowest mean connectivity and the highest model fit to scale-free topology. Adjacency matrices were transformed to topological overlap matrices (TOM) to determine co-regulated genes, using a minimal module size of 20 genes. Effects of noise and spurious association were reduced by transforming TOM to the corresponding dissimilarity. Modules with co-expression similarity higher than 75% were merged. Eigengene features gene significance (GS) and module membership (MM) were used in correlation analyses to environmental data.
Gene ontology was investigated using GO Enrichment analysis and redundant terms in the lists were reduced using REVIGO (Supek et al. 2011). Promoter enrichment analyses were carried out using ShinyGO v0.61 (Ge and Jung 2020) and protein–protein interactions were determined using STRING databases (Szklarczyk et al. 2019). Metabolic pathways were represented according to KEGG mappings using pathview (Luo and Brouwer 2013) in R.
Quantitative real time polymerase chain reaction (RT-qPCR)
RNA seq expression profile was confirmed by RT-qPCR for fourteen randomly selected genes (Fig. S3, Table S3). Primers were designed for coding sequences (CDs) of the candidate reference genes of Vitis vinifera genome 12X v.2 assembly and its structural annotation (VCost.v3), available at https://urgi.versailles.inra.fr/Species/Vitis/Annotations (Canaguier et al. 2017), using the default parameters of the software Primer3Plus (Untergasser et al. 2007) and amplicons ranging from 151 to 240 base pairs in size (Table S3). Expression was validated for wild and vinifera genotypes unrelated to those submitted to RNASeq, namely, Vitis riparia (wild) and ‘Weisser Riesling’ (V. vinifera).
Independent RNA extractions were carried out in triplicate for cultivated and wild material, using a modified CTAB protocol (Gambino et al. 2008). RNA quality and quantity were analyzed by spectrophotometric absorbance ratio at A260/A280 and A260/A230. The integrity of the total RNAs was confirmed in 1% (w/v) agarose gel electrophoresis, stained with SYBRSafe (Invitrogen, CA, USA). Genomic DNA was eliminated by RNase free DNAse I (ThermoFischer Scientific) treatment, as recommended by the manufacturer, and confirmed by qPCR. Synthesis of cDNA was carried out from 1 μg of total RNA using Oligo d(T) primers and SuperScript® III First-Strand Synthesis SuperMix (ThermoFischer Scientific), according to the manufacturer recommendations.
Amplifications were performed at a StepOne Real Time PCR System (Applied Biosystems) and the SYBR Green PCR Master Mix (Applied Biosystems). The reactions were performed in a total volume of 15 μL, consisting of 7.5 μL of the mix, 30 ng of cDNA and 300 nM each, forward and reverse, primer. Amplifications started with a denaturation step at 95 °C for 10 min, followed by 40 cycles consisting of 15 s at 95 °C and 1 min at 60 °C, finalized by the dissociation curve with denaturation at 95 °C for 15 s, cooling at 60 °C for 1 min and gradual heating, at 0.3 °C steps, up to 95 °C. Control reactions in the absence of template were used to confirm the absence of DNA contamination. The amplification curve for an equimolar pool of cDNAs from all tested samples at five distinct concentrations was used to validate the primers by dissociation curves and peak analyses. Relative expression levels were determined employing four reference genes (Table S3) using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Results
In the current work, we compared the transcriptome of commercially cultivated and wild grapes, grown in standard vineyards in tropical and sub-tropical conditions. The plants consist in phenotypically characterized germplasm accessions of vinifera cultivars, labrusca hybrids and wild grape species. We studied quantitative differences in gene expression and transcriptome organization among the genotypes. The consensus gene expression profile between vinifera and wild genotypes was determined, functionally classified, and related to environmental conditions.
Phenotypic and transcriptional plasticity of grapevines
Thirteen cultivated grapevine cultivars and four wild grape genotypes were characterized phenotypically for their phenological behavior and disease incidence responses, for at least 10 years, in experimental vineyards.
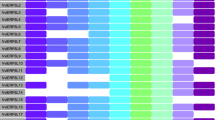
The ten V. vinifera cultivars exhibited similar phenological and disease incidence responses during long-term evaluations (Fig. 1, Fig. S4, Table S1). Among the considered phenological events in vinifera genotypes, flowering and bud burst timing were more widely distributed in time, ranging from 58 to 41, and from 89 to 76 days (Δ = 17 and 13), respectively, whereas, berry ripening ranged from 46 to 35 (Δ = 11) days in the investigated cultivars (Fig. 1, Table S1). V. vinifera cultivars exhibited visible viral infection symptoms, and most of them (6/10) were also highly susceptible to fungal pathogens, whereas, the incidence of fungal diseases and presence of viral infection symptoms were lower in V. labrusca hybrids (Fig. 1). The timing of the investigated phenological events was less disperse among the investigated V. labrusca cultivars, even though a late cultivar (‘Tardia de Caxias’) was included in the study. The timing of bud burst of ‘Tardia de Caxias’ was on average 10 days later in comparison to ‘Isabella’ genotypes in temperate and tropical conditions, and 10 days earlier, for berry ripening, (Fig. 1, Table S1).
Phenotypic analyses of the investigated V. vinifera, V. labrusca hybrids and wild grape genotypes. Phenological, disease response, and berry color data (a). Phenology and disease symptom incidence are represented as heatmaps. Values are presented in Table S1. Grey squares represent non-available data. Berry color is represented schematically by blue (noir), green (blanc), red (rouge) and pink (rosé). Hierarchical agglomerative clustering of the investigated genotypes based on their phenotypes (b). Quantitative variable multiple factor analyses (MFA) of the phenotypic data grouped in phenology and disease response (c)
The investigated wild grapes, including three Vitaceae species: V. flexuosa, V. gigas and V. tiliifolia, and P. heterophylla, exhibited no symptoms of viral infection and low incidence of fungal diseases (Fig. 1, Table S1). The timing of bud burst ranged from 74 (V. gigas and V. tiliifolia) to 95 days (P. heterophylla), whereas flowering was more condensed in time, ranging from 60 (V. tiliifolia) to 49 (V. flexuosa) days (Fig. 1, Table S1). Except for V. gigas, berry ripening data was not determined for the wild grapes due to inconsistencies in the classical ‘ripening’ features of berries from wild grapes, such as firmness, color, and sugar contents. Phenotypically, the investigated genotypes were clustered in seven groups, consisting of P. heterophylla, the wild species, ‘Isabella’ labrusca hybrids, and three clusters of vinifera cultivars, also including the hybrid ‘Tardia de Caxias’ (Fig. 1, Fig. S4).
The transcriptional profile of Vitis cultivars and wild grapes was determined for plants grown in germplasm vineyards located in distinct environmental conditions (Fig. 2a, Table S2). Sequencing depth was variable among the sequences, due to distinct technical and experimental conditions, such as library preparation and sequencing efficiency, rRNA carry-over, and biological contamination (Table 1). Contaminating sequences were removed by bioinformatic approaches. The read depth after cleaning was superior to the recommended for gene expression profiling experiments (5–25 million reads per sample) in all samples (Liu et al. 2014). Three major clusters of transcriptional profiles and one outlier (‘Pirovano 65’ syn. ‘Italia’) were detected for V. vinifera. For V. labrusca, ‘Isabella’ accessions cultivated in distinct locations exhibited similar whole transcriptome, whereas the global gene expression profile of ‘Tardia de Caxias’ was more divergent (Fig. 2b). In wild grape species, the transcriptome of V. tiliifolia was the most distinct, whereas the expression profiles of V. flexuosa, V. gigas and Parthenocissus heterophylla formed a single cluster (Fig. 2c).
Hierarchical clustering of whole transcriptome in V. vinifera (a), V. labrusca hybrids (b) and wild grape (c) genotypes. Environmental conditions of the vineyards are schematically represented as heatmap. Temperature (°C) and rainfall (mm) are show as maximum, average, and minimum yearly averages. Photoperiod is represented as maximum and minimum daylength (h), and subjective dawn (ZT0) and dusk (ZT12). Solar radiation (Irr) and photosynthetically active radiation (PAR) are represented as yearly maximum and minimum. Local ultra-violet radiation indices are shown. Heatmap is scaled independently for each environmental condition (row), according to the highest and lowest observed values. Values are shown in Table S2
The percentage of aggregated transcripts mapped to genes in V. vinifera reference genome ranged from 48.05% for V. labrusca to 59.73% to V. vinifera genotypes (Fig. 3a, Table 1). Wild grapes shared 49.18% of the annotated genes with the V. vinifera reference transcriptome (Fig. 3a, Table 1). A list of shared and genotype group-specific transcripts, along with their functional annotation and gene ontology classification, are presented in Table S4. The expressed genes of the investigated wild grapes exhibited 82.34% qualitative similarity to cultivated vinifera genotypes, although, with distinct regulation patterns (Fig. 3).
Transcriptome analyses of V. vinifera, V. labrusca and wild vine genotypes. Number of genes with mapped transcripts in comparison to the reference V. vinifera 12 X transcriptome (a). Venn diagram transcriptome comparisons (b). Total of mapped genes (c). Number of analyzed samples and detected co-expressed gene modules (d). Expression profile clustering and module detection in V. vinifera (e), V. labrusca hybrids (f) and wild grape (g) genotypes. Modules exhibiting more than 75% similarity were merged and are represented as Merged dynamic
The complexity of the transcriptome organization was not determined by the number of reads nor samples, since a difference of 17.67% in mapped reads resulted in a 40% difference in co-regulated gene modules between V. vinifera and wild grapes (Fig. 3b, c–g). In V. labrusca, the number of detected gene products corresponded to approximately half of the reference genome, organized in three co-regulated modules (Fig. 3b, d). The number of expressed genes detected in V. labrusca was 2.39% smaller than in wild grapes, whereas the number of identified co-regulated modules was approximately 2.3-fold higher (Fig. 3).
Differential gene expression between wild grapes and V. vinifera
To further investigate the differences between the transcriptome of cultivated and wild grapes, we performed differential gene expression analyses between subsets of cultivated and wild genotypes. The classification in vinifera and non-vinifera materials (labrusca plus wild) explained 60% of the variation in gene expression (Fig. S5A, B). The comparison between the transcriptomes of vinifera cultivars and labrusca hybrids revealed similar differences, although the global transcriptome of ‘Tardia de Caxias’ was like that of vinifera cultivars, specially ‘Cabernet Franc’ (Fig. S5C, D).
Most of the variance (80%) observed in differential expression analyses was explained by classification of the genotype in wild (Pathernocissus heterophylla, V. flexuosa and V. tiliifolia) versus vinifera (cultivars ‘Syrah’, ‘Tempranillo’ and ‘Trajadura’), regardless of the vineyard location (Fig. 4a, Fig. S5). The transcriptional profile of vinifera genotypes was similar, whereas wild grapes exhibited more divergent profiles among each other and, in comparison with vinifera (Fig. 4a). Approximately 9% (8.901%, 166 genes) of the transcripts were differentially expressed between wild grapes and vinifera cultivars (Fig. 4b–d). The overall transcriptome of V. tiliifolia was the most divergent among the investigated genotypes, and P. heterophylla was the wild grape the closest transcriptional profile to vinifera (Fig. 4a, Fig. S5A). Among vinifera cultivars, transcriptional regulation was more similar between ‘Tempranillo’ and ‘Syrah’, from tropical vineyards, in comparison to ‘Trajadura’ (Fig. S5A). Differential upregulation between wild species and vinifera cultivars ranged from 22.8- to 2.9-fold change, whereas downregulation was less divergent, going from − 8.7 to − 2.3 (Table S5). The genes with expression induced in wild vines in comparison to vinifera were enriched in photosynthesis light reactions, electron transport and chromoprotein assembly (Table S5). Among the most differentially regulated genes between wild grapes and vinifera cultivars were VIT_18s0001g05470, coding for a terpene synthase, that was more than 15-fold upregulated in all wild vines and VIT_05s0049g00770, which encodes a proline-rich extensin-like protein EPR1, approximately eightfold repressed in wild grapes (Fig. 4d, Table S5). Other genes significantly induced in wild vines, such as VIT_15s0046g02730, VIT_01s0011g03430, VIT_09s0018g00240, VIT_19s0093g00320, VIT_16s0039g01070 and VIT_01s0011g00960, are functionally associated with defense to biotic factors (Table S5). No significant ontology enrichment was detected for the downregulated DEGs between wild vines and vinifera cultivars, although a large portion (68.78%, 130/189) is involved in biosynthetic processes and their regulation (Table S5).
Differential gene expression between wild grapes and V. vinifera cultivars. Principal component analysis (PCA) of the whole transcriptome (a). Summary of the DEG between vinifera and wild genotypes (b). Gene ontology (GO) (c). Differential expression summary for the top 20 most significant DEGs between vinifera and wild grapes (d). Gene expression log-fold change is represented as heatmap and genotypes and vineyard locations are color-coded. Network representation of photosynthesis-enriched GO in the DEGs (e) and chlorophyll contents in cultivated and wild grapes (f). Letters represent statistically significant differences (P > 0.01) within each pigment class
Clustering analyses demonstrated two distinct groups of global transcription regulation between wild vines and vinifera cultivars (Fig. 4c, Fig. S5A). Gene ontology enrichment analyses revealed that genes associated to regulatory processes were repressed in wild grapes in comparison to vinifera, whereas those associated to metabolism were upregulated (Fig. 4b, c). The upregulated genes in wild grapes were significantly enriched in photosynthesis (Fig. 4e) and protein biosynthesis (Fig. S5B) ontologies. Accordingly, wild grapes exhibited higher chlorophyll contents under distinct temperature conditions (Fig. 4f). The promoters of the genes repressed in wild grapes are enriched in MADS box, Cold Shock Domain (CSD) and Homeodomain binding motifs (Fig. S5C), whereas those of the induced genes are significantly enrichment in APETALA2 (AP2) and basic Helix-Loo-Helix (bHLH) transcription factor binding sites (Fig. S5D).
Distinct functional classification of co-regulated gene modules wild grapes and V. vinifera
The transcriptome organization was also distinct between V. vinifera and wild grapes (Fig. 3e, g). Transcripts from V. vinifera were grouped in 18 co-regulated modules, whereas in wild grapes a simpler organization, consisting of seven modules, was detected (Fig. 5a, b). The representative (summary) eigengene for each module in vinifera and wild samples was used to investigate the association between gene expression and local environmental conditions using correlation analyses (Langfelder and Horvath 2012). In V. vinifera, three modules (magenta, tan, and turquoise) were significantly correlated to external cues (Fig. 5a). The significance of the modules was confirmed by intramodular connectivity, that is the strength of the associations between all genes (nodes) in the same module, was higher than 0.4 and is shown in Fig. 5c and d for the largest modules (turquoise) in vinifera and wild grapes, respectively. The smaller modules are shown in Fig. S6 for vinifera (A, B) and wild grapes (C, D). Functionally, module turquoise was enriched in genes associated to metabolism and regulatory processes and response to abiotic stimuli (Fig. 5e), whereas module tan had high frequency of genes located intracellularly (Fig. S6A). V. vinifera module tan exhibits significant enrichment in transcripts encoding proteins associated with hormone signaling pathways, including auxin, brassinosteroids, abscisic acid, and ethylene (Fig. 6a). No significant enrichment was detected for module magenta, although interaction is predicted for most genes (Fig. S6B). In wild grapes, modules turquoise, pink, red and black were significantly correlated to local environmental conditions, ranging from 0.95 (turquoise) to 0.37 (red) (Fig. 5d, Fig. S6C-D). Intramodular connectivity in module black was low (< 30%), indicating loose co-regulation among the genes, thus, it was excluded from further analyses. Modules turquoise and red were enriched in genes involved in metabolic processes (Fig. 5d, f and Fig. S6D). Gene ontology classification of the genes in module red in wild grapes was significantly enriched in hormone signaling pathways, as seen for module tan in vinifera, although encompassing a higher number of hormone classes (Fig. 6b). No significant functional enrichment was detected for genes in module pink, although most genes were predicted to interact by experimental evidence or in silico analyses (Fig. S6C).
Correlation between module eigengenes and environmental conditions in V. vinifera (a) and wild grapes (b). The rows correspond to the modules eigengenes, and the columns to the environmental conditions. Correlation values are represented as heatmap. Intramodular and gene ontology (GO) enrichment analyses for the largest cluster of co-regulated genes showing significant association with the environment in vinifera (c, e) and wild (d, f) grape genotypes
Functional enrichment analysis in hormone signaling pathways in co-regulated gene modules exhibiting significant association with environmental conditions in vinifera genotypes (module tan) (a) and wild grapes (module red) (b). Hormone signaling pathways are represented according to KEGG Pathways 3. Environmental Information Processing (map 04075—Plant hormone signal transduction). Enrichment in genes corresponding to signaling components is represented as hypergeometric distribution with the associated false discovery rate (FDR)
Consensus analyses of wild grapes and V. vinifera gene expression networks
We investigated the consensus between transcriptional networks in V. vinifera and wild grapes by detecting shared modules of co-expressed genes using a dissimilarity measure (Langfelder and Horvath 2007). Despite the differences in expression regulation, gene modules common to wild grapes and V. vinifera cultivars were identified, and their correlation to environmental conditions, investigated (Fig. 7). After merging gene groups with similar (≥ 80%) regulation patterns, 47 co-regulated consensus gene modules were found between V. vinifera and wild grapes, representing 36.98% (11,081 genes) of the whole transcriptome (Fig. 7a). The number of co-regulated genes per module ranged from 2938 (turquoise) to 38 (darkslateblue). Grey blocks represent genes (2235, 16.78%) whose expression pattern do not exhibit statistically significant co-regulation. The heatmap plots show the eigengene adjacencies in each co-regulated network, with positive and negative correlations depicted as high (red) and low (blue) adjacencies, respectively (Fig. 7b). The rows correspond to the module eigengene, represented by the color of the consensus module. The preservation of the relationships between the modules of co-regulated genes in vinifera and wild vines is shown for the overall network (Fig. 7c). The overall preservation of the eigengene networks, represented by the aggregate measure of the adjacency preservation between the gene networks in vinifera and wild grapes, was high (73%), ranging from 0.78 (module turquoise) to 0.68 (module darkslateblue) (Fig. 7c).
Consensus analyses of V. vinifera and wild grape transcriptomes. Correlation analyses between vinifera and wild consensus genes and environmental conditions (a). Eigengene networks for V. vinifera and wild grapes are shown as heatmaps, and b the mean preservation of adjacency in each module is shown as barplot . The overall preservation between the vinifera and wild grapes networks (c) is presented
Network preservation in cultivated vinifera genotypes was high inside the modules and among them, whereas in wild grapes, the preservation was preferentially intramodular (Fig. 7b). The relationships among the modules of co-regulated genes were shown to be distinct in V. vinifera and wild grapes (Fig. 7b).
Four consensus modules of co-regulated genes (darkorange, orangered4, midnightblue and turquoise), conserved in vinifera and wild grapes, exhibited high correlation to environmental factors (Fig. 7a). Non-redundant functional enrichment analyses demonstrated that module turquoise is enriched in genes associated with biological regulation, response to abiotic stresses, including osmotic, water deprivation, salt stress and cold, and signaling to environmental factors, such as temperature stimulus and abscisic acid (Fig. 8a). The most frequent molecular functions in the module correspond to signal perception and transduction, including transcription regulation and protein modification (Fig. 8b). The remaining co-regulated modules are smaller, consisting of 236 (midnightblue), 98 (darkorange) and 57 (orangered4) genes (Fig. 8c–e). No significant functional enrichment was found in modules darkorange and orangered4 (Fig. 8c, d), whereas transcripts encoding enzymes involved in terpenoid backbone biosynthesis were highly represented in module midnightblue (Fig. 8e).
Functional characterization of the consensus gene modules between vinifera and wild vines by GO enrichment analyses. Biological process (a) and molecular function (b) are shown for module turquoise. Number of genes and GO in modules darkorange (c) and orangered4 (d). Functional enrichment in module midnightblue (e). Metabolism of terpenoids and polyketides pathway is represented according to KEGG Pathways 1. Metabolism (map 00908—Zeatin biosynthesis). Enrichment in genes corresponding to metabolic enzymes is represented as hypergeometric distribution with the associated false discovery rate (FDR)
Discussion
In the family Vitaceae, most genomic and post-genomic information is from the cultivated species V. vinifera and its hybrids. The commercial importance of the fruit drives most studies to grapevine reproductive development, notably berry ripening (Magris et al. 2019; Dal Santo et al. 2016, 2018; Ghan et al. 2017; Zenoni et al. 2017; Cramer et al. 2014). However, the reproductive cycle of grapevines spans a period of two consecutive years, and environmental conditions during inflorescence differentiation and flower development are critical for berry production (Carmona et al. 2008). In the current study, we investigated the global profiles of gene expression of the vegetative development of cultivated and wild grapes, growing under distinct environmental conditions. We compared the structure of the transcriptomes, differential gene expression, the consensus between vinifera and wild grapes and the correlation between gene expression patterns and local environmental conditions in the vineyards.
Cultivated and wild grapes exhibit distinct transcriptome organization
Most cultivated grapes belong to the Eurasian species V. vinifera, and, even though there are thousands of known genotypes, they share extensive ancestry (Laucou et al. 2018). Cultivated grapevines exhibit a simple population structure with three ancestral groups and substantial admixture, suggesting that a small number of cross-fertilized parents gave rise to distinct descents (Zhou et al. 2019). Accordingly, modern Italian cultivars were demonstrated to share extensive haplotype regions (Magris et al. 2019). Four distinct transcriptome profiles were detected among ten investigated V. vinifera genotypes, although not directly representative of parentage relationships. ‘Pirovano 65’ (syn. ‘Italia’) exhibited the most divergent transcriptome, despite sharing extensive parentage with Red May, whose global expression profile was also distant from their common parent, ‘Muscat Hamburg’. Promoter motif-enrichment in co-expressed gene networks has been shown for several stress and developmental process in grapevine (Wong et al. 2017). A study combining genome sequencing and transcriptional profiling analyses demonstrated that conserved cis-regulatory regions are responsible for co-regulated gene expression in Italian cultivars sharing parentage, rather than haplotype conservation (Magris et al. 2019). Accordingly, the differentially expressed genes between vinifera and wild grapes were shown to be enriched in distinct promoter motifs in our study.
The organization of the transcriptome in vinifera consisted of a larger number of co-regulated gene modules; however, the total number of expressed genes was higher than in labrusca hybrids and wild grapes. A two-step filtering procedure in the statistical analyses, namely, first removing genes and samples with low variance, and subsequently, removing outlier samples, eliminates the bias induced by different gene numbers (Langfelder and Horvath 2012). Further confirmation of the biological meaning of the distinct transcriptome organization is shown in the comparison between labrusca hybrids and wild vines, where the number of identified genes is similar, and yet the number of significant co-regulated modules is higher in wild species.
Genome-wide expression analyses of wild grapes is scarce, except for the rootstock species V. riparia (Khadka et al. 2019; Hopper et al. 2016; Fennell et al. 2015). The transcriptional profile of four phylogenetically distant wild grapes, representing two of the major genera in Vitacea, Vitis and Parthenocissus (Chu et al. 2018; Ickert-Bond et al. 2018) was determined. The total number of expressed genes in wild species was higher than detected in V. labrusca hybrids and the most similar global profiles were found between the Asian species V. flexuosa and the American V. gigas, indicating that geographic origin does not directly determine transcriptional regulation, as observed for cultivated grapes (Livigni et al. 2019; Magris et al. 2019). The similarity of the expression profiles in V. vinifera and V. tiliifolia was also shown in a phylogenetic study based on transcriptome (Wen et al. 2013).
Besides the structural transcriptome organization, differential gene expression analyses demonstrated significant variation between vinifera and wild genotypes. The genes involved in regulatory processes are repressed in wild grapes, as observed for pistachio, olive, and other crops (Olsen and Wendel 2013; Gros-Balthazard et al. 2019; Zeng et al. 2019). Differences in expression and transcription wiring with domestication have been demonstrated for other crop species (Guo et al. 2016; Sauvage et al. 2017; Zeng et al. 2019). The simpler organization of the transcriptome agrees with repression of regulatory genes in wild vines in comparison to vinifera. In tomato, the transcriptome of wild and domesticated genotypes also displayed distinct wiring, although conserved metabolic hubs were present (Zhu et al. 2018). A broad study of the transcriptome of wild and domesticated animals and plants suggests that domestication reduced transcriptional diversity, due to a decrease in genetic variation (Liu et al. 2019). However, in olives the reduction in nucleotide diversity during domestication is not followed by decreased expression diversity (Gros-Balthazard et al. 2019). The authors suggest that the weaker domestication syndrome in perennial plant species may contribute for the retained transcriptional diversity. In grapevine, local sequence conservation, rather than genome wide diversity, appears to be responsible for gene expression regulation (Magris et al. 2019).
Environmental conditions are associated to gene expression patterns in vinifera and wild grapes
A complex combination of abiotic and biotic factors is responsible for the environmental conditions affecting plant growth in the field (Mittler 2006). The simultaneous occurrence of several stresses can induce synergistic or antagonistic responses, so transcriptome studies in grapevine tend to investigate the effect of isolated contributors (Hopper et al. 2016; Haile et al. 2017; Khadka et al. 2019). In our work, quantitative abiotic factors were correlated to gene expression in vinifera cultivars and wild grapes. The limitation of the approach is that it does not allow determination of causal relationships, so the observed patterns of expression co-regulation could be caused by or consequences of the local environment.
In V. vinifera, co-regulated genes associated to the environment are enriched in regulatory process, in agreement the induction of this class of genes in comparison to wild grapes. In contrast, in wild grapes the gene modules associated to environmental conditions are enriched in metabolic processes, including carbohydrate and nitrogen metabolism. The distinct strategies employed by vinifera and hybrid genomic contexts of grapes in response to abiotic stresses was shown previously (Zha et al. 2018; Carvalho and Amâncio 2019). Wild vines are considered important sources of resistance to biotic and abiotic stresses (Daldoul et al. 2020). Accordingly, the expression of genes associated with resistance processes is induced in wild grapes, in comparison to vinifera. The repression of the expression of genes involved in regulatory processes and induction of those associated with metabolism may underly the differences in the timing of phenological events between wild grapes and vinifera cultivars, such as the expanded timing of bud burst and the condensed transition to flowering. The maintenance of metabolic processes, such as photosynthesis, would allow favorable carbohydrate and energy levels to sustain earlier bud burst and extended vegetation in wild grapes. The involvement of carbohydrate and energy metabolism in developmental transitions in grapevine has been demonstrated in previous works (Carmona et al. 2008; Fennell et al. 2015). Similarly, in wild grapes the expression of several genes encoding hormone-signaling partners involved in reproductive development is tightly correlated with the environment, which may also contribute to the more diverse phenological behavior. A study combining QTL mapping and transcriptional analyses identified gibberellin and brassinosteroid signaling genes among those inducing early flowering in grapevine (Kamal et al. 2019), as observed in module red in our study. The correlation between these candidates and environmental conditions may contribute to understanding how exogenous cues control flowering in grapevine. Distinct transcriptome wiring between wild and cultivated genotypes has been described for other perennial crop species, including olives (Gros-Balthazard et al. 2019), pistachio (Zeng et al. 2019), and Asian pear (Li et al. 2019).
Consensus transcriptome shared by wild and cultivated vines contains modules of co-regulated genes associated to the environment
Despite the differences in gene expression and transcriptome organization, vinifera and wild grapes share consensus modules of co-regulated genes. However, the relationships between the conserved modules was different in vinifera and wild grapes, with the later exhibiting low network preservation between modules. These findings demonstrate a higher order of differential wiring between the transcriptomes of V. vinifera and the wild grapes. In tomato, a study combining large scale gene expression and population genomics demonstrated that domestication has caused high order transcriptional re-wiring, affecting whole gene networks involved in carbohydrate metabolism and epigenetic regulation of gene expression (Sauvage et al. 2017). The selection for alleles associated to larger fruits resulted in altered metabolite profiles due to their linkage with co-expressed, neighboring genes (Zhu et al. 2018).
The consensus modules significantly correlated to vineyard local conditions are enriched in genes associated with biological regulation, response to abiotic stresses, including osmotic, water deprivation, salt stress and cold, and signaling to environmental factors, such as temperature stimulus and abscisic acid, and represent molecular functions involved in signal perception and transduction, including transcription regulation and protein modification. The consensus network algorithm does not use a priori information, so the results of the enrichment analyses work as an internal validation criterion, confirming the association of the co-regulated modules with environmental features. A similar approach was used to identify key regulators in the development of flowers and fruits in wild strawberry (Fragaria vesca) (Shahan et al. 2018). Consensus networks have a more robust overall performance in predicting gene association than networks inferred from single datasets, as shown for Arabidopsis (Wirojsirasak et al. 2019). Our consensus network analyses indicate the common involvement of defense, biological regulation, and catabolism in the responses of grapevines and wild grapes to the environment. The consensus analyses also suggested a role for the genes encoding zeatin biosynthesis enzymes in grape responses to abiotic conditions. A similar role of the hormone and its precursors has been demonstrated for Arabidopsis and other species (Schäfer et al. 2015; Osugi et al. 2017).
In conclusion, comparative transcriptome analyses revealed distinct patterns of gene expression regulation and whole-transcriptome organization between cultivated and wild grapes (Fig. 9). Differential expression analyses revealed that regulatory genes are repressed, whereas those involved in metabolic processes are induced in wild grapes in comparison to vinifera. Differentially expressed genes between vinifera and wild genotypes are enriched in distinct cis-regulatory motifs, expanding the previous reports of the importance of promoter elements in controlling transcription diversity in Vitacea. The organization of the transcriptome in V. vinifera is structurally more complex than in wild grapes. Gene networks associated with local environmental conditions are enriched in regulatory processes in V. vinifera and in metabolic pathways in wild grapes. Consensus analyses revealed conserved co-expressed gene networks between wild and cultivated grapes, although with divergent connections, indicating differences in higher orders of transcriptome organization.
Summary schematic representation of the transcriptional differences between V. vinifera and wild vines. Red and green arrows represent differentially up- and down-regulated genes between vinifera and wild vines, respectively. Heatmap plots represent the network of the genes in co-regulated modules, obtained from the topological overlap matrix (TOM). High and low adjacency (overlap) are represented in red and light yellow, respectively. Gene networks are represented as color-coded modules presented in Fig. 3
Data and material availability
Sequencing data is deposited at Short Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) under accession numbers SAMN14331022 to SAMN14331038 (BioProject PRJNA611044).
References
Bigard A, Berhe DT, Maoddi E, Sire Y, Boursiquot JM, Ojeda H, Péros JP, Doligez A, Romieu C, Torregrosa L (2018) Vitis vinifera L. fruit diversity to breed varieties anticipating climate changes. Front Plant Sci 9:455. https://doi.org/10.3389/fpls.2018.00455
Canaguier A, Grimplet J, Di Gaspero G, Scalabrin S, Duchêne E, Choisne N, Mohellibi N, Guichard C, Rombauts S, Le Clainche I, Bérard A, Chauveau A, Bounon R, Rustenholz C, Morgante M, Le Paslier MC, Brunel D, Adam-Blondon AF (2017) A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genomics Data 14:56–62. https://doi.org/10.1016/j.gdata.2017.09.002
Carmona MJ, Chaïb J, Martínez-Zapater JM, Thomas MR (2008) A molecular genetic perspective of reproductive development in grapevine. J Exp Bot 59:2579–2596. https://doi.org/10.1093/jxb/ern160
Carvalho LC, Amâncio S (2019) Cutting the Gordian knot of abiotic stress in grapevine: from the test tube to climate change adaptation. Physiol Plant 165:330–342. https://doi.org/10.1111/ppl.12857
Chu Z, Wen J, Yang Y, Nie Z, Meng Y (2018) Genome size variation and evolution in the grape family Vitaceae. J Syst Evol 56:273–282. https://doi.org/10.1111/jse.12310
Coombe BG (1995) Growth stages of the grapevine: adoption of a system for identifying grapevine growth stages. Aust J Grape Wine Res 1:104–110. https://doi.org/10.1111/j.1755-0238.1995.tb00086.x
Cramer GR, Ghan R, Schlauch KA, Tillett RL, Heymann H, Ferrarini A, Delledonne M, Zenoni S, Fasoli M, Pezzotti M (2014) Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol 14:370. https://doi.org/10.1186/s12870-014-0370-8
Dal Bosco D, Sinski I, Ritschel PS, Camargo UA, Fajardo TVM, Harakava R, Quecini V (2018) Expression of disease resistance in genetically modified grapevines correlates with the contents of viral sequences in the T-DNA and global genome methylation. Transgenic Res 27(4):379–396. https://doi.org/10.1007/s11248-018-0082-1
Daldoul S, Boubakri H, Gargouri M, Mliki A (2020) Recent advances in biotechnological studies on wild grapevines as valuable resistance sources for smart viticulture. Mol Biol Rep 47(4):3141–3153. https://doi.org/10.1007/s11033-020-05363-0
Dal Santo S, Tornielli GB, Zenoni S, Fasoli M, Farina L, Anesi A, Guzzo F, Delledonne M, Pezzotti M (2013) The plasticity of the grapevine berry transcriptome. Genome Biol 14(6):r54. https://doi.org/10.1186/gb-2013-14-6-r54
Dal Santo S, Fasoli M, Negri S, D’Incà E, Vicenzi N, Guzzo F, Tornielli GB, Pezzotti M, Zenoni S (2016) Plasticity of the berry ripening program in a white grape variety. Front Plant Sci 7:970. https://doi.org/10.3389/fpls.2016.00970
Dal Santo S, Zenoni S, Sandri M, De Lorenzis G, Magris G, De Paoli E, Di Gaspero G, Del Fabbro C, Morgante M, Brancadoro L, Grossi D, Fasoli M, Zuccolotto P, Tornielli GB, Pezzotti M (2018) Grapevine field experiments reveal the contribution of genotype, the influence of environment and the effect of their interaction (G×E) on the berry transcriptome. Plant J 93:1143–1159. https://doi.org/10.1111/tpj.13834
Fennell AY, Schlauch KA, Gouthu S, Deluc LG, Khadka V, Sreekantan L, Grimplet J, Cramer GR, Mathiason KL (2015) Short day transcriptomic programming during induction of dormancy in grapevine. Front Plant Sci 6:834. https://doi.org/10.3389/fpls.2015.00834
FNA Editorial Committee (eds) (2016) Flora of North America North of Mexico, vol 12. Magnoliophyta: Vitaceae to Garryaceae. Oxford University Press, New York
Gambino G, Perrone I, Gribaudo I (2008) A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal 19:520–525. https://doi.org/10.1002/pca.1078
Ge SX, Jung D (2020) ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36(8):2628–2629. https://doi.org/10.1093/bioinformatics/btz931
Ghan R, Petereit J, Tillett RL, Schlauch KA, Toubiana D, Fait A, Cramer GR (2017) The common transcriptional subnetworks of the grape berry skin in the late stages of ripening. BMC Plant Biol 17(1):94. https://doi.org/10.1186/s12870-017-1043-1
Gros-Balthazard M, Besnard G, Sarah G, Holtz Y, Leclercq J, Santoni S, Wegmann D, Glémin S, Khadari B (2019) Evolutionary transcriptomics reveals the origins of olives and the genomic changes associated with their domestication. Plant J 100:143–157. https://doi.org/10.1111/tpj.14435
Guo J, Liu R, Huang L, Zheng XM, Liu PL, Du YS, Cai Z, Zhou L, Wei XH, Zhang FM, Ge S (2016) Widespread and adaptive alterations in genome-wide gene expression associated with ecological divergence of two Oryza species. Mol Biol Evol 33:62–78. https://doi.org/10.1093/molbev/msv196
Haile ZM, Pilati S, Sonego P, Malacarne G, Vrhovsek U, Engelen K, Tudzynski P, Zottini M, Baraldi E, Moser C (2017) Molecular analysis of the early interaction between the grapevine flower and Botrytis cinerea reveals that prompt activation of specific host pathways leads to fungus quiescence. Plant Cell Environ 40:1409–1428. https://doi.org/10.1111/pce.12937
Hartwell LH, Hopfield JJ, Leibler S, Murray AW (1999) From molecular to modular cell biology. Nature 402(6761 Suppl):C47–C52. https://doi.org/10.1038/35011540
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57(12):1332–1334. https://doi.org/10.1139/b79-163
Hopper DW, Ghan R, Schlauch KA, Cramer GR (2016) Transcriptomic network analyses of leaf dehydration responses identify highly connected ABA and ethylene signaling hubs in three grapevine species differing in drought tolerance. BMC Plant Biol 16:118. https://doi.org/10.1186/s12870-016-0804-6
Ickert-Bond SM, Harris A, Lutz S, Wen J (2018) A detailed study of leaf micromorphology and anatomy of New World Vitis L. subgenus Vitis within a phylogenetic and ecological framework reveals evolutionary convergence. J Syst Evol 56:309–330. https://doi.org/10.1111/jse.12313
Jones DM, Vandepoele K (2020) Identification and evolution of gene regulatory networks: insights from comparative studies in plants. Curr Opin Plant Biol 54:42–48. https://doi.org/10.1016/j.pbi.2019.12.008
Kamal N, Ochßner I, Schwandner A, Viehöver P, Hausmann L, Töpfer R, Weisshaar B, Holtgräwe D (2019) Characterization of genes and alleles involved in the control of flowering time in grapevine. PLoS One 14(7):e0214703. https://doi.org/10.1371/journal.pone.0214703
Kassambara A, Mundt F (2019) Factoextra R package: easy multivariate data analyses and elegant visualization. STHDA Statistical tools for high-throughput data analysis. http://www.sthda.com/english/wiki/factoextra-r-package-easy-multivariate-data-analyses-and-elegant-visualization. Data accessed: Accessed in 23 May 2020
Khadka VS, Vaughn K, Xie J, Swaminathan P, Ma Q, Cramer GR, Fennell AY (2019) Transcriptomic response is more sensitive to water deficit in shoots than roots of Vitis riparia (Michx.). BMC Plant Biol 19:72. https://doi.org/10.1186/s12870-019-1664-7
Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1:54. https://doi.org/10.1186/1752-0509-1-54
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
Langfelder P, Horvath S (2012) Fast R functions for robust correlations and hierarchical clustering. J Stat Softw 46(11):i11
Laucou V, Launay A, Bacilieri R, Lacombe T, Adam-Blondon AF, Bérard A, Chauveau A, de Andrés MT, Hausmann L, Ibáñez J, Le Paslier MC, Maghradze D, Martinez-Zapater JM, Maul E, Ponnaiah M, Töpfer R, Péros JP, Boursiquot JM (2018) Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS One 13(2):e0192540. https://doi.org/10.1371/journal.pone.0192540
Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25(1):1–18. https://doi.org/10.18637/jss.v025.i01
Li X, Liu L, Ming M, Hu H, Zhang M, Fan J, Song B, Zhang S, Wu J (2019) Comparative transcriptomic analysis provides insight into the domestication and improvement of pear (P. pyrifolia) fruit. Plant Physiol 180:435–452. https://doi.org/10.1104/pp.18.01322
Liang Z, Duan S, Sheng J, Zhu S, Ni X, Shao J, Liu C, Nick P, Du F, Fan P, Mao R, Zhu Y, Deng W, Yang M, Huang H, Liu Y, Ding Y, Liu X, Jiang J, Zhu Y, Li S, He X, Chen W, Dong Y (2019) Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat Commun 10(1):1190. https://doi.org/10.1038/s41467-019-09135-8
Liu Y, Zhou J, White KP (2014) RNA-seq differential expression studies: more sequence or more replication? Bioinformatics 30(3):301–304. https://doi.org/10.1093/bioinformatics/btt688
Liu W, Chen L, Zhang S, Hu F, Wang Z, Lyu J, Wang B, Xiang H, Zhao R, Tian Z, Ge S, Wang W (2019) Decrease of gene expression diversity during domestication of animals and plants. BMC Evol Biol 19(1):19. https://doi.org/10.1186/s12862-018-1340-9
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Livigni S, Lucini L, Sega D, Navacchi O, Pandolfini T, Zamboni A, Varanini Z (2019) The different tolerance to magnesium deficiency of two grapevine rootstocks relies on the ability to cope with oxidative stress. BMC Plant Biol 19(1):148. https://doi.org/10.1186/s12870-019-1726-x
Lloyd G, McCown BH (1980) Commercially-feasible micropropagation of Mountain Laurel, Kalmia latifolia, by shoot tip culture. Proc Int Plant Propag Soc 30:421–427
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Luo W, Brouwer C (2013) Pathview: an R/bioconductor package for pathway-based data integration and visualization. Bioinformatics 29(14):1830–1831. https://doi.org/10.1093/bioinformatics/btt285
Maia JDG, Camargo UA, Tonietto J, Zanus MC, Quecini V, Ferreira ME, Ritschel PS (2015) Grapevine breeding programs in Brazil. In: Reynolds AG (ed) Grapevine breeding programs for the wine industry: tradition and molecular techniques, 1st edn, Ch. 11. Woodhead Publishing, Swaston, pp 247–271. https://doi.org/10.1016/B978-1-78242-075-0.00011-9
Magris G, Di Gaspero G, Marroni F, Zenoni S, Tornielli GB, Celii M, De Paoli E, Pezzotti M, Conte F, Paci P, Morgante M (2019) Genetic, epigenetic and genomic effects on variation of gene expression among grape varieties. Plant J 99:895–909. https://doi.org/10.1111/tpj.14370
Mallet J (2015) New genomes clarify mimicry evolution. Nat Genet 47:306–307. https://doi.org/10.1038/ng.3260
Marrano A, Micheletti D, Lorenzi S, Neale D, Grando MS (2018) Genomic signatures of different adaptations to environmental stimuli between wild and cultivated Vitis vinifera L. Hortic Res 5:34. https://doi.org/10.1038/s41438-018-0041-2
Maul E et al (2019) Vitis International Variety Catalogue—www.vivc.de. Date accessed: accessed 13 Oct 2019
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19. https://doi.org/10.1016/j.tplants.2005.11.002
Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R (2009) ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25:2607–2608. https://doi.org/10.1093/bioinformatics/btp450
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858. https://doi.org/10.1094/Phyto-69-854
Olsen KM, Wendel JF (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol 64:47–70. https://doi.org/10.1146/annurev-arplant-050312-120048
Osugi A, Kojima M, Takebayashi Y, Ueda N, Kiba T, Sakakibara H (2017) Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat Plants 3:17112. https://doi.org/10.1038/nplants.2017.112
Parker A, García de Cortázar-Atauri I, Chuine I, Barbeau G, Bois B, Boursiquot J-M, Cahurel J-Y, Claverie M, Dufourcq T, Gény L, Guimberteau G, Hofmann RW, Jacquet O, Lacombe T, Monamy C, Ojeda H, Panigai L, Payan JC, Rodriguez-Lovelle B, Rouchaud E, Schneider C, Spring J-L, Storchi P, Tomasi D, Trambouze W, Trought M, van Leeuwen C (2013) Classification of varieties for their timing of flowering and véraison using a modelling approach: a case study for the grapevine species Vitis vinifera L. Agric For Meteorol 180:249–264. https://doi.org/10.1016/j.agrformet.2013.06.005
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. https://doi.org/10.1038/nmeth.4197
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Sauvage C, Rau A, Aichholz C, Chadoeuf J, Sarah G, Ruiz M, Santoni S, Causse M, David J, Glémin S (2017) Domestication rewired gene expression and nucleotide diversity patterns in tomato. Plant J 91(631):645. https://doi.org/10.1111/tpj.13592
Schäfer M, Brütting C, Meza-Canales ID, Großkinsky DK, Vankova R, Baldwin IT, Meldau S (2015) The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J Exp Bot 66:4873–4884. https://doi.org/10.1093/jxb/erv214
Shahan R, Zawora C, Wight H, Sittmann J, Wang W, Mount SM, Liu Z (2018) Consensus coexpression network analysis identifies key regulators of flower and fruit development in wild strawberry. Plant Physiol 178:202–216. https://doi.org/10.1104/pp.18.00086
Soneson C, Love MI, Robinson MD (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. https://doi.org/10.12688/f1000research.7563.1
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6(7):e21800. https://doi.org/10.1371/journal.pone.0021800
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. https://doi.org/10.1093/nar/gkm306
VanBuren R, Man Wai C, Pardo J, Giarola V, Ambrosini S, Song X, Bartels D (2018) Desiccation tolerance evolved through gene duplication and network rewiring in Lindernia. Plant Cell 30(12):2943–2958. https://doi.org/10.1105/tpc.18.00517
Wang P, Yang Y, Shi H, Wang Y, Ren F (2019) Small RNA and degradome deep sequencing reveal respective roles of cold-related microRNAs across Chinese wild grapevine and cultivated grapevine. BMC Genomics 20(1):740. https://doi.org/10.1186/s12864-019-6111-5
Wellburn AF (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144(3):307–313. https://doi.org/10.1016/S0176-1617(11)81192-2
Wen J, Xiong Z, Nie ZL, Mao L, Zhu Y, Kan XZ, Ickert-Bond SM, Gerrath J, Zimmer EA, Fang XD (2013) Transcriptome sequences resolve deep relationships of the grape family. PLoS One 8(9):e74394. https://doi.org/10.1371/journal.pone.0074394
Wirojsirasak W, Kalapanulak S, Saithong T (2019) Pan- and core- gene association networks: integrative approaches to understanding biological regulation. PLoS One 14(1):e0210481. https://doi.org/10.1371/journal.pone.0210481
Wong DCJ (2020) Network aggregation improves gene function prediction of grapevine gene co-expression networks. Plant Mol Biol 103(4–5):425–441. https://doi.org/10.1007/s11103-020-01001-2
Wong DCJ, Lopez Gutierrez R, Gambetta GA, Castellarin SD (2017) Genome-wide analysis of cis-regulatory element structure and discovery of motif-driven gene co-expression networks in grapevine. DNA Res 24(311):326. https://doi.org/10.1093/dnares/dsw061
Zeng L, Tu XL, Dai H, Han FM, Lu BS, Wang MS, Nanaei HA, Tajabadipour A, Mansouri M, Li XL, Ji LL, Irwin DM, Zhou H, Liu M, Zheng HK, Esmailizadeh A, Wu DD (2019) Whole genomes and transcriptomes reveal adaptation and domestication of pistachio. Genome Biol 20(1):79. https://doi.org/10.1186/s13059-019-1686-3
Zenoni S, Dal Santo S, Tornielli GB, D’Incà E, Filippetti I, Pastore C, Allegro G, Silvestroni O, Lanari V, Pisciotta A, Di Lorenzo R, Palliotti A, Tombesi S, Gatti M, Poni S (2017) Transcriptional responses to pre-flowering leaf defoliation in grapevine berry from different growing sites, years, and genotypes. Front Plant Sci 8:630. https://doi.org/10.3389/fpls.2017.00630
Zha Q, Xi X, He Y, Jiang A (2018) Comprehensive evaluation of heat resistance in 68 Vitis germplasm resources. Vitis 57:75–81. https://doi.org/10.5073/vitis.2018.57.75-81
Zhou Y, Minio A, Massonnet M, Solares E, Lv Y, Beridze T, Cantu D, Gaut BS (2019) The population genetics of structural variants in grapevine domestication. Nat Plants 5:965–979. https://doi.org/10.1038/s41477-019-0507-8
Zhu G, Wang S, Huang Z, Zhang S, Liao Q, Zhang C, Lin T, Qin M, Peng M, Yang C, Cao X, Han X, Wang X, van der Knaap E, Zhang Z, Cui X, Klee H, Fernie AR, Luo J, Huang S (2018) Rewiring of the fruit metabolome in tomato breeding. Cell 172(1–2):249–261.e12. https://doi.org/10.1016/j.cell.2017.12.019
Acknowledgements
The authors would like to thank the financing agencies SEG-Embrapa and CNPq. We also would like to express our gratitude to the dedicated staff at Embrapa involved in germplasm phenotyping and data collection.
Funding
The current work was supported financially by Sistema Embrapa de Gestão (SEG) (Grant 02.13.14.002.00.00 to TVMF and Grant 02.11.01.014.00.00 to VQ) and Conselho National de Desenvolvimento Científico e Tecnológico (CNPq) (Grants 307031/2010-1 and 400120/2016-0 to VQ).
Author information
Authors and Affiliations
Contributions
TVMF selected the genotypes, extracted RNA, performed RNA-Seq experiments and revised the manuscript. VQ performed RT-qPCR and biochemical analyses, analyzed the data, and drafted the manuscript. The authors discussed and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
TVMF declares no conflict of interest. VQ declares no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fajardo, T.V.M., Quecini, V. Comparative transcriptome analyses between cultivated and wild grapes reveal conservation of expressed genes but extensive rewiring of co-expression networks. Plant Mol Biol 106, 1–20 (2021). https://doi.org/10.1007/s11103-021-01122-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01122-2