Abstract
The mechanism by which endogenous salicylic acid (SA) regulates leaf senescence remains elusive. Here we provide direct evidence that an enhancement of endogenous SA level, via chemical-induced upregulation of ISOCHORISMATE SYNTHASE 1 (ICS1), could significantly accelerate the senescence process of old leaves through mediation of the key SA signaling component NON EXPRESSOR OF PATHOGENESIS RELATED GENES 1 (NPR1) in Arabidopsis. Importantly, by taking advantage of this chemically induced leaf senescence system, we identified a mitogen-activated protein kinase (MAPK) cascade MKK4/5-MPK1/2 that is required for the SA/NPR1-mediated leaf senescence. Both MKK4/5 and MPK1/2 exhibited SA-induced kinase activities, with MPK1/2 being the immediate targets of MKK4/5. Double mutants of mkk4 mkk5 and mpk1 mpk2 displayed delayed leaf senescence, while constitutive overexpression of the kinase genes led to premature leaf senescence. Such premature leaf senescence was suppressed when they were overexpressed in an SA synthesis defective mutant (sid2) or signaling detective mutant (npr1). We further showed that MPK1, but not MPK2, could directly phosphorylate NPR1. Meanwhile, MPK1 also mediated NPR1 monomerization. Notably, induction of disease resistance was significantly compromised in the single and double mutants of the kinase genes. Taken together, our data demonstrate that the MKK4/5-MPK1/2 cascade plays a critical role in modulating SA signaling through a complex regulatory network in Arabidopsis.
Key message
We reveal a MAPK cascade, MKK4/5-MPK1, which not only phosphorylates the key SA signaling component NPR1 specifically, but also regulates its oligomer-to-monomer conformational change somehow via mediation of TRX-h3/5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf senescence is a genetically programmed active process for facilitating plant development via massively remobilizing nutrients from senescing leaves to rapidly developing organs and eventually to reproductive/storage organs (Lim et al. 2007; Masclaux-Daubresse and Chardon 2011). The process is regulated by both developmental and environmental cues, which are mostly mediated by phytohormones. Ethylene (ET), abscisic acid (ABA) and jasmonic acid (JA) are positive regulators, whereas cytokinins and auxins act to delay initiation/progression of leaf senescence (Gan and Amasino 1997; Graaff et al. 2006; Kusaba et al. 2013; Jibran et al. 2013). Salicylic acid (SA) is a key mediator of disease resistance in plants (Malamy et al. 1990; Metraux et al. 1990; Gaffney et al. 1993; Delaney et al. 1994; Wildermuth et al. 2001). However, its role in leaf senescence has been largely unclear (Raskin 1992; Vlot et al. 2009), mainly because that mutants or transgenic plants defective in SA biosynthesis/accumulation or signaling showed only a marginal alteration, if any, in their senescence phenotype, albeit a number of genes showed altered expression patterns (Morris et al. 2000). Recently, Zhang et al. reported that the null mutation of SA 3-HYDROXYLASE (S3H) gene, encoding an enzyme that converts SA to 2,3-dihydroxybenzoic acid, resulted in an elevated level of endogenous SA and precocious leaf senescence, suggesting that SA may positively regulate leaf senescence (Zhang et al. 2013).
SA biosynthesis and its signaling pathway during pathogen infection are well documented. Pathogen-induced SA synthesis undergoes predominantly the two sequential reactions catalyzed by isochorismate synthase (ICS) and putative isochorimate pyruvate lyase (IPL), respectively, with chorismate serving as the initial substrate (Wildermuth et al. 2001). In Arabidopsis, there are two isochorismate synthases called ICS1 (also named as SID2) and ICS2, with ICS1 acting as the major enzyme (Wildermuth et al. 2001). Pathogen-induced increase in endogenous SA level or exogenous application of SA leads to the growth-to-defense transition with changes in the expression of thousands of genes. A core component of the SA signaling pathway is NON EXPRESSOR OF PATHOGENESIS RELATED GENES 1 (NPR1), a transcriptional coactivator that plays a pivotal role in the plant systemic acquired resistance (SAR) (Cao et al. 1997; Mark et al. 2000). Under normal growth conditions, NPR1 exists as oligomers and resides in the cytoplasm. Upon pathogen infection or SA treatment, NPR1 oligomers are reduced to monomers, which relocate to the nucleus where they are subject to phosphorylation. Phosphorylation of NPR1 is important for its role in gene activation and also facilitates its nuclear import and turnover (Mou et al. 2003; Lee, et al. 2015). It has been shown that NPR1 phosphorylation occurs at residues Ser11/Ser15 (Spoel et al. 2009); however, the kinase(s) responsible for NPR1 phosphorylation at these residues has yet to be identified.
The mitogen-activated protein kinase (MAPK) cascade represents one of the most highly conserved signaling pathways and is involved in various biotic and abiotic stress responses, hormone signaling, cell proliferation, and other developmental processes (Nakagami et al. 2005). In Arabidopsis, there are 60–80 predicted MAP kinase kinase kinases (MAPKKKs), 10 MAP kinase kinases (MAPKKs), and at least 20 MAP kinases (MAPKs), which can theoretically generate twenty to thirty thousand distinct MAPKKK-MAPKK-MAPK modules. A battery of MAPK, including MAPKKK8 (MEKK1), MKK1/2/4/5 and MPK3/4/6, have been characterized to play key roles in plant immune responses, possibly through activation of WRKY transcription factors (TFs) (Asai et al. 2002; Gao et al. 2008). Moreover, genetic analyses have demonstrated that MAPKKK8, MKK9 and MPK6 are involved in regulation of leaf senescence (Zhou et al. 2009; Miao et al. 2007), suggesting the complexity and overlapping functions of the MAPK-signaling network in mediating diverse biological processes.
Probenazole (3-allyloxy-1,2-Benzisothiazole-1,1-dioxide, PBZ), an active ingredient of a widely used agrochemical Oryzemate, can activate SA/NPR1 signaling pathway to efficiently trigger SAR (Yoshioka et al. 2001). We previously showed that PBZ enhances endogenous SA level via ICS1 dependent biosynthesis pathway (Yu et al. 2010). In this study, we demonstrate that PBZ can also induce leaf senescence through the SA signaling pathway. Importantly, we reveal a MAPK signaling module MKK4/5-MPK1/2 that is responsible for NPR1 phosphorylation and monomerization, which is critical for both the PBZ-induced leaf senescence and disease resistance.
Results
PBZ accelerates the senescence of old leaves via SA biosynthesis and NPR1 signaling in Arabidopsis
Although SA has been shown to regulate the expression of many genes during leaf senescence, the fact that the mutants defective in SA biosynthesis and signaling have negligible senescence-related phenotypes makes it difficult to evaluate the exact role of SA in regulating leaf senescence (Morris et al. 2000). We have previously shown that PBZ is able to enhance the levels of both free and total endogenous SA via ICS1-mediated SA biosynthesis when applied by root drenching (Yu et al. 2010). Intriguingly, we also detected an accelerated senescence of old leaves about 2 weeks after PBZ treatment (Fig. 1a, b). To examine if the accelerated leaf senescing phenotype was caused by the enhanced SA biosynthesis, we treated Arabidopsis plants grown under short day-growth conditions for 5 weeks with SA of different concentrations through root drenching and indeed observed a similar yellowing phenotype on older leaves three days after treatment when 1.5 mM SA was applied (Supplementary Fig. 1). Considering the ease and a more typical symptom obtained with PBZ, as well as being convinced by the following genetic data, we continued to use PBZ to treat Arabidopsis plants in further experiments.
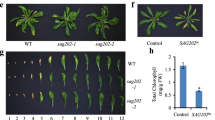
PBZ promotes the senescence process of old leaves. a Plants of indicated genotypes were grown under short day-growth conditions for 5 weeks and treated with or without 0.5 mM PBZ through root drenching. Pictures were taken at 12 DAP (days after PBZ treatment). b Leaf phenotypes of indicated genotypes at 12 DAP. Unless stated otherwise, the 5th through the 8th leaves were taken for further physiological and molecular analyses. Chl contents (c) and ion leakages (d) of Col-0, sid2, npr1, and 35S::NPR1 plants at 12 DAP. Error bars indicate SD (n = 3), Fisher’s least significant difference (LSD) ANOVA test, P < 0.05. Changes in the expression of a leaf senescence marker gene SAG12 (e) and a photosynthesis marker gene CAB (f) in the leaves of indicated genotypes at 12 DAP. ACTIN2 was included as an internal control. Error bars indicate SD (n = 3), LSD ANOVA test, P < 0.05. Fv/Fm ratios (g) and Fv′/Fm′ ratios (h) in the leaves of indicated genotypes at 12 DAP. Error bars indicate SD (n = 3), LSD ANOVA test, P < 0.05
The 5th through the 8th true leaves were chosen for physiological and molecular analyses as they exhibited the most significant leaf yellowing phenotype (Fig. 1b). In addition to a reduced content of chlorophyll (Chl), other typical changes of senescence-associated parameters, including an increased level of ion leakage, induction of a senescence marker gene (SAG12), down-regulation of a photosynthetic gene (CAB), and reduced levels of photo-chemical quantum efficiency of photosystem II (Fv/Fm ratio and Fv′/Fm′ ratio), were also observed at 12 DAP (days after 0.5 mM PBZ treatment), demonstrating that PBZ can efficiently promote the senescence process of old leaves in our system (Fig. 1c–h). Importantly, no obvious signs of senescence symptoms were observed in an SA biosynthesis mutant sid2 (sid2-2) and SA signaling mutant npr1 (npr1-1), while constitutive overexpression of NPR1 (35S::NPR1) led to an accelerated leaf yellowing, confirming that PBZ promotes the senescence of old leaves through SA biosynthesis and signaling (Fig. 1).
MKK4/5 and MPK1/2 are required for the SA-accelerated senescence of old leaves
The MAPK cascade signaling plays critical roles in various signal transduction pathways of plants, including disease resistance and leaf senescence (Asai et al. 2002; Zhou et al. 2009; Zhao et al. 2014). Our microarray and qPCR analyses revealed that the expressions of one MAPKKK gene (MEKK3), five MAPKK genes (MKK1, MKK2, MKK4, MKK5 and MKK9) and five MAPK genes (MPK1, MPK2, MPK4, MPK7 and MPK11) were significantly up-regulated at 3 DAP, as compared with those by mock treatment (Fig. 2a; Supplementary Table 1). To determine if these genes were involved in mediating the PBZ-accelerated senescence, we identified T-DNA insertion mutants of these up-regulated genes (Supplementary Fig. 2; Supplementary Table 1). Single knock-down or knock-out mutants of MKK4, MKK5, MPK1 and MPK2 showed moderate to slight compromises in senescence symptom, as compared to wild-type plants (WT) (Fig. 2b–f). Previous studies showed that MKK4 and MKK5 are grouped together, and so are MPK1 and MPK2, based on a phylogenetic analysis, indicating a potential redundancy of their functions (Kazuya et al. 2002). We thus generated a double mutant (mpk1 mpk2) of MPK1 and MPK2 with their single knock-out mutants (SALK_063847C, SALK_047422C) and a double mutant (mkk4 mkk5) of MKK4 and MKK5 with their single knock-down mutants (SALK_018804C, SALK_050700) since loss-of-function of both MKK4 and MKK5 (mkk4* and mkk5* mutants were kindly provided by Dr. Dingzhong Tang) causes lethality (Wang et al. 2007). In our mkk4 mkk5, ~ 80% and ~ 60% of their expressions were eliminated for MKK4 and MKK5, respectively (Supplementary Fig. 2). As expected, both mkk4 mkk5 and mpk1 mpk2 exhibited more significant reductions in the extent of the PBZ-accelerated senescence (Fig. 2b–f). Notably, loss-of-function mutation of either MKK9 or MPK6, both of which are involved in a MAPK cascade shown to play an important role in natural leaf senescence (Zhou et al. 2009), had no effect on the PBZ-accelerated senescence (Supplementary Fig. 3).
Involvements of MKK4/5 and MPK1/2 in the regulation of PBZ-induced leaf senescence. a Expressions of MKK4/5 and MPK1/2 in response to PBZ treatment. Error bars indicate SD (n = 4), paired Student’s t tests, **P < 0.01. b Leaf phenotypes of indicated genotypes at 12 DAP. Chl contents (c) and ion leakages (d) of indicated genotypes at 12 DAP. Error bars indicate SD (n = 4), LSD ANOVA test, P < 0.05. Changes in the expressions of SAG12 (e) and CAB (f) in the leaves of indicated genotypes at 12 DAP. Expressions of SAG12 (e) and CAB (f) in water treated plants were arbitrarily set to 1. ACTIN2 was included as an internal control. Error bars indicate SD (n = 3), LSD ANOVA test, P < 0.05
SA biosynthesis is required for the phosphorylation of MKK4/5 and MPK1/2
To understand the action modes of MKK4/5 and MPK1/2 and their molecular links to SA biosynthesis and signaling, we established a protoplast-based transient protein expression system to monitor the dynamics of protein phosphorylation upon SA treatment. As a preliminary experiment, we transiently expressed NPR1-GFP in WT protoplasts, which were subject to 0.04 mM SA treatment 12 h after transfection. Total proteins were extracted at indicated time points and subject to a Phos-tag mobility shift assay. As expected, a quick induction of NPR1-GFP phosphorylation was detected 1 h after SA treatment, which became more prominent 2 h after treatment (Supplementary Fig. 4a). In contrast, no effect on the phosphorylation of NPR1-GFP was observed upon mock treatment, indicating that this system is reliable for monitoring SA dependent MAPK signaling (Yoo et al. 2007) (Supplementary Fig. 4a).
Since phosphorylation of MKKs and MPKs is a prerequisite for their activation, we subsequently examined the phosphorylation status of MKK4/5 and MPK1/2 in response to SA treatment. As in the case of NPR1, a gradual increase of phosphorylated forms of MKK4-4 × MYC/MKK5-4 × MYC and MPK1-4 × FLAG/MPK2-4 × FLAG were obviously observed 2 h after SA treatment (Fig. 3a, b; Supplementary Fig. 4b). We also observed that fractions of MKK5-4 × MYC and MPK1-4 × FLAG were phosphorylated before SA treatment, which could be induced by the background level of SA likely accumulated during protoplast preparation and/or culture or due to that these kinases may also be able to respond to certain signal molecules other than SA (Fig. 3a, b).
Phosphorylation of MKK4/5 and MPK1/2 depends on in vivo SA biosynthesis. MKK4-4 × MYC/MKK5-4 × MYC (a) and MPK1-4 × FLAG/ MPK2-4 × FLAG (b) were phosphorylated upon SA treatment in a protoplast-based assay. Respective MYC tagged MKK or FLAG tagged MPK proteins were transiently expressed in WT protoplasts for 12 h and then treated with 0.04 mM SA. Proteins were extracted at indicated time points and subject to immunoblot (IB) analysis by an anti-MYC antibody (a) or an anti-FLAG antibody (b) with (top panel) or without (middle panel) the addition of the Phos-tag reagent. IB of ACTIN2 (ACT2) (bottom panel) was used as a control. P, phosphorylated; UP, unphosphorylated. c, d Overexpression of MKK4-4 × MYC, MKK5-4 × MYC, MPK1-4 × FLAG or MPK2-4 × FLAG in Col-0 but not in sid2 caused an obvious leaf yellowing phenotype at 12 DAP. c Leaf phenotypes of indicated genotypes at 12 DAP. d Chl contents of indicated genotypes at 12 DAP. Error bars indicate SD (n = 3), LSD ANOVA test, P < 0.05. e, f, g In vivo phosphorylation of MPK1 or MPK2 in response to PBZ treatment depends on the biosynthesis of endogenous SA. e MPK1 and MPK2 were phosphorylated in WT + MPK1-4 × FLAG and WT + MPK2-4 × FLAG transgenic plants upon PBZ treatment. Phosphorylation status was examined at 6 DAP. f MPK1 and MPK2 were not phosphorylated in sid2 + MPK1-4 × FLAG and sid2 + MPK2-4 × FLAG transgenic plants upon PBZ treatment. g MPK1 and MPK2 were phosphorylated in sid2 + MPK1-4 × FLAG and sid2 + MPK2-4 × FLAG transgenic plants treated with 0.5 mM SA for 24 h. P phosphorylated; UP unphosphorylated. Similar results were obtained from three independent experiments
To determine whether in vivo SA biosynthesis is required for MKK4/5 and MPK1/2 to mediate PBZ-accelerated senescence, we individually overexpressed MKK4, MKK5, MPK1 or MPK2 under the constitutive control of 35S promoter in WT or in sid2. As shown in Fig. 3c, d, transgenic plants overexpressing MKK4/5 or MPK1/2 in WT, but not in sid2, exhibited an accelerated leaf yellowing phenotype, suggesting that in vivo SA biosynthesis is critical for the MAPK cascade mediation of PBZ-accelerated senescence (Fig. 3c, d; Supplementary Figs. 5a–d, 6a, b). We subsequently examined the in vivo phosphorylation status of MPK1 and MPK2 in response to PBZ or SA treatment. Indeed, we detected an accumulation of phosphorylated MPK1 at 6 DAP in the WT + MPK1-4 × FLAG but not in the sid2 + MPK1-4 × FLAG transgenic plants (Fig. 3e, f). However, when we treated the sid2 + MPK1-4 × FLAG leaves with 0.5 mM SA, we could recover a similar phosphorylated band of MPK1-4 × FLAG (Fig. 3g). Similar results were obtained with MPK2 (Fig. 3e–g). Isochorismate Synthase 1 (ICS1) encodes a key enzyme in SA production. We extracted and quantified total salicylic acid (free and sugar-conjugated) in WT, mkk4 mkk5 and mpk1 mpk2 by HPLC as described (Yu et al. 2010), and found there was no significant difference among them (Supplementary Fig. 12). Taken together, these results suggest that PBZ activates the MAPK signaling pathway through biosynthesis of endogenous SA.
Phosphorylation of MPK1/2 requires the full function of MKK4 and MKK5
We then examined whether the activation of MPK1/2 depends on MKK4/5. To this end, MPK1-4 × FLAG or MPK2-4 × FLAG was introduced into the protoplasts prepared from the leaves of mkk4 mkk5. As shown in Fig. 4a, b, neither the background- nor exogenous SA-induced phosphorylation could be detected 2 h after treatment, whereas co-introduction of MKK4-4 × MYC and MKK5-4 × MYC with MPK1-4 × FLAG or MPK2-4 × FLAG into mkk4 mkk5 protoplasts reestablished the phosphorylation of MPK1/2 (Fig. 4a, b), indicating that the phosphorylation of MPK1 and MPK2 depends on the full function of both MKK4 and MKK5. To confirm that the band shift was due to phosphorylation, we treated the protoplasts with λ-PPase, a protein phosphatase that can efficiently remove phosphate groups from phosphorylated residues. As expected, the shifted bands disappeared after λ-PPase treatment (Fig. 4c, d, lane 2), and addition of Na3VO4, a phosphatase inhibitor, abolished the effect of λ-PPase action (Fig. 4c, d, lane 3).
Phosphorylation of MPK1/2 requires the full function of MKK4 and MKK5. Partial loss-of-functions of MKK4 and MKK5 abolished SA-induced phosphorylation of MPK1 (a) and MPK2 (b). Presence of MKK4-4 × MYC and MKK5-4 × MYC recovered the phosphorylation of MPK1-4 × FLAG (c) or MPK2-4 × FLAG (d) in vitro. MKK4-4 × MYC/MKK5-4 × MYC and MPK1-4 × FLAG were individually or together introduced into mkk4 mkk5 protoplasts, which were subsequently subject to phosphorylation analysis. For λ-PPase and Na3VO4 treatments, 10 units µl−1 λ-PPase and/or 0.1 mM Na3VO4 were added to the reaction and incubated at 30 °C for 30 min. Similar results were obtained from three independent experiments
MPK1 can phosphorylate NPR1
Phosphorylation of NPR1 is a key step in SA signaling (Spoel et al. 2009). In light of that NPR1 is critical for PBZ-induced leaf senescence and phosphorylation of NPR1 represents a key event of its functionality during SAR, we hypothesized that NPR1 might be a core downstream component of the MKK4/5-MPK1/2 signaling cascade. If this was the case, we would expect that loss-of-function mutation of NPR1 could block the MKK4/5-MPK1/2 signaling in vivo. Indeed, we found that individual overexpressions of MKK4, MKK5, MPK1 and MPK2 in npr1 failed to exhibit their capability to accelerate leaf senescence upon PBZ treatment, indicating that MKK4/5 and MPK1/2-mediated leaf senescence requires NPR1 (Fig. 5a, b; Supplementary Fig. 5a–h, 6c).
MPK1 phosphorylates NPR1 and mediates NPR1 monomerization. a, b Overexpression of MKK4-4 × MYC, MKK5-4 × MYC, MPK1-4 × FLAG or MPK2-4 × FLAG in npr1 didn't cause an obvious leaf yellowing phenotype at 12 DAP. a Leaf phenotypes of indicated genotypes at 12 DAP. b Chl contents of indicated genotypes at 12 DAP. Error bars indicate SD (n = 3), LSD ANOVA test, P < 0.05. c Phosphorylation status of NPR1-GFP was examined in the protoplasts prepared from indicated genotypes. d NPR1 was phosphorylated in vitro by MPK1 in the presence of ATP. MPK1/MPK2-His and NPR1-GST were expressed in E. coli and purified. The plus and minus signs indicated the presence and absence of ATP and respective proteins. Aliquots of the samples were separated by SDS-PAGE with Phos-tag reagent and subject to IB analysis using a GST antibody. e Oligomer-to-monomer transition of NPR1-GFP in response to SA treatment was blocked in mpk1. For the examination of the oligomer-to-monomer transition of NPR1-GFP, reduced reagent (DTT) was omitted during protein extraction and IB assay. Similar results were obtained from three independent experiments. f Cytoplasmic-to-nucleus translocation of MPK1 after SA treatment. Bar = 15 μm. Similar results were obtained from two independent experiments
NPR1, as a transcriptional coactivator, acts together with TGA transcription factors to regulate PR expression (Després et al. 2003). We examined whether MPK1 or MPK2 directly regulates upstream regulators of plant immunity response, such as NPR1. Yeast two-hybrid assays showed that only MPK1 interacted strongly with NPR1 (Supplementary Fig. 11). We then examined whether MPK1 or MPK2 is required for phosphorylation of NPR1 in response to SA treatment. Protoplasts were prepared from the leaves of WT + NPR1-GFP, mpk1 + NPR1-GFP and mpk2 + NPR1-GFP plants, and treated with 0.04 mM SA. Instead of a quick induction of NPR1 phosphorylation in WT, the SA-induced NPR1 phosphorylation was nearly abolished in mpk1 background (Fig. 5c). Intriguingly, loss-of-function of MPK2 didn’t show an obvious effect on NPR1 phosphorylation. An in vitro phosphorylation assay was carried out with MPK1, MPK2 and NPR1 proteins expressed in and purified from E.coli, and it was also shown that NPR1 could be phosphorylated by MPK1 in the presence of 1 mM ATP (Fig. 5d). These results suggest that MPK1, but not MPK2, is critical for NPR1 phosphorylation.
MPK1 mediates NPR1 monomerization
The oligomer-to-monomer transition of NPR1 is an indication of its activation, which leads to its nuclear translocation and phosphorylation, and subsequently the expression of Pathogenesis-related (PR) genes. Consistent with previous reports (Mou et al. 2003; Spoel et al. 2009), SA treatment decreased the level of NPR1-GFP oligomers, accompanied by an increase of its monomers in WT + NPR1-GFP (Fig. 5e). Intriguingly, the monomerization of NPR1 was largely blocked in mpk1, indicating that MPK1 is also somehow critical for NPR1 activation (Fig. 5e). Unlike in the case of MPK1, loss-of-function of MPK2 alone had negligible effect on NPR1 monomerization (Fig. 5e). Notably, we also observed a cytoplasmic-to-nucleus translocation of MPK1 in the leaf after SA treatment (Fig. 5f). Furthermore, we found that the expression of both TRX-h3 and TRX-h5, known to regulate cellular redox balance and facilitate NPR1 oligomer-to-monomer transition (Tada et al. 2008), was significantly reduced in mpk1 (Supplementary Fig. 7).
MKK4/5 and MPK1/2 are involved in induction of disease resistance
SA biosynthesis and signaling have been largely implicated in the mediation of systemically acquired disease resistance in plants (Malamy et al. 1990; Metraux et al. 1990; Gaffney et al. 1993; Delaney et al. 1994; Wildermuth et al. 2001). In Arabidopsis (Col-0), SA content at 6 DAP reaches a similar level to that detected at 6 hpi [hours post inoculation with Pseudomonas syringae pv. maculicola ES4326 (Psm. ES4326) at an OD600 of 0.0001] (Yu et al. 2010; Wang et al. 2015). Notably, NPR1-GFP phosphorylation were also detected in WT + NPR1-GFP plants at 6 DAP and 6 hpi (Supplementary Fig. 8). To determine whether MKK4/5 and MPK1/2 are also involved in the induction of disease resistance, we inoculated 5-weeks-old plants with Psm. ES4326 at an OD600 of 0.001. The leaf chlorosis and growth of Psm. ES4326 in mkk4 mkk5, mkk4* and mpk1 mpk2 were more prominent than those in WT, comparable to those in npr1, indicating that their resistance to Psm. ES4326 was greatly impaired (Fig. 6a, b). Consistently, the expression of disease resistance marker genes, e.g. PR1, PR2, PR5, and NPR1, was more or less compromised in these mutants (Fig. 6c). These results suggest critical roles of MKK4/5 and MPK1/2 in the induction of disease resistance.
MKK4/5 and MPK1/2 are involved in the induction of disease resistance. a Disease symptoms of Col-0, npr1, mkk4 mkk5, mkk4* and mpk1 mpk2 plants at 3 dpi (days post inoculation). Five-week-old plants grown in short day-growth conditions were pressure-infiltrated with Psm.ES4326 at an OD600 of 0.001. b Measurements of Psm.ES4326 growth in the leaves of indicated genotypes at 3 dpi. Error bars indicate SD (n = 8), LSD ANOVA test, P < 0.05. c Transcript levels of PR1, PR2, PR5, and NPR1 genes in the leaves of indicated genotypes at 3 dpi. Error bars indicate SD (n = 3), LSD ANOVA test, P < 0.05
Discussion
Exogenous SA, ABA, JA and ET have long been shown to be able to induce leaf senescence; but, except in the case of ET, decreases in their levels and/or mutations in their signaling components don’t normally cause obvious senescence phenotypes under normal growth conditions (Gan and Amasino 1997; Graaff et al. 2006; Kusaba et al. 2013; Jibran et al. 2013). It therefore remains largely unclear whether and how these hormones are involved in the regulation of leaf senescence (Gan and Amasino 1997; Graaff et al. 2006; Kusaba et al. 2013; Jibran et al. 2013). On the other hand, genetically increasing the level of endogenous hormones often disturb whole plant development and is therefore not an ideal approach to determine the exact role of the major hormones in regulating leaf senescence (Park et al. 1998). In this study, by taking advantage of PBZ-induced biosynthesis of endogenous SA, we demonstrate that increasing the endogenous level of SA can significantly accelerate the senescence of old leaves in Arabidopsis. It has been reported that the expression of many senescence related genes is affected in the mutants of SA biosynthesis and signaling genes, but no obvious effects on senescence phenotype are observed in its signaling mutant npr1 and in nahG plants where SA is converted to catechol (Morris et al. 2000). Consistently, we didn’t observe any senescence phenotype in mutants defective in SA biosynthesis (sid2) and signaling (npr1) or in NPR1 constitutively overexpressed plants (35S::NPR1) without PBZ treatment. However, upon PBZ treatment, sid2 and npr1 plants exhibited a significantly delayed senescence phenotype, whereas 35S::NPR1 plants displayed precocious leaf yellowing. All the physiological and molecular parameters examined were consistent with the observed senescence phenotypes. These data may imply that SA-triggered leaf senescence has been evolved to strategically sacrifice parts of the leaf tissue that are severely affected by environmental stresses, e.g. a heavy pathogen infection that induces a dramatic increase in SA level (Pajerowska-Mukhtar et al. 2012). This hypothesis is supported by our observation that basal resistance shares a MAPK cascade with SA acceleration of leaf senescence, as will be discussed below.
MKK4 and MKK5 are widely implicated in plant abiotic and biotic stresses (Asai et al. 2002; Zhao et al. 2014; Kim et al. 2011), whereas MPK1 and MPK2 were reported to be involved in response to wounding (Ortiz-Masia et al. 2007). In this study, we revealed an involvement of MKK4/5-MPK1/2 cascade in SA signaling. We found that single knock-down or knock-out mutants of MKK4, MKK5, MPK1 or MPK2 inhibited PBZ-accelerated leaf senescence to a subtle but detectable degree, with plant growth and development not being obviously affected under normal conditions. This inhibition was more obvious in their double mutants (mkk4 mkk5 and mpk1 mpk2) (Fig. 2b–f). Consistently, overexpression of MPK1 or MPK2 produced a precocious senescence phenotype. Importantly, the precocious senescence was blocked in the mutants of ICS1 or NPR1 (Figs. 3c, d, 5a, b). These data suggest that the MKK4/5-MPK1/2 cascade plays an important role in PBZ-accelerated leaf senescence and its functionality requires SA/NPR1. The MKK9-MPK6 cascade, as well as MAPKKK8, has been reported to be involved in the regulation of developmental leaf senescence (Zhou et al. 2009; Miao et al. 2007). However, inconsistent with a recent report (Chai et al. 2014), our data indicate that MKK9-MPK6 cascade may not be involved in SA signaling-dependent leaf senescence (Supplementary Fig. 3). Our results imply that different MAPK cascades may be required to mediate leaf senescence in different scenarios. Meanwhile, we found that MPK1/2 also play an essential role in basal resistance (Fig. 6). These observations suggest that there is a common MAPK cascade regulating both SA-induced disease resistance and leaf senescence.
Remarkably, we showed that MPK1 phosphorylated NPR1, whereas MKK4/5 phosphorylated MPK1/2. NPR1 protein was recently reported to be required for SA-induced senescence of detached leaves in Arabidopsis (Chai et al. 2014), and its phosphorylation at Ser589/Thr373 by SNF1-RELATED PROTEIN KINASE 2.8 (SnRK2.8) mediated SAR induction in distal tissues by facilitating its nuclear import (Lee et al. 2015). Importantly, it was previously shown that NPR1 needs to be phosphorylated at Ser11/Ser15 in the nucleus for full induction and subsequent establishment of SAR (Spoel et al. 2009). However, which kinase(s) phosphorylates NPR1 protein at the residues remains unknown. Chai et al. recently reported that MPK6 could affect the expression of NPR1 gene via mediation of WRKY6 and TRX-h5 and consequently the monomerization of NPR1 protein during leaf senescence (Chai et al. 2014). Here, by in vitro and in vivo phosphorylation assays, we found that MPK1, but not MPK6, directly phosphorylated NPR1 (Fig. 5c–g; Supplementary Fig. 9). Unexpectedly, MPK2 showed only a trivial, if any, activity of NPR1 phosphorylation, although it was found to regulate SA/NPR1-dependent leaf senescence redundantly with MPK1 (Fig. 5c). It therefore remains unknown for how MPK2 is exactly involved in the regulation of SA/NPR1-dependent leaf senescence. In light of that the function of NPR1 is dependent on W-boxes in its promoter (Yu et al. 2010), and a large number of WRKY TFs have been reported to be involved in the regulation of plant disease resistances and leaf senescence (Wang et al. 2006; Eulgem and Somssich 2007; Ishihama and Yoshioka 2012; Singh et al. 2014), we hypothesize that MPK2 might be responsible for the phosphorylation of some of these WRKY TFs. The hypothesis is supported by the findings that a large number of WRKYs are phosphorylated by MPKs (Popescu et al. 2009; Hiroaki et al. 2015). In addition, we found that MPK2 can interact with WRKYs, which played an important role in leaf senescence signaling pathway by regulating the expression of ICS1 and NPR1 in our another article (Gao et al. 2019, under review).
Mou et al. discovered that exogenous application of SA can activate NPR1 conformational change, and, importantly, NPR1 monomerization is both necessary and sufficient for its function (Mou et al. 2003). Interestingly, we found that NPR1 monomerization was also inhibited when MPK1 was mutated (Fig. 5e), indicating that MPK1 is crucial not only for NPR1 phosphorylation but also for its conformational change. This finding is corroborated by our further detection that the expression of TRX-h3/5 was significantly reduced in mpk1 compared with Col-0 plants (Supplementary Fig. 7). Both TRX-h3 and TRX-h5 are required for full induction of PR genes, and TRX-h5 is known to facilitate NPR1 oligomer-to-monomer transition via regulating cellular redox balance (Tada et al. 2008). It would therefore be interesting to reveal the molecular link between MPK1 phosphorylation activity and its effect on TRX-h3/5 expression. These results indicated that MPK1 was involved in regulating the activation of NPR1, possibly by enhancing the expression of TRX-h3 and TRX-h5.
By taking advantage of PBZ-induced leaf senescence, we reveal a MAPK cascade, MKK4/5-MPK1, which not only phosphorylates the key SA signaling component NPR1 specifically, but also regulates its oligomer-to-monomer conformational change somehow via mediation of TRX-h3/5. Both of the functions depend on endogenous SA biosynthesis. Our revelation of this novel regulatory module suggests that there exists a hierarchical regulatory network in the NPR1-mediated SA signaling (Supplementary Fig. 10).
Materials and methods
Plant materials and growth conditions
Plants were grown under a short-day photoperiod (8 h light/16 h dark) at 23 ± 2 °C, with approximately 70% relative humidity and 100 μmol m−2 s−1 light intensity. Five-week-old plants were applied with 0.5 mM probenazole (PBZ) via root drenching to induce endogenous SA biosynthesis. Transgenic plants were obtained via vacuum infiltration (Bechtold et al. 1993) and were selected on ½ Murashige and Skoog medium containing 25 mg l−1 kanamycin (for pSKM36), or 25 mg l−1 hygromycin (for pCAMBIA1302). Generally, more than ten independent transformants were obtained for each construct, and three lines were randomly chosen for gene expression and chlorophyll content analyses (Supplementary Figs. 5, 6). T-DNA insertion lines for mkk4 (SALK_018804C), mkk5 (SALK_050700), mpk1 (SALK_063847C) and mpk2 (SALK_047422C) were obtained from Arabidopsis Biological Resource Center (ABRC). sid2-2 was kindly provided by Dr. Frederick Ausubel, npr1-1 and 35S::NPR1 (35S-NPR1-GFP) by Dr. Xinnian Dong, and mkk4* (mkk4-18) and mkk5* (mkk5-18) by Dr. Dingzhong Tang. A substantial but not complete loss-of-function double mutant of MKK4 and MKK5 was generated by crossing mkk4 with mkk5 and genotyping accordingly. T-DNA insertion sites were determined by directly sequencing PCR products amplified with T-DNA left border and gene-specific primers. Primers for genotyping were listed in Supplementary Table 2.
Measurements of senescence-associated parameters
For the measurement of senescence-associated physiological parameters, the 5th through the 8th leaves (~ 0.1 g fresh weight) of five-week-old plants were used. Chl contents were quantified with a spectrometer (TU-1900, Persee) according to Benedetti and Arruda (Benedetti and Arruda 2002). Fluorescence in leaves was measured using LI-6400 (LI-COR) under a fixed LED light source (500 mmol m−2 s−1) at 25 °C and 300 μmol s−1 flow speeds, as specified in the manufacturer’s instruction. For measuring ion leakages, leaves were immersed in deionized distilled water, shaken in a 25 °C water bath for 40 min, and the conductivity was measured using a digital conductivity meter (Waterproof ECTestr11 + , MultiRange). Samples were boiled for 15 min and the conductivity then monitored. The ratio of the first measurement over the second measurement was used as an indicator of membrane leakage.
RNA analyses
RNA was extracted from 100 mg of respective leaf samples using Trizol (Invitrogen) and dissolved in 10 μL of DEPC-treated water. cDNA was synthesized using Superscript first-strand synthesis kit. qPCR (quantitative PCR) was performed using SYBR Green I PCR kit (Toyobo) on an iCycler according to the manufacturer’s instruction, with ACTIN2 as a reference (Bio-Rad). The reaction conditions for qPCR were as follows: 95 °C for 2 min, 40 cycles of 95 °C for 30 s, 60 °C for 25 s, and 72 °C for 20 s. For microarray analysis, two biological replicates of each sample were collected before and after 72 h PBZ treatment. Biotinylated cDNAs were prepared according to the standard Affymetrix protocol from 10 μg of total RNA. GeneChips were scanned using the GeneArrayTM scanner 3000. Microarray data were shown in GEO: GSE72636.
Transient expressions of proteins in protoplasts
Transient expression assays were performed with the protoplasts isolated from Arabidopsis mesophyll cells. Protoplasts from WT or mutants were transiently transformed by adding 10 mg polyethylene glycol. Twelve hours after transformation, the protoplasts were collected by centrifugation and then re-suspended in protein extraction buffer for further analyses. Details in the isolation, transformation, and cultivation of protoplasts were described by Sheen (https://genetics.mgh.harvard.edu/sheenweb/).
Plasmid construction
To construct overexpressing vectors, coding sequences of MKK4 and MKK5 were amplified and inserted into the cloning vector pSKM36 after digestion with AscI and SpeI. A 4 × MYC tag was amplified and inserted into the resultant vectors at the AscI site to generate MKK4-4 × MYC and MKK5-4 × MYC constructs, respectively. Coding sequences of MPK1 and MPK2 were amplified and inserted into pCAMBIA1302 to generate MPK1-4 × FLAG and MPK2-4 × FLAG constructs, respectively. For the protein expression in E. coli, coding sequences of NPR1 and MPKs were amplified and inserted into pET28 and pGEX-4T-1, respectively. Primers used in the plasmid construction were listed in Supplementary Table 2.
Protein analyses
For Western blot assay, total proteins were extracted from leaf tissues using 1 × SDS sampling buffer, or from protoplasts using protein isolation buffer (50 mM HEPES, pH 7.5; 0.1 mM Na2EDTA, 5 mM DTT, 0.01% Brij35, 2 mM MnCl2, 0.1% Triton X-100, and 1/100 protease inhibitor cocktail). About 10 μg proteins were separated in a 12% SDS-PAGE gel and transferred onto a nitrocellulose membrane. MYC-tagged, FLAG-tagged, and GFP-tagged proteins were detected using anti-MYC (1:2000, M20002M, Abmart), anti-FLAG (1:2000, F1804, Sigma), and anti-GFP antibodies (1:2000, 632569, Clontech), respectively. DTT was omitted for the examination of the oligomer-to-monomer transition of NPR1-GFP. Phosphorylated proteins were separated in a gel with 100 μM Phos-tag reagent (NARD Institute) and 200 μM MnCl2. For λ-PPase and Na3VO4 treatments, λ-PPase (10 units ml−1) and/or Na3VO4 (0.1 mM) were added, followed by incubating at 30 °C for 30 min. Reactions were stopped by adding ¼ volume of 5 × SDS-PAGE sampling buffer. For the in vitro kinase assay (Peck 2006), 0.5 μg MBP-MPK1 or MBP-MPK2 and 0.25 μg GST-NPR1 were added to kinase assay solution [20 mM Tris (pH 7.4), 100 mM NaCl, 12 mM MgCl2, 1 mM DTT, and 1 mM ATP]. Recombinant proteins were incubated or co-incubated in kinase reaction solution at 37 °C for 60 min.
Change history
13 July 2020
Due to an unfortunate turn of events, the second co-corresponding author, Dr. Benke Kuai, was omitted from the original publication. The corrected authors��� list and author contribution statement are published here and should be treated as definitive.
References
Asai T et al (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983
Bechtold N, Ellis J, Pelletier G (1993) In-planta agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis-thaliana plants. Cr. Acad. Sci. III-Vie 316:1194–1199
Benedetti CE, Arruda P (2002) Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol 128:1255–1263
Cao H, Jane G, Joseph DC, Sigrid V, Dong XN (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88:57–63
Chai J, Liu J, Zhou J, Xing D (2014) Mitogen-activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. J Exp Bot 65:6513–6528
Delaney TP et al (1994) A central role of salicylic acid in plant disease resistance. Science 266:1247–1250
Després C et al (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15:2181–2191
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Gaffney T et al (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261:754–756
Gan SS, Amasino RM (1997) Making sense of senescence (molecular genetic regulation and 584 manipulation of leaf senescence). Plant Physiol 113:313–319
Gao MH et al (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18:1190–1198
Hiroaki A et al (2015) WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27:2645–2663
Ishihama N, Yoshioka H (2012) Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol 15:431–437
Jibran R, Hunter DA, Dijkwel PP (2013) Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol Biol 82:547–561
Kazuya I et al (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7:301–308
Kim SH et al (2011) Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem Biophys Res Commun 412:150–154
Kusaba M, Tanaka A, Tanaka R (2013) Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth Res 117:221–234
Lee HJ et al (2015) Systemic immunity requires SnRK28-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 27:3425–3438
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250:1002–1004
Mark K, Weihua F, Dong XN (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12:2339–2350
Masclaux-Daubresse C, Chardon F (2011) Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J Exp Bot 62:2131–2142
Metraux JP et al (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250:1004–1006
Miao Y, Thomas ML, Anja S, Ulrike Z (2007) Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol 65:63–76
Morris K et al (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23:677–685
Mou Z, Fan WH, Dong XN (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113:935–944
Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10:339–346
Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ (2007) Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. Febs Lett 581:1834–1840
Pajerowska-Mukhtar KM et al (2012) The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22:103–112
Park JH, Oh SA, Kim YH, Woo HR, Nam HG (1998) Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol Biol 37:445–454
Peck SC (2006) Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J 45:512–522
Popescu SC et al (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Gene Dev 23:80–92
Raskin I (1992) Role of salicylic acid in plants. Annu Rev Plant Biol 43:439–463
Singh V, Roy S, Singh D, Nandi AK (2014) Arabidopsis FLOWERING LOCUS D influences systemic-acquired-resistance-induced expression and histone modifications of WRKY genes. J Bioscience 39:119–126
Spoel SH et al (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137:860–872
Tada Y et al (2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321:952–956
van der Graaff E et al (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792
Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206
Wang D, Amornsiripanitch N, Dong XN (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. Plos Pathog 2:e123
Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19:63–73
Wang XY et al (2015) TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. Plant J 82:151–162
Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562–565
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versitile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yoshioka K, Nakashita H, Klessig DF, Yamaguchi I (2001) Probenazole induces systemic acquired resistance in Arabidopsis with a novel type of action. Plant J 25:149–157
Yu J et al (2010) The pathway and regulation of salicylic acid biosynthesis in probenazole-treated Arabidopsis. J Plant Biol 53:417–424
Zhang KW, Halitschke R, Yin C, Liu CJ, Gan SS (2013) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA 110:14807–14812
Zhao C et al (2014) EDR1 physically interacts with MKK4/5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. Plos Genet 10:e10043895
Zhou CJ, Cai ZH, Guo YF, Gan SS (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150:167–177
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31700246 to J.G.) and Science and Technology Commission of Shanghai Municipality (15JC1400800).
Author information
Authors and Affiliations
Contributions
J.Z. and J.G. conceived and designed the experiments. J.Z., J.G., Z.Z. and Y.S. performed the experiments, X.W., X.W. and X.Z. contributed new materials, J.G. and J.Z. analyzed the data and wrote the paper. All authors read and improved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Gao, J., Zhu, Z. et al. MKK4/MKK5-MPK1/MPK2 cascade mediates SA-activated leaf senescence via phosphorylation of NPR1 in Arabidopsis. Plant Mol Biol 102, 463–475 (2020). https://doi.org/10.1007/s11103-019-00958-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00958-z