Abstract
Key message
Differences in FAE1 enzyme affinity for the acyl-CoA substrates, as well as the balance between the different pathways involved in their incorporation to triacylglycerol might be determinant of the different composition of the seed oil in Brassicaceae.
Abstract
Brassicaceae present a great heterogeneity of seed oil and fatty acid composition, accumulating Very Long Chain Fatty Acids with industrial applications. However, the molecular determinants of these differences remain elusive. We have studied the β-ketoacyl-CoA synthase from the high erucic feedstock Thlaspi arvense (Pennycress). Functional characterization of the Pennycress FAE1 enzyme was performed in two Arabidopsis backgrounds; Col-0, with less than 2.5% of erucic acid in its seed oil and the fae1-1 mutant, deficient in FAE1 activity, that did not accumulate erucic acid. Seed-specific expression of the Pennycress FAE1 gene in Col-0 resulted in a 3 to fourfold increase of erucic acid content in the seed oil. This increase was concomitant with a decrease of eicosenoic acid levels without changes in oleic ones. Interestingly, only small changes in eicosenoic and erucic acid levels occurred when the Pennycress FAE1 gene was expressed in the fae1-1 mutant, with high levels of oleic acid available for elongation, suggesting that the Pennycress FAE1 enzyme showed higher affinity for eicosenoic acid substrates, than for oleic ones in Arabidopsis. Erucic acid was incorporated to triacylglycerol in the transgenic lines without significant changes in their levels in the diacylglycerol fraction, suggesting that erucic acid was preferentially incorporated to triacylglycerol via DGAT1. Expression analysis of FAE1, AtDGAT1, AtLPCAT1 and AtPDAT1 genes in the transgenic lines further supported this conclusion. Differences in FAE1 affinity for the oleic and eicosenoic substrates among Brassicaceae, as well as their incorporation to triacylglycerol might explain the differences in composition of their seed oil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most plant derived seed oils are enriched in 18 carbon (C18) fatty acids, being oleic (18:1) and linoleic (18:2) acids, the most abundant ones in major oil commodities like soybean, sunflower, olive or even in seed oils from model plants like Arabidopsis thaliana (Harwood 1996). However, some other plant species accumulate unusual fatty acids that include atypical fatty acid chain length (from C8 to C14, and from C20 to C22), or presence of functional groups like hydroxyl or epoxy ones, among others (Harwood 1996; Jaworski and Cahoon 2003; Singh et al. 2005). Because of their chemical properties, some of these seed oils containing unusual fatty acids have industrial applications for the obtention of lubricants, paints, cosmetics or biofuels (Thelen and Ohlrogge 2002). Among these fatty acids, erucic acid (22:1Δ13) has an enormous interest as a precursor of biological compounds like waxes or sphingolipids (Post-Beittenmiller 1996; Merril et al. 1997) or due to its accumulation in triacylglycerol (TAG), (Taylor et al. 1993). In addition, erucic acid is an industrial precursor for the synthesis of erucamide, which is essential in the manufacturing industry of slip-promoting agents, surfactants, plasticizers, nylon or surface coatings (Sonntag 1995). More recently, erucic acid enriched oil has attracted the interest of the biofuel industry because of its outstanding low-temperature and high-energy properties, essential for the obtention of biofuel for aviation (Moser 2012).
Very Long Chain Fatty Acids (VLCFA), like eicosenoic acid (20:1 Δ11) or erucic acid, can be found in seed oils from many different plant species including members of the Brassicaceae and Tropalaceae families (Harwood et al. 1996; Post- Beittenmiller 1996; Ghanevati and Jaworski 2001). However, their content and distribution is highly variable even among species of the same family or genus. Thus, while the erucic acid content in the model plant Arabidopsis thaliana, which belongs to Brassicaceae, is rather low (< 2.5%), other members of the family can reach total erucic acid content in the seed oil ranging from 40 to 60%, as is the case of Brassica napus, Crambe abyssinica or Thlaspi arvense (Sun et al. 2013; Claver et al. 2017). The reason of these differences on the erucic acid content in the seeds from Brassicaceae remains unclear. VLCFA like erucic acid are synthesized in the endoplasmic reticulum through the sequential elongation of C18 acyl-CoA substrates by the Fatty Acid Elongase (FAE) complex. The process involves a series of reactions that include: i) condensation of malonyl-CoA to a C18 acyl-CoA substrate to produce a β-ketoacyl-CoA; ii) reduction to β-hydroxyacyl-CoA; iii) dehydration to an enoyl-CoA; and iv) reduction to form the elongated acyl-CoA (Ghanevati and Jaworski 2001), which is later incorporated to the triacylglycerol biosynthetic pathway in the seed (Bates et al. 2013). This FAE complex is composed of four integral membrane enzymes, but the activity of the first condensing enzyme, the β-ketoacyl-CoA synthase (KCS), is the rate-limiting step determining VLCFA production (Millar and Kunst 1997; Katavic et al. 2001). In vivo and in vitro transformation assays in yeast using the Arabidopsis FAE1 gene, which is the seed-specific isoform responsible for the elongation of C18 fatty acids to be incorporated to triacylglycerol, demonstrated that this enzyme alone is capable to direct the synthesis of VLCFA in yeast (Millar and Kunst 1997; Blacklock and Jaworski 2002). Similar experiments expressing either the Arabidopsis or Brassica napus genes in yeast showed that although both FAE1 enzymes were capable to elongate oleyl (18:1Δ9) substrates, the Arabidopsis FAE1 enzyme produced eicosenoic acid as major VLCFA, while the B. napus enzyme used eicosenoic acid substrates to produce erucic acid (Han et al. 2001; Katavic et al. 2002). Complementation analysis of a fae1-1 mutant from Arabidopsis with the Teesdalia nudicaulis FAE1 protein showed efficient production of eicosenoic acid but no production of erucic acid, (Mietkiewska et al. 2007). It is worth mentioning that Teesdalia nudicaulis accumulates up to 47% of eicosenoic acid, with very low levels of erucic one in its seed oil (Mietkiewska et al. 2007). Similarly, expression of a Nasturtium FAE1 gene in Arabidopsis also resulted in an increase of erucic acid in its seed oil (Mietkiewska et al. 2004). However, given the high amount of eicosenoic acid already present in the Arabidosis seed oil (around 20%), together with the lack of data in an appropriate mutant background, these results were not sufficient to demonstrate differences in FAE1 substrate affinities among plant species.
TAG is the major fraction in plant seed oils, representing an 80–90% of total seed lipids (Bates et al. 2013). Two pathways contribute to TAG biosynthesis in the seed. On one hand, TAG biosynthesis is performed by a series of enzymes (glycerol 3-phosphate acyltransferase, GPAT; lyso-phosphatidyl-acyltransferase LPAT; phosphatidic acid phosphatase, PAP) that perform the sequential acylation of the sn-1, sn-2 and sn-3 position of the glycerol backbone through the Kennedy pathway (Ohlrogge and Browse 1995). On the other hand, acyl-editing reactions convert PC into lysoPC via phosphatidyl choline acyl transferase (PDAT1). LysoPC can be converted again into PC through the action of lysophosphatidyl choline acyl transferase (LPCAT). This PC/lysoPC pool acts as a reservoir for PC/DAG interconversion by phosphatidilcoline diacylglycerol coline phosphotransferase (PDCT), being another pathway of diacylglycerol (DAG) supply for TAG synthesis in Arabidopsis (Bates and Browse 2012; Wang et al. 2012; Bates et al. 2013). In both pathways, the final acylation is performed by an acyl-CoA:diacylglycerol acyltransferase (DGAT), whose activity has been shown to determine the carbon flow into TAG (Weselake et al. 2009; Liu et al. 2012; Bates et al. 2013). Not only differences in elongase activity, but also in the incorporation of VLCFA to TAG through both pathways have been pointed out as possible bottlenecks responsible of the different erucic acid content in plant seed oils (Guan et al. 2014). In that sense, differences in substrate affinity among specific BnDGAT2 isoforms to incorporate erucyl-CoA to TAG have been recently identified in Brassica cultivars with different erucic acid content, suggesting a relevant role of this enzyme in the fatty acid composition of plant seed oils (Demski et al 2019).
In the search for new FAE enzymes for biotechnological applications, our group became interested in the elongase from Thlaspi arvense L. (Pennycress). Pennycress belongs to the Brassicaceae family and is closely related to the model plant Arabidopsis thaliana, as well as other agronomical important plants such as rapeseed (Brassica napus) or camelina (Camelina sativa). In addition to its extreme cold tolerance or over-wintering growth, Pennycress seeds have unique characteristics in terms of seed oil content (32–36% w/w) and composition, with a high content (38–40%) of erucic acid. This has attracted the interest on Pennycress as biofuel feedstock because of the exceptional low-temperature behaviour and energetic capacity characteristics of its seed oil (Moser et al. 2009; Moser 2012; Fan et al. 2013). In addition to this, advances in the study of Pennycress at the molecular level including transcriptome (Dorn et al. 2013), metabolite fingerprint (Tsogtbaatar et al. 2015), translational model (Chopra et al. 2018) and transformation and gene editing (McGinn et al. 2018), have increased the attraction of Pennycress as a model organism for oilseed biotechnology. However, although Pennycress could be an attractive alternative feedstock in Europe, particularly in the Mediterranean region, it is not cultivated in the EU territory. We have recently characterized Pennycress germplasm of European origin and some interesting agronomical traits were detected. Genes involved in triacylglycerol, or erucic acid biosynthesis were identified, and their regulation was studied during Pennycress seed maturation (Claver et al. 2017). In addition, our previous results indicated that triacylglycerol was the major reservoir of erucic acid in Pennycress seeds (Claver et al. 2017). Furthermore, our data indicated that it was incorporated to TAG in very high amounts at the early stages of seed maturation (Claver et al. 2017). To further characterize the elongase from Pennycress and, in parallel, investigate the molecular and biochemical elements responsible for the great heterogeneity in VLCFA content and composition among Brassicaceae, we have performed a functional analysis of the TaFAE1 gene, encoding the seed-specific Pennycress β-ketoacyl-CoA synthase, in Arabidopsis. Two genetic backgrounds were used, Col-0 plants that accumulate very low levels (less than 2.5%) of erucic acid and other VLCFA and a fae1-1 mutant, deficient in FAE1 activity that does not accumulate VLCFA. Results obtained in both mutant backgrounds suggested that the Pennycress FAE1 enzyme showed higher affinity for eicosenoyl substrates than the endogenous AtFAE1 one. Lipid fraction analysis of their seed oil also suggested that DGAT1 was the major responsible of erucic acid incorporation to TAG. Our data point to differences in β-ketoacyl-CoA synthase affinity for the oleyl and eicosenoyl substrates among brassicaeae, as well as their pathway of incorporation to TAG to explain differences in seed oil composition in Brassicaceae. These data can be useful for the manipulation of the erucic acid trait in oilseed plants.
Materials and methods
Plant materials
Arabidopsis thaliana Col-0 ecotype was used as wild-type. A fae1-1 mutant was also used. Seeds from the fae1-1 mutant were obtained from NASC (N6345). fae1-1 was a EMS mutant deficient in fatty acid elongase activity (Katavic et al. 1995). Both lines were used as genetic backgrounds for the obtention of the different transgenic lines. Arabidopsis seeds were sterilized and germinated in MS medium or directly in pots. Seeds were vernalized for 3 days at 4 °C and then moved to a growth chamber for 14 days. For gene expression or lipid analysis, seeds from the different Arabidopsis lines were collected after 5–6 weeks of growth in the culture chamber. Growth conditions were, light intensity of 130 µmol m−2 s−1, 16/8 h light/dark photoperiod, 22/18 °C, and a relative humidity of 60/65%.
Generation of transgenic Arabidopsis lines expressing the Pennycress FAE1 gene
Pennycress genomic DNA was isolated by the CTAB method (Doyle and Doyle 1990). The TaFAE1 gene (Genbank accession number: KT223024) was amplified from genomic DNA using Phusion High-Fidelity DNA Polymerase (Thermo) and the primers described in Supplementary Table S1. Two NcoI and BamHI sites were added to the primers to favor directional cloning. The PCR fragment of 1521 bp, corresponding to the complete TaFAE1 gene sequence, was cloned in a pGEM-T-Easy vector. Both the Arabidopsis and Pennycress FAE1 genes contain no introns (Claver et al. 2017). Then, the 1521 bp NcoI-BamHI TaFAE1 gene fragment was cloned in a modified pFGC5941 vector obtained from Dr. F. Vastiij (CNAP-University of York; UK). The pFGC5941 binary vector was modified by eliminating the constitutive 35S promoter and substitution by an OLE2 promoter to ensure seed-specific expression of the transgene and to avoid over-expression or tissue-unspecific artefact results in our analysis. All constructs were analyzed to verify the sequence before plant transformation.
Agrobacterium mediated transformation (GV3101 strain) of Arabidopsis plants was performed by floral dip (Clough and Bent 1998) either in Col-0 or fae1-1 mutant lines. Positive transformants were selected for Basta® resistance and genotyped by Phire® Plant direct PCR kit (Thermo). Lines producing 100% resistant plantlets were selected (homozygous lines, single insertion locus) and used for further analysis. Ten independent transgenic homozygote T3 lines were selected and, after lipid analysis, at least three independent transgenic events on each Col-0 and fae1-1 backgrounds were used for further analysis.
Sequence analyses
All sequences were obtained from GENBANK. Protein alignment was performed using the CLUSTALW2 multiple alignment tool. Phylogenetic trees were generated using the PHYML software (Guindon and Gascuel 2003; www.atgc-montpellier.fr/PHYML), with bootstraps 100. The names and GENBANK accession numbers of the sequences used for the analysis can be found in the legend of supplementary Fig. 1.
Quantitative PCR analysis
Total RNA was extracted from 0.5 g of seeds with Trizol (Life Technologies) according to manufacturer’s instructions. First-strand cDNA was synthesized from 3 µg of DNAase-treated RNA with M-MLV reverse transcriptase (Promega) and oligodT. Quantitative PCR (qRT-PCR) of target genes was performed using a 7500 Real Time PCR System (Applied Biosystems), SYBR Green Master Mix (Applied Biosystems), and specific primers (Supplementary Table S1). The Ct values were calculated relative to ACT-2 reference gene (At3g618780) using 2−ΔΔCt method (Livak and Schmittgen 2001). Data were obtained from the analysis of at least three biological samples with three independent technical repeats for each sample. In all cases, the expression values obtained for Col-0 were used as calibrator and fixed to 1.
Lipid and fatty acid composition analysis
Total lipids were extracted from Arabidopsis thaliana seeds (250–300 seeds) with chloroform:methanol (2:1, v/v) as described by Bligh and Dyer (1959). Fatty acid methyl esters of total lipids or individual lipid classes were produced by acid-catalyzed transmethylation (Garcés and Mancha 1993), and analyzed by gas chromatography (GC), using a 7890A (Agilent, Santa Clara, CA USA) fitted with a capillary column (60-m length; 0.25-mm inner diameter; 0.2-µm film thickness) of fused silica (Supelco, Bellafonte, PA, USA) and a FID detector. Helium was used as a carrier gas with a linear rate of 1.2 ml min−1 and split ratio of 1/50. The injector temperature was 250 °C and the detector temperature was 260 °C. The oven temperature was modified as follows: 170 °C for 30 min, then raising the temperature by 5 °C /min to 200 °C. 17:0 (1 μg μl−1) was used as an internal standard. Data from fatty acid analysis were obtained from three independent groups of five plants that were pooled for further analysis. At least three technical replicates were performed for each determination. Analysis of variance (ANOVA) was applied to compare treatments. Statistical analyses were carried out with the program Statgraphics Plus for Windows 2.1, using a level of significance of 0.05.
For lipid class analysis, TLC plates (Silica Gel 60, Merck) were activated by heating to 110 °C for at least one hour prior to the analysis in order to drive off any moisture. Then, total lipids extracted from the seeds corresponding to the different transgenic lines were loaded onto the TLC plates. Triacylglycerol (TAG) and diacylglycerol (DAG) fractions were separated in TLC plates developed with a mixture of heptane:diethylether:acetic acid (70:30:1, v/v/v), following the method described by Li-Beisson et al. (2013). Detection of lipids was performed by short exposure to iodine vapour. Bands were marked, scraped off and extracted with a methanol:chloroform:water (100:50:40, v/v/v) solution followed by an additional separation in a chloroform:water (50:50, v/v) mixture. Commercial standards of TAG and DAG (Sigma) were used as a reference for band identification. 17:0 (1 μg μl−1) was used as an internal standard for quantification purposes. Fatty acid methyl esters of the different TAG and DAG fractions were obtained as described above.
Results
Brassicaceae are characterized by a heterogeneous erucic acid content in their seed- oil
Figure 1 shows the erucic acid content in the seed oil of 23 lines of Brassicaceae. Data were obtained from the literature (Han et al. 2001; Mietkiewska et al. 2004, 2007; Sun et al. 2013) and from our own analysis in the case of Thlaspi arvense (Claver et al. 2017), Camelina sativa and Arabidopsis thaliana (Román et al. 2015). Overall, the erucic acid content ranged from 0% in Cardamine parviflora to 70% in Tropaelum majus, which belongs to Tropalaceae, a family with high erucic acid content in their seed oil (Mietkiewska et al. 2004). Distribution of plant species according to their erucic acid content could be classified into three groups (Fig. 1). A group of plants (Group I) with very low erucic acid content, currently in the range of 0–5%, such as Arabidosis thaliana, Camelina sativa, or Capsella bursa-pastoris (Fig. 1). A second group (Group II) corresponds to plant species, like Isatis tinctoria, Lepidium campestre or Thlaspi arvense, with erucic acid in the range of 25–40% (Fig. 1). Finally, plant species with erucic acid content higher than 40% (Group III), correspond to species like Crambe abyssinica, Crambe hispanica or Brassica napus (Fig. 1). It is worth mentioning that in some cases, the distribution of erucic acid correlated with the same genus, such as Crambe or Brassica in Group III or Arabidopsis in Group I. In other cases, some closely related species presented great differences in erucic acid content in their seed oil. This is the case of Thlaspidae with T. arvense or T. perfoliatum classified in Group II while T. caereulens was classified in Group I (Fig. 1). To further illustrate the heterogeneity of the distribution of the erucic acid trait, Fig. 1 also shows three species, Teesdalia nudicaulis, Lymnanthes douglasii, that belong to Brassicaceae and Simondsia chinensis (jojoba), that belongs to Simondsiaceae, all of them with very low erucic acid content (similar to that of Group I) but with a very high content (40–60%) of eicosenoic acid in their seed oil.
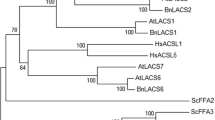
Since the FAE1 gene encodes the enzyme responsible for the biosynthesis of erucic acid and VLCFA, we carried out a phylogenetic tree that included the FAE1 genes of 22 accessions represented in Fig. 1 that belonged to the three groups. As shown in Supplementary Fig. S1, the phylogenetic analysis was consistent with the grouping of plant species in relation to the erucic acid trait shown in Fig. 1. Species like Lymnanthes douglasii or Simondsia chinensis, which accumulate high levels of eicosenoic acid but lower ones of erucic acid, were grouped in a different and separated clade from those of Group I or Groups II and III (Supplementary Fig. S1). Plant species with low erucic acid content (Group I), like Arabidopsis thaliana, Arabidopsis lyrata, Camelina sativa or Capsela bursa-pastoris were separated from those species with high erucic acid content in their seed oil (Groups II and III), (Supplementary Fig. S1). In fact, plants containing high amounts of erucic acid like Crambe abyssinica or Brassica napus (Group III) were grouped in a different clade with respect to those of Group I and II, like Thlaspi arvense or Isatis tinctoria, containing medium levels of erucic acid (Supplementary Fig. S1). Most Group II plants were found grouped in an intermediate branch of the tree between those from Group I and those from Group III, consistent with their erucic acid content (Supplementary Fig. S1). Interestingly, Orychofragmus violaceous was grouped in a single branch alone in the same clade of Group III species (Supplementary Fig. S1). Despite the high identity of the Orychofragmus violaceous FAE1 enzyme with high-erucic accumulating species (> 87% protein identity), it has very low erucic acid levels (Wu et al. 2009; Sun et al. 2013). Yeast complementation experiments suggested that this low erucic acid levels would be related with a loss of enzyme activity (Wu et al. 2009; Sun et al. 2013).
At this point, we decided to analyse into more detail the CLUSTAL multiple alignment of 19 plant species from all three Groups to identify motifs or residues that could be behind these differences in the erucic acid content and FAE1 activity. For a better illustration of the results of the analysis, an alignment of three species, A. thaliana, T. arvense, and C. abyssinica, representative of Groups I, II and III, respectively, is shown in Fig. 2 while the complete alignment is shown in Supplementary Fig. S2. The FAE1 enzymes from all Groups showed a high degree of identity at the protein sequence level, with values higher than 85% (Fig. 2; Supplementary Fig. S2). Residues of the active site like Cys223, His391 or Asn424 (Ghanevati and Jaworski 2001, 2002) as well as Ser282 (Han et al. 2001; Katavic et al. 2002), were conserved in all the FAE1 sequences. In their yeast complementation studies, Ghanevati and Jaworski (2002) also pointed to the specific role of residue 92 as a determinant of substrate specificity of the FAE1 enzyme. As shown in Fig. 2 and Supplementary Fig. S2, all plant species belonging to Groups II and III, with the exceptions of Isatis tinctoria and Lepidium campestre, showed an Arg residue in this position while those with low erucic acid content like Arabidopsis or Camelina (Group I) showed a Lys (Supplementary Fig. S2). Overall, 67 polymorphisms were detected in the analysis from FAE1 proteins from Group I, II and III species, representing a 13% of the complete FAE1 sequence. These changes were distributed in 28 in which group III were identical to group II but different to Group I (Fig. 2). These changes would be representative of high erucic containing species. On the contrary, 18 changes in group II species were identical to Group I, but different to group III and 13 ones in which group III were identical to group I, but different to group II (Fig. 2). Sun et al (2013) performed a phylogenetic study with 62 accessions in an attempt to identify protein motifs involved in high erucic acid accumulation and identified 7 protein motifs that could be related with this trait. In our analysis with 19 different plants from all three groups, with the exception of motif 1 (residues 182–183: KE in groups II and III/ RE in group I) and motif 3 (residues 256–257: YN in groups II and III/ QG in group I and 262: D in groups II and III/ E in group I; Fig. 2), the rest of the motifs were not conserved in plant species belonging to any of the three groups.
Comparison of the amino acid sequences of the Thlaspi arvense FAE1 gene (GENBANK accession no. KT223024) with KCS-FAE1 proteins from other plant species (Arabidopsis thaliana and Brassica napus) with different erucic acid content. Residues are shown in coloured lines. Residues identified as part of the active site are marked with a red box. Other residues for which differences were identified among groups are also marked within boxes of different colours: black, conserved in Groups II and III; blue, conserved in Groups I and II and green, conserved in Groups I and III. Position of the motifs identified in the previous analysis from Sun et al. (2013) is also indicated in the figure with circled numbers above their position in the sequence
Seed-specific transformation of the Pennycress FAE1 gene in Arabidopsis
To gain further information about the functional identity of the Pennycress FAE1 enzyme, and to evaluate its substrate preference and affinity, the Pennycress FAE1 gene was linked to a seed-specific OLE2 promoter and expressed in two different genetic backgrounds. On one hand, a Col-0 background was used in which the endogenous AtFAE1 gene was present. On the other hand, we transformed a fae1-1 mutant background (Katavic et al. 1995). This mutant, (NASC N6345), obtained by chemical mutagenesis with EMS, is defective in elongase activity and the typical acyl composition of its seed oil and the acyl-CoA pools is completely devoid of VLCFA, being oleyl-CoA and oleic acid the major species (Katavic et al. 1995). Nevertheless, both the Col-0 and the fae1-1 mutant have, a priori, oleyl-CoA enough to act as a substrate of the successive elongation to eicosenoic acid and erucic acid by the Pennycress FAE1 enzyme. Genotyping of the fae1-1 mutant used in this work, obtained from NASC, revealed a deletion at nt 848 of the coding sequence of the AtFAE1 gene that induced a frameshift in the ORF that later generated a stop codon (Fig. 3). It is worth mentioning that the fae1-1 mutant presented an unexpected dwarf phenotype upon germination and further plant development (Fig. 3). The reasons of this dwarf phenotype are not clear. However, our data are consistent with previous reports in which the over-expression of the AtFAE1 gene in Arabidopsis produced alterations in plant morphology (Millar and Kunst 1997). Since certain VLCFA, like eicosenoic acid, act as precursors for the synthesis of waxes and ceramides necessary for leaf cuticle function (von Wettstein-Knowles 1982; Han et al. 2001), the dwarf phenotype of the fae1-1 mutant could be related with this.
a Photograph showing the dwarf phenotype of the fae1-1 mutant used as an additional genetic background in our complementation analysis. The photograph shows the dwarf phenotype either in plates or in growing pots. The photograph also shows some representative complemented lines (4.3.1) that recovered the wild-type phenotype. b Genotyping of the fae1-1 mutant used in this study. Single nucleotide mutations are indicated in red boxes. The deletion at nt 848 that induced a shift in the ORF that generated an stop codon downstream is indicated in a green box
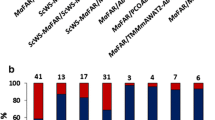
Twenty five BASTA-resistant T1 plants were selected from Agrobacterium-mediated Arabidopsis transformed plants from each genetic background. The T2 progeny was collected individually and genotyped for both BASTA and TaFAE1 genes. Positive lines were fully segregated to obtain a T3 progeny. At least 10 different T3 transgenic lines from each background were further analysed for the fatty acid composition of their seed lipids. As shown in Fig. 4a, transgenic lines obtained from the Col-0 background showed systematically two different types of results. On one hand, transgenic lines like 8.2.1 or 8.2.2 that showed a fatty acid composition of total seed lipids almost identical to that of Col-0 plants, with eicosenoic acid and erucic acid content of 20.8/20.9% and 2.61/2.54%, respectively (Fig. 4a). Without precluding any other options, differences in transgene expression due co-suppression with the endogenous AtFAE1 gene or insertion of the transgene in a silenced region of the genome might explain the absence of phenotype. On the other hand, other T3 lines, like 7.1.1. or 8.1.1, showed a relevant increase in their erucic acid content with values of 8.56 and 7.43%, respectively, which represent a 3–fourfold increase with respect to Col-0 plants (2.5%; Fig. 4a). Interestingly, these lines with higher erucic acid levels showed also a significant reduction of eicosenoic acid when compared to Col-0 or 8.2.1/8.2.2 lines (Fig. 4a). This result, together with the absence of relevant changes in oleic acid content in the different transgenic lines, suggested that the TaFAE1 enzyme used more efficiently the eicosenoyl-CoA pool for elongation to produce erucic acid than the oleyl-CoA one in Arabidopsis.
Fatty acid composition of total seed-lipids from Arabidopsis transgenic lines expressing the Pennycress FAE1 gene under the control of the seed-specific OLE2 promoter. a Fatty acid composition of Arabidopsis transgenic lines obtained in the Col-0 background. b Fatty acid composition of Arabidopsis transgenic lines obtained in the fae1-1 mutant background. Data from four different T3 homozygous lines from each background representative from ten lines analysed are shown. e.v. indicates data from Col-0 lines transformed with the empty vector. The data were obtained from three independent pools of seeds from five plants of each line
Ten different T3 homozygous lines obtained in the fae1-1 mutant background were analysed. The fatty acid composition of some representative transgenic lines is shown in Fig. 4b. As occurred in the Col-0 background, two different groups of results were obtained. On one hand, transgenic lines like 4.1.1, which showed similar fatty acid composition in the total seed lipid fraction to that of the fae1-1 mutant, characterized by a high oleic acid levels (24%) with respect to Col-0 plants (11.59%), (Fig. 4b). On the other hand, another group of transgenic lines, like 4.3.1, 4.4.1 or 4.1.2 showed slight but significant increases of either eicosenoic acid and erucic acid of different extent. Indeed, the 4.3.1 line showed a 1.8% of eicosenoic acid and 0.3% of erucic acid. When compared with the fatty acid composition of the fae1-1 mutant (0.4% of eicosenoic acid and 0.04% of erucic acid), the increase in VLCFA in the 4.3.1. transgenic line represented a 4.5- and 7.5-fold increase, respectively (Fig. 4b), with respect to the fae1-1 parental line. The 4.4.1 line also showed an increase of both eicosenoic acid (0.91%) and erucic acid (0.1%) levels with respect to the fae1-1 mutant. Finally, the 4.1.2 line showed increases only of erucic acid (0.12%) with no changes of eicosenoic acid levels with respect to the fae1-1 mutant (Fig. 4b).
Lipid class analysis of the Arabidopsis transgenic lines expressing the Pennycress FAE1 gene
We were interested in determining whether the erucic acid produced in the transgenic lines was efficiently incorporated to TAG and secondly, in which step of TAG biosynthesis was this erucic acid being incorporated. To that end, total lipids from seeds of T3 homozygous lines were extracted and subjected to TLC to separate TAG and DAG fractions. Fatty acid composition of both fractions was further analysed. In the transgenic lines obtained from the Col-0 background, the results were consistent with those obtained in the analysis of the total lipid fraction. Thus, 7.1.1 and 8.1.1 lines showed a 9.32% and 8.15% of erucic acid in TAG, which represented a threefold increase with respect to that of the control Col-0 line (3.22%), (Fig. 5a). In the 7.1.1 line, the increase in erucic acid occurred together with a decrease of eicosenoic acid levels as well as an increase of oleic acid with respect to Col-0 (Fig. 5a). On the contrary, in 8.1.1 plants, the increase in erucic acid occurred without changes in eicosenoic acid levels, although an increase of oleic acid ones could also be observed (Fig. 5a). The 8.2.2 line showed no substantial changes in fatty acid composition in TAG with respect to Col-0, consistent with the results of total seed-lipids.
Fatty acid composition of the TAG fraction obtained from total seed-lipids from Arabidopsis transgenic lines expressing the Pennycress FAE1 gene under the control of the seed-specific OLE2 promoter. a Fatty acid composition of TAG seed oil fraction from Arabidopsis transgenic lines obtained in the Col-0 background. b Fatty acid composition of TAG seed oil fraction from Arabidopsis transgenic lines obtained in the in the fae1-1 mutant background. The inset shows detailed data from the VLCFA fraction of the TAG composition from the fae1-1 mutant and two representative transgenic lines. Data from four different T3 homozygous lines from each background representative from ten lines analysed are shown. The data were obtained from three independent pools of seeds from five plants of each line
The analysis of the TAG fraction from the transgenic lines obtained in the fae1-1 mutant background was also consistent with those obtained in the total lipid fraction. Again, the extent of the change was very small in absolute terms, but increased relatively when compared to the fae1-1 data. Thus, the 4.3.1 line, which showed the most important increase in erucic acid levels in the total lipid fraction also showed the highest accumulation (sevenfold) of erucic acid in TAG, with a 0.35% compared to 0.05% in the control fae1-1 mutant (Fig. 5b; see inset). This increase was accompanied by an important increase in eicosenoic acid levels (1.85% in 4.3.1 vs 0.4% in the fae1-1 mutant; Fig. 5b, see inset), representing a 4.6-fold increase relative to fae1-1. This increase in eicosenoic acid was accompanied by a decrease in oleic acid levels in both transgenic lines that was higher in the 4.3.1 line which showed the highest erucic and eicosenoic acid increases (Fig. 5b). These results indicated that the erucic acid, as well as the eicosenoic acid, produced by the Pennycress FAE1 enzyme in the transgenic lines, were efficiently incorporated to TAG in Arabidopsis.
Arabidopsis shows high ratios of PC/DAG interconversion (Bates and Browse 2012; Wang et al. 2012). It has been suggested that this pathway might be a major route for DAG accumulation and subsequent TAG biosynthesis in the seed (Bates and Browse 2012; Wang et al. 2012; Bates et al. 2013). We were interested in analysing in which part of the pathway erucic acid, produced by the TaFAE1 enzyme, was being incorporated to TAG. To that end, we analysed the fatty acid composition of the DAG fraction in the transgenic lines. The analysis was focused in the transgenic lines from the Col-0 background since DAG is present in lower amounts (10%) with respect to TAG (80%) in total seed lipids and, secondly, because the low eicosenoic and erucic acid levels in the transgenic lines derived from the fae1-1 mutant made difficult to detect them in this minor seed-lipid fraction. The fatty acid composition of the DAG fraction from lines 7.1.1 or 8.1.1, that showed a relevant increase in erucic acid in total seed lipids or TAG fraction, depicted no significant changes in the fatty acid composition of DAG with respect to the control Col-0 plants or the non-erucic accumulating 8.2.2 line (Fig. 6). In all cases, palmitic (16:0) and oleic acids were the major fatty acids in DAG.
Fatty acid composition of the DAG fraction obtained from total seed-lipids from Arabidopsis transgenic lines expressing the Pennycress FAE1 gene under the control of the seed-specific OLE2 promoter. The data were obtained from Arabidopsis transgenic lines from the Col-0 background. Data from three different T3 homozygous lines from each background representative from ten lines analysed are shown. The data were obtained from three independent pools of seeds from five plants of each line
Expression analysis of genes of the triacylglycerol biosynthetic pathway in the transgenic lines
We then monitored the expression of different genes from the triacylglycerol biosynthetic pathway in the transgenic lines. The study was focused in the transgenic lines obtained from the Col-0 background since the fae1-1 mutation could introduce additional effects in the expression of triacylglycerol biosynthetic genes and secondly, because these lines showed bigger increases in VLCFA/erucic acid content. Both 7.1.1 and 8.1.1 lines, which showed higher erucic acid content, showed higher expression of the FAE1 gene (Fig. 7). However, expression of FAE1 in the 8.2.2 plants was not significantly altered (Fig. 7), consistent with the lack of significant changes in the seed oil composition in these lines. It is worth mentioning that given the high sequence identity in the coding regions of both Pennycress and Arabidopsis FAE1 genes, as well as the elimination of UTR regions in the Pennycress transgene used for transformation, it was hardly difficult to differentiate the expression of both genes in our transgenic lines. To solve this problem, at least partially, we used 3′-UTR primers to monitor the expression of the endogenous AtFAE1 gene. No significant changes in AtFAE1 mRNA levels was detected in our transgenic lines, suggesting that the higher erucic acid content correlated well with the expression of the TaFAE1 transgene.
qRT-PCR expression analysis of genes (FAE1, AtFAE1, AtPDAT1, AtLPCAT1 and AtDGAT1) involved in the biosynthesis of VLCFA and triacylglycerol in Arabidopsis transgenic lines expressing the Pennycress FAE1 gene under the control of the seed-specific OLE2 promoter in the Col-0 background. The data were obtained from three independent pools of seed from five plants of each line. Data represent means of at least three biological replicates
Our expression analysis showed reduced expression of the AtPDAT1 gene in the transgenic lines with higher erucic acid content that was not observed in Col-0 or the 8.2.2 transgenic lines, that showed erucic acid levels similar to those from the wild-type (Fig. 7). This reduction of AtPDAT1 mRNA levels was accompanied by a small but significant reduction of AtLPCAT1 mRNA levels in the transgenic lines (Fig. 7). These results suggest that this pathway showed reduced activity in the Arabidopsis transgenic lines showing higher erucic acid levels.
DAG is converted to TAG by DGAT1. Our data indicated a small (less than twofold) but significant increase in the expression of the AtDAGT1 gene in 7.1.1 and 8.1.1 lines, but not in 8.2.2 ones (Fig. 7), that could be consistent with the higher erucic acid content in 7.1.1 and 8.1.1 lines.
Discussion
Brassicaceae represent a very heterogeneous family in terms of erucic acid content in their seed oil, with values ranging from 2.5% (Arabidopsis thaliana), to higher than 50% (like Brassica napus or Crambe abyssinica). This different erucic acid content can even occur within members of the same genus. It is the case of Thlaspidae, where T. arvense shows a 40% average of erucic acid in its seed oil while T. caereulens, has very low (2.5%) erucic acid content, similar to that from Arabidopsis (Claver et al. 2017). The reasons of this heterogeneity in VLCFA content remain still unclear. Of the four enzymes that form part of the elongase complex, both the reductases and the dehydratase are expressed in the whole plant, and are common to other microsomal systems including those involved in the synthesis of waxes or sphingolipids (Post-Beittenmiller 1996; Millar and Kunst 1997). Since the FAE1 enzyme has seed-specific expression and catalyses the rate-limiting step in the elongation of oleyl-CoA to eicosenoyl-CoA (Millar and Kunst 1997; Katavic et al. 2001), the most obvious explanation for the differences in VLCFA content in Brassicaceae should be related with differences at the gene sequence/protein structure that modify FAE1 activity. Consistent with this hypothesis, the phylogenetic analysis was capable to separate low erucic acid containing species (Arabidopsis thaliana) from high accumulating ones (Thlaspi arvense, Brassica napus, Crambe abyssinica; Supplementary Fig. S1). Furthermore, the phylogenetic analysis was capable to group in a different clade two species like Lymnanthes and Jojoba that accumulate eicosenoic acid as major VLCFA species instead of erucic acid (Supplementary Fig. S1; Mietkiewska et al. 2004; Sun et al. 2013). These results suggested the existence of differences in the gene sequence/protein structure of the FAE1 enzyme behind the high erucic acid trait. However, when these differences were analysed into more detail (Fig. 2 and Supplementary Fig. S2), they were limited to a reduced number of residues, not located or being part of regions involved in the activity of the enzyme. In fact, all the residues involved in KCS-FAE1 activity previously identified by Ghanevati and Jaworski (2001; 2002), were conserved in all the FAE1 sequences analysed in this work, independently of their low/high erucic acid content. This included Ser282 that has been related with the low erucic acid trait in B. napus (Han et al. 2001; Katavic et al. 2002). Furthermore, only two out of the seven motifs proposed by Sun et al. (2013) as representative of the high erucic acid trait, motif 1 (KE) and motif 3 (YN), were also consistent with our analysis. These results suggest that although certain residues could be behind a higher β-ketoacyl-CoA synthase activity, other factors might also be involved in the high heterogeneity of erucic acid content among brassicaceae. In that sense, the FAE1 gene has been sequenced in many plant species including low (Arabidopsis) or high erucic ones (Brassica, Crambe, Pennycress), and, in some of these cases (Arabidopsis or Brassica), functional analysis of the FAE1 genes was also performed (Millar and Kunst 1997; Katavic et al. 2001; Blacklock and Jaworski 2002). These analyses have suggested that the substrate specificities of the FAE1 enzyme might play a role in determining the length and unsaturation degree of the VLCFA pool (Lassner et al. 1996; Millar and Kunst 1997). To gain further insight into the different activity and the different acyl-substrate specificity among FAE1 enzymes, we carried out a functional analysis of the Pennycress FAE1 enzyme in Arabidopsis. The Pennycress FAE1 enzyme was a good choice because of its similarities in growth cycle, plant development or gene structure with respect to Arabidopsis (Claver et al. 2017), apart from its interest as a new biofuel feedstock (Moser 2012). The utilization of two different Arabidopsis genetic backgrounds like Col-0 and the fae1-1 mutant was key to understand the different substrate specificities between the Arabidopsis and the Pennycress FAE1 enzymes. The acyl-CoA pool of Col-0 plants is enriched in palmitoyl (15%), linoleyl (20%) and eicosenoyl (22%) CoA moieties (Li-Beisson et al. 2013; Yurchenko et al. 2014). Oleyl-CoA pool represents a 7–10% of total acyl-CoA moieties in Col-0 plants (Li-Beisson et al. 2013; Yurchenko et al. 2014). On the other hand, in the fae1-1 mutant, the acyl-CoA pools is completely devoid of VLCFA, being oleyl-CoA and oleic acid the major species, representing a 25% of total acyl-CoA moieties (Katavic et al. 1995). Nevertheless, both the Col-0 and the fae1-1 mutant have, a priori, oleyl-CoA enough to act as a substrate of the successive elongation to eicosenoic and erucic acid by the Pennycress FAE1 enzyme. Our data showed that seed-specific expression of the Pennycress FAE1 gene in Col-0 or fae1-1 mutant backgrounds resulted in an increase in the erucic acid content in the seed oil from the transgenic lines from 2.5 to 7–9% (threefold), in the case of the Col-0 derived transgenic lines, or from 0.04 to 0.3 (sevenfold), in the case of the fae1-1 mutant derived lines (Figs. 4 and 5). The results obtained in the Col-0 derived transgenic lines were in the same range of those obtained with 35S overexpression of the Arabidopsis FAE1 gene (Millar and Kunst 1997) or the seed-specific expression of the Nasturtium FAE1 gene in Arabidopsis (Mietkiewska et al. 2004). The fact that there were no great differences between the 35S constitutive promoter used by Millar and Kunst (1997) and our data obtained with the seed-specific OLE2 promoter, suggests that there is a limit in the amount of erucic acid that can be synthesized in Arabidopsis that might not be directly related with the activity of the FAE1 enzyme, but probably with the own capacity of each plant species to accumulate TAG in its seed oil and to esterify acyl-CoA moieties to TAG. It is worth mentioning that in the transgenic lines derived from the Col-0 background, the increase of erucic acid was concomitant with a decrease of eicosenoic acid levels, which are already high in Col-0 (i.e., 20% of total fatty acids in seed oil; Fig. 4a). Interestingly, no changes in oleic acid levels were detected in the same lines, suggesting that the Pennycress FAE1 enzyme was capable to elongate efficiently the Arabidopsis eicosenoyl-CoA pool much better than the oleyl one. This conclusion was supported by the observation that in the fae1-1 derived lines, the extent of oleic acid elongation to eicosenoic acid, although occurred, was rather low (Figs. 4b and 5b). Given the huge amount of oleyl-CoA available for elongation in this mutant, there was substrate enough for efficient elongation of oleyl-CoA to eicosenoyl-CoA and then to erucoyl-CoA. These results suggested that the Pennycress FAE1 enzyme showed higher substrate affinity for eicosenoyl-CoA than the Arabidopsis one, confirming in planta previous yeast complementation studies (Lassner et al. 1996; Millar and Kunst 1997). The intriguing question is why in Pennycress plants, the FAE1 enzyme is perfectly capable to efficiently use the oleyl-CoA pool to synthetize eicosenoic acid and then erucic acid and very early during seed maturation (Claver et al. 2017). Since the chemical nature of the acyl-CoA moieties is essentially the same in both species, other factors, like the inefficient interaction of the Pennycress FAE1 β-ketocacyl-CoA synthase with the other three enzymes that participate in the elongation reaction could also be determinant for an efficient elongation of both oleyl and eicosenoyl-CoA pools. However, these three enzymes are the same for the elongation of oleic acic to eicosenoic acid and from then to erucic acid. Other possible explanation could be related with the fact that FAE1 is going to compete with other enzymes like GPAT, LPAT, DGAT or LPCAT for the oleyl-CoA substrate while such competition is not stablished for eicosenoyl-coA that is directly channelled to the elongase complex. Understanding this will be of great help for the biotechnological manipulation of the erucic acid trait in the seed oil in Brassicaceae.
Lipid class analysis of oilseed fraction in our transgenic lines showed that erucic acid was incorporated to TAG without a significant accumulation of VLCFA in DAG (Fig. 6). Without precluding the possibility of a rapid conversion of DAG to TAG that might keep constant the erucic acid content in DAG, our data suggested that the incorporation of erucic acid to TAG may occur preferentially through the Kennedy pathway rather than from PC/DAG interconversion. Our data suggest that this incorporation may occur during the third acylation from DAG to TAG via DGAT1 and not till the very beginning of the Kennedy pathway through the acylation of glycerol-3-phosphate or LPA through GPAT and LPAT activities (Bates et al. 2013). The preference of GPAT and LPAT enzymes for 18 carbon substrates has been already reported in Arabidopsis (Wang et al. 2012; Li-Beisson et al. 2013; Guan et al. 2014). In that sense, the DGAT1 enzyme from certain varieties of rapeseed with high erucic acid content were reported to have higher affinity for VLCFA than low erucic acid ones (Bernerth and Frentzen 1990; Weselake et al. 1991). More recently, it has been suggested that erucic acid accumulation in TAG from Crambe abyssinica should occur through the activation of specific DGAT isozymes specialized in the incorporation of these VLCFA (Guan et al. 2014). However, the gene search in the Crambe genome for these isozymes did not identify such isoforms (Guan et al. 2014). Moreover, in Arabidopsis, PC/DAG interconversion is the major route for DAG biosynthesis in the seed (Bates and Browse 2012). However, the fact that i) no erucic acid was accumulated in the DAG fraction in our transgenic lines and ii) expression of genes responsible of acyl-editing like AtPDAT1 and AtLPCAT1 decreased in the transgenic lines accumulating erucic acid, suggests that the contribution of the PC/DAG interconversion pathway would be modified in our transgenic lines to favour the incorporation of erucic acid to TAG via DGAT1. This hypothesis was further strengthened by the observation that AtDGAT1 expression increased in the transgenic lines accumulating erucic acid in their seed oil. Consistent with this, Guan et al. (2014) demonstrated that Crambe seeds (high erucic trait) have low PC/DAG interconversion rates and that erucic acid was incorporated to TAG through the Kennedy pathway/DGAT1, as occurred in our transgenic lines. A possible explanation for this DGAT preference could be related with the fact that the erucoyl-CoA synthetized by the elongase complex can be immediately incorporated to TAG via DGAT1, but not from PDAT activity, in order to avoid that unusual fatty acids, like erucic acid, might be incorporated into membrane lipids. Future labelling studies will help to quantify the fluxes between both pathways either in Pennycress or in the transgenic lines expressing the Pennycress FAE1 enzyme. Nevertheless, these results suggest that not only the size of the acyl-CoA pools, but also the pathway used for TAG biosynthesis would determine the extent of VLCFA incorporation to TAG in plant seed oils and explain the differences among Brassicaceae.
References
Bates PD, Browse J (2012) The significance of different diacylglycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci 3:147–150
Bates PD, Stymne S, Ohlrogge J (2013) Biochemical pathways in seed oil synthesis. Curr Op Plant Biol 16:358–364
Bernerth R, Frentzen M (1990) Utilization of erucoyl-CoA by acyltransferases from developing seeds of Brassica napus (L.) involved in tryacylglycerol biosynthesis. Plant Sci 67:21–28
Blacklock BJ, Jaworski J (2002) Studies into factors contributing to substrate specificity of membrane-bound 3-ketoacyl-CoA synthases. Eu J Biochem 269:4789–4798
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Chopra R, Johnson EB, Daniels E, McGinn M, Dorn KM, Esfahanian M, Folstad N, Amundson K, Altendorf K, Betts K, Frels K, Anderson JA, Wyse DL, Sedbrook JC, Marks MD (2018) Translational genomics using arabidopsis as a model enables the characterization of pennycress genes through forward and reverse genetics. Plant J 96:1093–1105
Claver A, Rey R, López MV, Picorel R, Alfonso M (2017) Identification of target genes and processes involved in erucic acid accumulation during seed development in the biodiesel feedstock Pennycress (Thlaspi arvense L.). J Plant Physiol 208:7–16
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Demski K, Jeppson S, Lager I, Mistak A, Jasieniecka-Gazarkiewicz K, Waleron M, Stymne S, Banas A (2019) Isoforms of Acyl-CoA:diacylglycerol acyltransferase2 differ substantially in their specificities toward erucic acid. Plant Physiol 181:1468–1479
Doyle JJ, Doyle JL (1990) Isolation of fresh DNA from fresh tissue. Focus 12:13–15
Dorn KM, Fankhauser JD, Wyse DL, Marks MD (2013) De novo assembly of the pennycress (Thlaspi arvense) transcriptome provides tools for the development of a winter cropland biodiesel feedstock. Plant J 75:1028–1038
Fan J, Shonnard DR, Kalnes TN, Johnsen PB, Rao S (2013) A life cycle assessment of pennycress (Thlaspi arvense L.)–derived jet fuel and diesel. Biomass Bioener 55:87–100
Garcés R, Mancha M (1993) One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal Biochem 211:139–143
Ghanevati M, Jaworski JG (2001) Active-site residues of a plant membrane-bound fatty acid elongase beta-ketoacyl- CoA- synthase, FAE1 KCS. Biochim Biophys Acta 1530:77–85
Ghanevati M, Jaworski JG (2002) Engineering and mechanistic studies of the Arabidopsis FAE1 beta-ketoacyl- CoA- synthase, FAE1 KCS. Eur J Biochem 269:3531–3539
Guan R, Lager I, Li X, Stymne S, Zhu L-H (2014) Bottlenecks in erucic acid accumulation in genetically engineered ultrahigh erucic acid Crambe abyssinica. Plant Biotech J 12:193–203
Guindon S, Gascuel O (2003) A simple, fast and accurate algorithm to estimate large phylogenies by maximum likehood. System Biol 52:696–704
Han J, Lühs W, Sonntag K, Zähringer U, Borchardt DS, Wolter FP, Heinz E, Frentzen M (2001) Functional characterization of beta-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol Biol 46:229–239
Harwood JL (1996) Recent advances in the biosynthesis of plant fatty acids. Biochim Biophys Acta 1301:7–56
Jaworski J, Cahoon EB (2003) Industrial oils from transgenic plants. Curr Op Plant Biol 6:178–184
Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou JT, Mackenzie SL, Covello PS, Kunst L (1995) Alteration of seed fatty acid composition by an ethylmethanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108:399–409
Katavic V, Friesen W, Barton DL, Gossen KK, Giblin EM, Luciw T, An J, Zou J-T, Mackenzie SL, Keller WA, Males D, Taylor DC (2001) Improving erucic acid content in rapeseed through biotechnology: what can the Arabidopsis FAE1 and the yeast SLC1-1 genes contribute. Crop Sci 41:739–747
Katavic V, Mietkiewska E, Barton DL, Giblin EM, Reed DW, Taylor DC (2002) Restoring enzyme activity in non-functional low erucic Brassica napus fatty acid elongase 1 by a single aminoacid substitution. Eur J Biochem 269:5625–5631
Lassner MW, Lardizabal K, Metz JG (1996) A jojoba beta-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8:281–292
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durret TP et al (2013) Acyl-lipid metabolism. Arabidopsis book 11:161
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time PCR and the 2(-delta Delta C(T) method. Methods 25:402–408
Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ (2012) Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog Lipid Res 10:350–377
McGinn M, Phippen WB, Chopra R, Bansal S, Jarvis BA, Phippen ME, Dorm KM, Esfahanian M, Nazarenus TJ, Cahoon EB, Durrett TP, Marks MD, Sedbrook JC (2018) Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnol J. https://doi.org/10.1111/pbi.13014
Merril AHJ, Schmelz EM, Wang E, Dillehay DL, Rice LG, Meredith F, Riley RT (1997) Importance of sphingolipids and inhibitors of sphingolipid metabolism as components of animal diets. J Nutr 127:830–833
Mietkiewska E, Giblin EM, Wang S, Barton DL, Dirpaul J, Brost JM, Katavic V, Taylor DC (2004) Seed specific heterologous expression of a Nastortium FAE gene in Arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol 136:2665–2675
Mietkiewska E, Brost JM, Giblin EM, Barton DL, Taylor DC (2007) Cloning and functional characterization of the fatty acid elongase 1 (FAE1) gene from high erucic Crambe abyssinica cv Prophet. Plant Biotechnol 5:636–645
Millar AA, Kunst L (1997) Very-long chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J 12:121–131
Moser BR, Knothe G, Vaughn SF, Isbell TA (2009) Production and evaluation of biodiesel from field Pennycress (Thlaspi arvense L.) oil. Energy Fuels 23:4149–4155
Moser RB (2012) Biodiesel from alternative oilseed feedstocks: camelina and filed pennycress. Biofuels 3:193–209
Post-Beittenmiller D (1996) Biochemistry and molecular biology of wax production in plants. Ann Rev Plant Physiol Plant Mol Biol 47:405–430
Román A, Hernández ML, Soria-García A, López-Gomollón S, Lagunas B, Picorel R, Martínez-Rivas JM, Alfonso M (2015) Non-redundant contribution of the plastidial FAD8 omega-3 desaturase to glicerolipid unsaturation at different temperatures in Arabidopsis. Mol Plant 8:1599–1611
Singh SP, Zhou X-R, Liu Q, Stymne S, Green A (2005) Metabolic engineering of new fatty acids in plants. Curr Op Plant Biol 8:197–203
Sonntag NOV (1995) Industrial utilization of long-chain fatty acids and their derivatives. In: Kimber DS, McGregor DI (eds) Brassica oilseeds. CAB International, Oxon, pp 339–352
Sun X, Pang H, Guo H, Yan Q, Hang Y (2013) Evolutionary pattern of the FAE1 gene in Brassicaceae and its correlation with the erucic acid trait. PLoS ONE 8:1–12
Taylor DC, Kunst L, MacKenzie SL (1993) Bioassembly of storage lipids in oil-seed crops; Target: Trierucin. In: Janick J, Simon JE (eds) Progress in New Crops. Wiley, New York, pp 181–191
Thelen JJ, Ohlrogge JB (2002) Metabolic engineering if fatty acid biosynthesis in plants. Met Eng 4:12–21
Tsogtbaatar E, Cocuron JC, Corchado Sonera M, Alonso AP (2015) Metabolite fingerprinting of pennycress (Thlaspi arvense L.) embryos to assess active pathways during oil synthesis. J Exp Bot 66:4267–4277
Von Wettstein-Knowles PM (1982) Elongase and epicuticular wax biosynthesis. Physiol Vegetal 20:797–809
Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, Stymne S, Weselake RJ, Zou J (2012) Metabolic interactions between the lands Cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 24:4652–4669
Weselake R, Taylor DC, Pomeroy MK, Lawson SL, Underhill EW (1991) Properties of diacylglycerol acyltransferase from microspore-derived embryos of Brassica napus. Phytochemistry 30:3533–3550
Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PBE, Harwood JL (2009) Increasing the flow of carbon into seed oil. Biotechnol Adv 27:866–878
Wu YH, Wu G, Xiao L, Cao YL, Lu CM (2009) Discovery of low erucic acid wild species and functional characterization of FAE1 genes in crucifer species. Sci Agric Sin 42:3819–3827
Yurchenko O, Singer SD, Nykiforuk CL, Gidda S, Mullen RT, Moloney MM, Weselake RJ (2014) Production of a Brassica napus low-molecular mass Acyl-Coenzyme A-binding protein in Arabidopsis alters the acyl-Coenzyme A pool and acyl composition of oil in seeds. Plant Physiol 165:550–560
Acknowledgements
This research was supported by grants RTC2017-6461-3 from Ministerio de Economía y Competitividad (MINECO) and RTI2018-097755-B-100 from Ministerio de Ciencia, Innovación y Universidades (MICINN) to MA. Funding from Gobierno de Aragón (A09-20R; Function of genes and plant proteins) is also acknowledged. Authors wish to thank Patricia Lorente for skilful technical assistance.
Author information
Authors and Affiliations
Contributions
MA, MVL and RP conceived the research; AC, RRG, MAL and MdlV, performed the experiments; MA, RRG and MVL analysed the data; MA wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Research involving human and animal rights
This study did not involve human and animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2020_1042_MOESM1_ESM.pptx
Supplementary Fig. 1 Phylogenetic tree of the FAE1 gene from different accessions representing species with different erucic acid content. The tree was obtained with boot-straps 500 using the PHYML software. The names and GENBANK accession numbers of the sequences used for the analysis were: Arabidopsis lyrata (GU929425.1); Arabidopsis thaliana (U29142.1); Brassica carinata (KF664166); Brassica napus (U50771.1); Brassica oleracea (GU325726.1); Brassica rapa (GU325723.1); Camelina sativa (KF030545); Capsella bursa-pastoris (KF664171); Cardamine parviflora (KF664172); Crambe abyssinica (KC565742); Crambe hispanica (KF664157); Isatis tinctoria (AY888038.1); Lepidium apetala (KF030527); Lepidium campestre (FJ907545.1); Lymnanthes douglasii (AF247134); Orychophragmus violaceus (AY888042.1); Raphanus raphanistrum (KF664162); Simondsia chinensis (U37088); Sinapsis arvense (AY888041.1); Sinapsis alba (AY888040.1); Thlaspi arvense (KT223024) and Thlaspi perfoliatum (KF030530). Supplementary file1 (PPTX 57 kb)
Rights and permissions
About this article
Cite this article
Claver, A., de la Vega, M., Rey-Giménez, R. et al. Functional analysis of β-ketoacyl-CoA synthase from biofuel feedstock Thlaspi arvense reveals differences in the triacylglycerol biosynthetic pathway among Brassicaceae. Plant Mol Biol 104, 283–296 (2020). https://doi.org/10.1007/s11103-020-01042-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01042-7