Abstract
Key message
Rubisco activase of plants evolved in a stepwise manner without losing its function to adapt to the major evolutionary events including endosymbiosis and land colonization.
Abstract
Rubisco activase is an essential enzyme for photosynthesis, which removes inhibitory sugar phosphates from the active sites of Rubisco, a process necessary for Rubisco activation and carbon fixation. The gene probably evolved in cyanobacteria as different species differ for its presence. However, the gene is present in all other plant species. At least a single gene copy was maintained throughout plant evolution; but various genome and gene duplication events, which occurred during plant evolution, increased its copy number in some species. The exons and exon–intron junctions of present day higher plant’s Rca, which is conserved in most species seem to have evolved in charophytes. A unique tandem duplication of Rca gene occurred in a common grass ancestor, and the two genes evolved differently for gene structure, sequence, and expression pattern. At the protein level, starting with a primitive form in cyanobacteria, RCA of chlorophytes evolved by integrating chloroplast transit peptide (cTP), and N-terminal domains to the ATPase, Rubisco recognition and C-terminal domains. The redox regulated C-terminal extension (CTE) and the associated alternate splicing mechanism, which splices the RCA-α and RCA-β isoforms were probably gained from another gene in charophytes, conserved in most species except the members of Solanaceae family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Photosynthesis plays a vital role in converting light energy into chemical energy, which supports energy needs of the most living organisms. Ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco, EC 4.1.1.39) is the key enzyme in fixing atmospheric CO2. During carbon assimilation, Rubisco must first be carbamylated by an activator CO2, in addition to the substrate CO2, and must bind to Mg2+ before binding the five-carbon substrate, ribulose-1,5-bisphosphate (RuBP) (Lorimer and Miziorko 1980; Lorimer 1981). Various naturally occurring sugar phosphates, including RuBP, may bind to the active sites of Rubisco and block further carbamylation or catalysis (Badger and Lorimer 1981; Jordan and Chollet 1983; Brooks and Portis 1988). Rubisco activase (RCA), a catalytic chaperone of Rubisco, removes the inhibitory sugars from the Rubisco active sites by remodeling the enzyme’s conformation (Andersson 2008; Stotz et al. 2011).
As a member of the AAA+ family of ATPases associated with diverse cellular activities, RCA uses energy from ATP hydrolysis to modify Rubisco (Neuwald et al. 1999). Crystal structure suggests that RCA may function as a hexamer (Stotz et al. 2011). In the higher plants, RCA is a nuclear-encoded chloroplast protein, generally consisting of two immunologically related polypeptides named as RCA larger or RCA-α and RCA smaller or RCA-β isoform. The plant RCA is composed of five domains: chloroplast transit peptide (cTP), N-terminal, ATPase domain, Rubisco recognition domain and C-terminal (Salvucci 2004; Portis et al. 2008). The RCA-α contains an additional C-terminal extension region (CTE) that allows for the redox regulation of RCA in vivo (Zhang and Portis 1999). The size of the mature RCA protein, which lacks the cTP region, varies from 41 to 43 kDa for RCA-β and 45 to 47 kDa for RCA-α. In many plant species, the two RCA isoforms (RCA-α and RCA-β) are a result of an alternate splicing of the Rca mRNA. Both isoforms are capable of activating Rubisco (Shen et al. 1991; Salvucci et al. 2003). Although both RCA isoforms are capable of Rubisco activation at in vivo conditions (Zhang et al. 2002), the RCAα isoform with CTE domain functionally differs from the RCAβ isoform by integrating the light activated redox signaling and sensitive to physiological ratios of ADP/ATP into Rubisco activation. The two cysteine (Cys) residues located at the CTE domain of RCAα form a disulfide bridge during the oxidizing conditions and become susceptible to ADP inhibition (Zhang and Portis 1999). In plants expressing both RCAα and RCAβ isoforms, the redox regulation of the RCAα isoform is conferred to the RCAβ when they form multimeric holoenzyme. There are differences in the RCAα and RCAβ isoform expression levels in some species and these variations could influence the regulatory function of the multimeric RCA enzyme. The RCAβ isoform of most plant species appears to be insensitive to changes in the ratios of ADP/ATP and redox regulation; however, the RCAβ isoform of tobacco showed an acute sensitivity to ADP and responded similarly to the Arabidopsis RCAβ when in association with its corresponding RCAα isoform (Carmo-Silva and Salvucci 2013).
The RCA function appears to be essential for photosynthesis in all photoautotrophs; however, the evolution of this function from a simpler prokaryotic system to highly complex plant systems present in angiosperms is still not known. Characterized Rca mutants of filamentous cyanobacterium Anabaena variabilis, green algae Chlamydomonas reinhardtii, and Arabidopsis showed reduction in Rubisco activity and very slow growth (Salvucci et al. 1985; Li et al. 1993; Pollock et al. 2003; Kurek et al. 2007). Search for RCA isoforms in different organisms using RCA specific antibodies resulted in its identification in many photosynthetically active higher plants, algae, and even in some prokaryotic organisms including cyanobacteria (Salvucci et al. 1987; Li et al. 1993). Studies have shown that not all the plants express both RCA-α and RCA-β isoforms. Plants species that express both RCA-α and RCA-β were Arabidopsis, spinach, soybean, kidney bean, pea, celery, oat, (Salvucci et al. 1987), apple (Watillon et al. 1993), rice (To et al. 1999), barley (Rundle and Zielinski 1991b), wheat (Law and Crafts-Brandner 2001), cotton (Feller and Crafts-Brandner 1998), creosote bush and Antarctic grass (Salvucci and Crafts-Brandner 2004), red maple (Weston et al. 2007), and sweet potato (Xu et al. 2010). Initial studies on maize suggested the presence of only RCA-β isoform (Salvucci et al. 1987; Ayala-Ochoa et al. 2004), but later studies (Ristic et al. 2009; Yin et al. 2014) showed presence of both RCA-α and RCA-β isoforms. In tobacco, only the RCA-β isoform was identified (Salvucci et al. 1987). Heat stress changes the expression levels of RCA-α and RCA-β isoforms in rice (Wang et al. 2010), wheat (Law and Crafts-Brandner 2001), and maize (Ristic et al. 2009). Inter-varietal variation for expression levels was also observed for these two isoforms in wheat and maize (Ristic et al. 2009). In most cases, it is not clear whether these isoforms are product of alternate splicing or from the expression of separate genes. Similarly, the gene number and structure of RCA coding genes vary greatly among photosynthetic organisms, especially in the higher plants. Various cyanobacteria species differ for the presence of Rca, as the gene was found mainly in the species with β carboxysomes-containing form 1B carboxysomal Rubisco (Zarzycki et al. 2013). Only a single Rca gene is present in these cyanobacteria genomes (Li et al. 1993; Zarzycki et al. 2013). The chlorophyte model Chlamydomonas reinhardtii contains a single Rca gene (Roesler and Ogren 1990), where an another green alga Chlorococcum littorale contains up to two Rca gene copies (Beuf et al. 1999). Arabidopsis genome has one gene, AtRca (AT2G39730) on chromosome 2, the loss of which causes deleterious effects on plant growth by seriously compromising photosynthesis (Salvucci et al. 1985; Zhang et al. 2002; Kurek et al. 2007). In soybean, the two Rca genes along with three other Rca-like genes are present on five different chromosomes (Yin et al. 2010). Reports on the number of Rca genes in the rice genome were not consistent, one study claimed a single gene (To et al. 1999), whereas the other study reported up to two Rca genes (Zhang and Komatsu 2000). Barley has two tightly linked tandemly oriented genes, one expresses a single transcript, and the other expresses two transcripts by alternate splicing, thereby making three different peptides (Rundle and Zielinski 1991b). In polyploid species including wheat and cotton, the number of Rca genes changed with the change in ploidy level with each sub-genome containing Rca genes (Salvucci et al. 2003; Carmo-Silva et al. 2015).
Although almost all the studied photosynthetic organisms contain RCA, a detailed analysis of the gene number, duplications, deletions, exon–intron structure and their evolution in important plant species is still lacking. Similarly, the information about the presence of light sensitive CTE domain, alternate splicing mechanism of Rca transcripts, and immunologically related peptides was reported mostly at species level. We hypothesized that a systematic analysis on the whole genome sequences of different model plant species could provide more information about the Rca gene evolution. In this study, we investigated the RCA coding genes from cyanobacteria to higher plants to address (a) the copy number variation (b) structural evolution of Rca gene in plants (c) structure and expression pattern of Rca genes of Poaceae and Solanaceae species, and (d) evolution of different RCA domains. Additionally, we have identified when the redox regulation was integrated into RCA and its putative source sequence and investigated about the evolution of alternate splicing mechanism of Rca genes.
Materials and methods
Identification of true Rca ortholog sequences from different plants
Gene models and predicted protein sequences of RCA coding genes from different organisms were collected from multiple databases using the characterized 474 aa sequence of AtRCA (AT2G39730.1) (https://www.arabidopsis.org/) as the query sequence in the translated nucleotide sequence database (TBLASTN) with default settings. The species were selected based on its genomic sequence availability, taxonomic position, and evolutionary significance. The criteria used for searching true ortholog in this study were described elsewhere (Dhaliwal et al. 2014). In brief, the ortholog sequences were selected based the high sequence identity, maximum query coverage, presence of all domains and motifs, similarity to relative size and distance among the domains to that of the AtRCA sequence. The predicted protein sequences from the collected gene models were manually analyzed for fulfilling all the above mentioned true ortholog criteria. The databases used were: Phytozome-11 (http://phytozome.jgi.doe.gov/); NCBI (http://www.ncbi.nlm.nih.gov); Ensembl plants (http://plants.ensembl.org/index.html); TAIR (https://www.arabidopsis.org); MaizeGDB (http://www.maizegdb.org); Rice Genome annotation project (http://rice.plantbiology.msu.edu); Klebsormidium flaccidum genome project (http://www.plantmorphogenesis.bio.titech.ac.jp); Solgenomics network (http://solgenomics.net), and Congenie (http://congenie.org). The last searches made on the database were before July 2016. Complete details of the search results along with its corresponding database is provided in Supplementary Table S1.
Gene model prediction
Gene models were predicted by aligning multiple cDNA and EST sequences with genomic DNA sequences. NCBI EST and non-redundant databases were used for collecting the EST and cDNA sequences. Sequences with maximum query coverage and 100% sequence identity were collected. The collected sequences were aligned against genomic DNA sequence using multiple sequence alignment program Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Exon and intron structures were marked based on alignment, and open reading frames (ORFs) were predicted using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) to get the UTRs and protein sequence information. Multiple EST sequences were assembled using CAP3 (http://biosrv.cab.unina.it/webcap3/) to get full-length coding sequences. For some species, the ESTs and the cDNA prediction did not cover the whole sequence. In these cases, the corresponding genomic DNA region was screened for possible matches and manually added to the model if possible. GenBank accession number for all the cDNA and EST sequences used in gene model prediction is provided in the Supplementary Tables S2, S3 and S4.
RCA domain analysis
We used the domain information from previously characterized RCA of Arabidopsis (Kumar et al. 2009), tobacco (Stotz et al. 2011) and wheat (Carmo-Silva et al. 2015) for RCA domain analysis. For new gene models, protein sequences were used in ChloroP1.1 database (http://www.cbs.dtu.dk/services/ChloroP/) to identify the transit peptide; and other domains were marked based on the previously characterized RCA domains. NCBI database was also used to get the putative RCA sequences of cyanobacteria, chlorophyte and charophyte species. Previously characterized RCA sequences were used as queries and their corresponding GenBank accession numbers were provided on the legend. Collected sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) with default settings for further analysis.
Results
Copy number variation for RCA coding genes in plants
We performed genome-wide surveys for the RCA coding genes in the genomic sequence databases of 51 sequenced plant species including chlorophytes, bryophytes, lycophytes and higher plants using Arabidopsis Rubisco activase protein sequence (GenBank ID: AEC09714.1) as a query. The criteria used to identify Rca orthologous genes in different species are described in “Materials and methods”. Of the six sequenced chlorophyte species, Chlamydomonas reinhardtii, Coccomyxa subellipsoidea C-169, Micromonas sp. RCC299 and Ostreococcus lucimarinus showed a single Rca gene in each of their genomes; whereas the partially sequenced Micromonas pusilla CCMP1545 and Volvox carteri genomic sequence databases did not contain any Rca like sequence or gene model. However, the NCBI-EST (expressed sequence tag) database contains Rca EST sequences for Micromonas pusilla CCMP1545 and Volvox carteri suggesting the presence of an Rca gene in the genome. Additionally, the NCBI’s nucleotide database showed presence of Rca sequences in five other chlorophyte species (Supplementary Fig. S1). Similar to chlorophytes, the charophyte Klebsormidium flaccidum genome also contains a single RCA coding gene KfRca1 (Supplementary Table S1).
The genomes of early land plants, including bryophytes and lycophytes, showed a variable number of Rca gene copies. The bryophyte Liverwort (Marchantia polymorpha) genome contains a single RCA coding gene (MpRca1), whereas multiple copies were identified in mosses Physcomitrella patens and Sphagnum fallax (Supplementary Table S1). Five RCA coding genes PpRca1-PpRca5 were found in the P. patens genome, of which PpRca4 and PpRca5 were tandemly oriented (Fig. 2). The Sphagnum fallax genome showed four RCA coding genes, suggesting multiple gene duplication events after divergence of liverworts and mosses from a common bryophytic ancestor. The lycophyte Selaginella moellendorffii, a model organism for an ancient vascular system, showed four Rca genes in its genome (Supplementary Table S1).
In the higher plant genomes, the number of Rca gene copies among species ranged from one to six, depending on the ploidy and number of genome duplication events that occurred during evolution (Supplementary Table S1). The gymnosperm Norway spruce (Picea abies) genome contains a single Rca gene. Interestingly, the genome of a close relative of ancient angiosperm Amborella trichopoda, which is always positioned near the base of the flowering plant’s divergence in phylogenetic studies (Amborella Genome Project 2013), also showed a single Rca gene (Supplementary Table S1). Genomes of Aquilegia coerulea, a model for the eudicot evolutionary studies, which is placed nearly equidistantly between Arabidopsis and rice evolution (Kramer 2009), contains two Rca genes. Genomes of monocot species contain two to three Rca gene copies. Spirodela polyrhiza has two copies, whereas banana (Musa acuminate) and pineapple (Ananas comosus) have three Rca genes each in their genomes. These genes are present mostly on different chromosomes, except in species of Poaceae family (grasses), where two Rca genes were found in a tandem orientation with their reading frames pointing in the same direction (Supplementary Table S1). In dicot species, variation for the gene copy number was observed within a genus. For example, most of the Brassicaceae members including Arabidopsis thaliana, Arabidopsis lyrata, Boechera stricta, Capsella grandiflora, Capsella rubella and Eutrema salsugineum have a single gene each, whereas Brassica rapa and Brassica oleracea contain up to three Rca genes in their respective genomes. The Solanaceae species potato and tomato contain two Rca genes; pepper contains three and the cultivated tetraploid tobacco showed six genes in its genomes. Similarly, genomes of some of the Rosaceae and Fabaceae members showed three to six copies of RCA coding genes (Supplementary Table S1). These results suggest that at least a single Rca gene copy is necessary for photosynthesis, and appeared to be maintained in the genomes from lower photosynthetic plants to the flowering plants. Gene copy numbers might have changed by the whole genome, segmental, and tandem gene duplication events occurred during evolution at various levels of plants speciation.
Evolution of higher plant Rca gene: exons
Although the size of introns of Rca genes varies from species to species, and between duplicated copies of the same species, the exon sizes and the exon–intron junctions were highly conserved. To study the conservation, we color coded exons of Arabidopsis AtRca1 (Fig. 1). The corresponding exonic regions of Rca genes of various species were marked based on the sequence similarity (Figs. 2, 3, 4). The putative untranslated region (UTR) of the first and last exons of all the Rca genes were not included in the analysis. The AtRca1-α contains seven exons (45, 317, 106, 86, 473, 282 and 116 bp in size), and transcribe an mRNA to code for the 474 amino acids (aa) isoform. The alternately spliced AtRca1-β and AtRca1-β2 transcripts code for 446 and 441 aa long isoforms respectively (DeRidder et al. 2012). The AtRCA1-α isoform contains N-terminal (N), ATPase (AAA+), Rubisco recognition (Rubisco), C-terminal (C) and a redox sensitive C-terminal extension (CTE) domains. Alternate splicing of the seventh exon results in the loss of CTE domain in the AtRCA1-β and AtRCA1-β2 isoforms (Fig. 1). These exons originated in the lower photosynthetic organisms, especially in the charophytes and bryophytes, and the exon size and exon–intron boundaries were retained in most of the higher plants (Fig. 2). The first four exons of AtRca1 and other Rca coding genes of bryophytic and higher plants were also found in the KfRca1 of charophyte K. flaccidum as first (45 bp), third (315 bp), fourth (103 bp) and fifth (86 bp) exons. The 47 bp second exon of KfRca1 is not present in higher plants and some lower plants including the bryophytes. The fifth (473 bp), sixth (282 bp) and seventh (116 bp) exons of AtRca1 were found in the bryophytes P. patens PpRca5 as fourth (473 bp), fifth (282 bp) and sixth exons (127 bp) respectively (Table 1). The RCA coding genes of gymnosperm P. abies and angiosperms A. trichopoda and A. coerulea show exon sizes and exon–intron junctions similar to that of Arabidopsis, suggesting that the exon sizes and splice junctions were determined in the early land plants and were conserved throughout the higher plant evolution (Fig. 2). In rice and other grasses, two RCA coding genes are tandemly oriented and separated by a small intergenic sequence. Unlike other grass Rca genes (Fig. 3), the OsRca1 of rice accumulated multiple deletions in the coding sequence and appears to be non-functional (Supplementary Fig. S2). The gene doesn’t seem to express as no EST sequence, nor RNA-seq read is available in any of the databases.
Structure of Arabidopsis Rubisco activase gene, CDS and protein. Gene model for AtRca1 was redrawn using the information from AT2G39730 locus (http://www.arabidopsis.org). The dotted line represents chromosome, gray bars represent untranslated regions (UTRs) and thick horizontal black lines represent introns. Green, yellow, brown, sky blue, red, navy blue and purple color bars represent first to seventh exons of AtRca1 gene. The gene models AtRca1-β and AtRca1-β2 below the AtRca1-α show alternatively spliced mRNA versions. The thin vertical lines connecting coding DNA sequence (CDS) and protein structures show the corresponding regions of translated exonic sequences in RCA protein domains. RCA domains are shown as cTP, N, AAA+, Rubisco, C and CTE boxes. Two cysteine residues forming disulfide bond in CTE domain is shown as C::C. The gene and protein structure were drawn on nucleotide and amino acid scales separately
Structure of Rubisco activase genes in evolutionarily important model plants. Structure of RCA coding genes of chlorophyte, charophyte, bryophyte, and higher plants. The color bars on and below the gray dotted lines represent gene models and its corresponding alternate mRNA splicing models. The α and β in gene model names denote the type of isoforms coded from the gene models. Species with two or more gray dotted lines represent RCA coding genes on different chromosomes. In gene models, the thin black bar represents introns and light gray bars represent UTR regions. The green, yellow, brown, sky blue, red, navy blue and purple color bars represent corresponding sequences of Arabidopsis exons 1 to 7. The gene models were drawn to scale
Evolution of Rubisco activase gene in monocots. The color bars on and below the gray dotted lines represent gene models and its corresponding alternate mRNA splicing models. Species with multiple gray lines represent RCA coding genes on different chromosomes. The α and β symbols in gene model names denote the type of isoforms coded from the gene models. In gene models, the thin black bar represents introns and thick light gray bars represent UTR regions. The green, yellow, brown, sky blue, red, navy blue and purple color bars represent corresponding sequences of Arabidopsis exons 1 to 7. The exons of non-functional rice OsRca1 is shown as white bars. The gene model in the box is putative common grass ancestor Rca gene and predicted evolutionary changes based on the structure of other Rca genes
Structure of Rubisco activase genes in Solanaceae crops. Structure of RCA coding genes of tomato (Solanum lycopersicum), potato (Solanum tuberosum), capsicum (Capsicum annuum), and tobacco (Nicotiana tabacum). The color bars on and below the gray dotted lines represent gene models and its corresponding alternate mRNA splicing models. The a and b in gene model names denote the type of isoforms coded from the gene models. Species with two or more gray dotted lines represent RCA coding genes on different chromosomes. In gene models, the thin black bar represents introns and light gray bars represent UTR regions. The green, yellow, brown, sky blue, red, navy blue and purple color bars represent corresponding sequences of Arabidopsis exons 1 to 7. The orange triangle on fifth intron of CaRca3-β indicates a 3.6 kb transposable element insertion. The gene models were drawn to scale
Evolution of higher plant Rca gene: introns
The RCA coding genes from lower unicellular photosynthetic organisms to higher plant species showed a very high level of conservation for the exonic sequences and exon–intron junctions during evolution; whereas the number, size, and sequence of introns were highly variable. The intron numbers varied from one to ten (Table 1). Although the intron numbers were stably maintained between five to six in the RCA coding genes of bryophytes and higher plants, further loss of introns occurred in the tandemly duplicated Rca genes of the grass family members. The highest level of variations for intron numbers was observed in chlorophytes. The Rca gene of Bathycoccus prasinos did not contain any intron, whereas the Rca gene of Auxenochlorella protothecoides contains 11 introns (Supplementary Fig. S1). In C. reinhardtii, the CrRca1-β contains up to nine introns (Fig. 2). Similar to CrRca1-β, 6516 bp KfRca1-α transcript of charophyte K. flaccidum splices ten introns, and produces a mature 1782 bp long mRNA (GenBank ID: HO431775.1). The MpRca1 and PpRca1 to PpRca5 of bryophytes M. polymorpha and P. patens showed five and between three to five introns respectively (Fig. 2). The exon–intron structure and sequence similarities of PpRca3 are closely related to PpRca4, whereas the PpRca2 and PpRca5 resembled PpRca1 (Fig. 2). The single Rca gene of gymnosperm species P. abies (PaRca1) has seven exons. The Rca genes of the flowering plants A. trichopoda, A. coerulea, and A. thaliana contain six introns, and this number appeared to be conserved among most higher plants. Interestingly, the tandemly oriented OsRca1 and OsRca2 genes of the grass model plant rice showed loss of introns in both the genes (Fig. 2). Most of the RCA-β coding transcripts of flowering plants splice an additional intron at the 3′ UTR and share the same splice end site of the larger RCA-α coding transcripts, but with a novel intron splice site after the RCA-β transcript stop codon (Fig. 2).
Expression pattern of RCA isoforms in the model organisms
Comparative analysis of RCA isoform expression patterns from lower to higher organisms provided additional information about the Rca gene evolution and its possible role in plant adaptation. In this study, the term “expression” is used to show the type of isoform i.e. RCA-α or RCA-β and its corresponding transcripts expressed by the gene, but not the quantitative levels of the transcripts or the translated protein expressed. The CrRca1 of chlorophyte C. reinhardtii code for 408aa peptide, forms a 41.54 kDa mature RCA-β isoform after cleaving the 33aa long cTP domain. Similarly, predicted RCA isoforms from the putative Rca genes of nine other chlorophytes from NCBI database also showed high sequence similarity with CrRCA1, and molecular weight around 41–42 kDa (Supplementary Fig. S3). The charophyte model organism K. flaccidum expresses two RCA isoforms of 594 and 494 aa long with molecular weights of 58.24 and 47.96 kDa via alternate splicing of KfRca1 transcripts (Table 1). The 47.96 kDa KfRCA1-α2 isoform was more similar to the RCA-α isoform of higher plants in terms of sequence similarity and domain structure; whereas the larger KfRCA1-α1 contains additional sequences at the C terminal, which share similarity with CRISPR-associated RAMP protein (Supplementary Fig. S4). The EST sequence analyses of other charophyte organisms such as Chlorokybus atmophyticus, showed putative expression of both RCA-α and RCA-β isoforms, Nitella hyalina expressed the RCA-β isoforms, and Penium margaritaceum expressed an RCA-α isoform (Supplementary Fig. S4). These charophyte RCA isoforms were highly similar to the RCA isoforms of higher plants. Additionally, P. margaritaceum also showed expression of two other isoforms with extended CTE sequences similar to the KfRCA1-α1. The single MpRca1 gene of bryophytic liverwort Marchantia polymorpha expresses both RCA-α and RCA-β isoforms via alternate splicing of the pre-mRNA, whereas one of the five Rca gene copies (PpRca1) of P. patens expresses RCA-α and RCA-β isoforms by alternate splicing. Three copies (PpRca3, PpRca4, and PpRca5) express only the RCA-β isoform and the transcripts of PpRca2 have an in-frame stop codon on the second exon, which could potentially terminate the translation, or code a smaller 35 kDa peptide sharing similarity with RCA-β isoform via alternate translation start site (Table 1). In higher plants, many analyzed model species including the gymnosperm P. abies express RCA-α and RCA-β isoforms via alternate splicing of RCA coding transcripts. The OsRca1 of rice did not show any expression, but the other copy OsRca2 expresses both RCA-α and RCA-β isoforms. Both the Rca genes of Aquilegia coerulea express both the isoform via alternate splicing (Table 1). However, there are few known exceptions, such as Panicoideae species and cotton (Salvucci et al. 2003), in which the RCA-α and RCA-β isoforms are coded from different genes and not by the alternate splicing of mRNA.
Evolution of Rca gene structure and expression pattern in monocot
For this analysis, we collected the DNA sequence of genomic regions coding Rca genes in rice, Brachypodium, barley, wheat, maize, sorghum and foxtail millet, and developed gene models using cDNA and EST sequences, and information from previous publications (Rundle and Zielinski 1991b; To et al. 1999; Yin et al. 2014; Carmo-Silva et al. 2015). We compared the Rca gene models of the species mentioned above, and we found two Rca genes in tandem orientation separated by a small inter-genic region. Based on the structure of these tandemly duplicated Rca genes, a model for the grass Rca evolution is proposed in Fig. 3. As found in Rca genes of the A. trichopoda and A. coerulea, the common monocot ancestor Rca gene might have contained seven exons and been capable of coding both larger and smaller RCA isoforms via alternate splicing. By comparing the pattern of intron loss and exon fusion from the duplicated Rca genes, it is evident that the duplication event probably occurred before the divergence of Poaceae sub families Oryzoideae, Pooideae, and Panicoideae, which produced two Rca coding genes in a tandem orientation. These duplicated genes evolved differently by losing or gaining introns, accumulating deletions in the coding sequences, and acquiring premature stop codons.
In the tandemly duplicated Rca genes, the promoter sequences of Rca1, located at the 5′ side, has comparatively longer than Rca2, mainly because of the 3′ untranslated region of Rca1 flanks upstream promoter region of the Rca. Thus, the promoter of Rca2 or the intergenic sequences ranges from 681 bp in maize and up to 3055 bp in sorghum (Fig. 3). The ancestral Rca1 gene underwent significant changes in terms of losing third, fifth and sixth introns before the divergence, and the loss of the sixth intron affected the alternate splicing. Subfamily and genus specific intron loss was identified in the Rca1 gene as, the nonfunctional OsRca1 of rice retained the possible first, second, and fourth intron regions; Pooideae members (Brachypodium, barley, and wheat) retained only the first intron; and Panicoideae member maize retained the first and the fourth introns, whereas sorghum retained only the fourth intron. The SiRca1 of Foxtail millet did not contain any intron. Novel intron and its alternate splicing were also identified in wheat Rca1. Although there was no alternate splicing observed in the coding region, an 83 bp intron was spliced in the 5′ UTR of some transcripts of TaRca1 A and B genome copies (GenBank accessions: HX114853, HX114528, and HX086211) (Fig. 3). Based on the available ESTs, this specific intron splicing was observed for the A and the B copies, but not for the D copy. Variation for intron lengths was also observed as the first intron of the Pooideae members varied from 1418 bp in case of BdRca1 to 2222 bp in HvRca1, whereas in maize it was only 76 bp (Table 2).
Unlike the grass Rca1 gene, the Rca2 exon–intron structure was relatively unchanged, and resembled more closely the ancestral Rca gene until the divergence of the subfamilies (Fig. 3). The Rca2 gene of rice lost the third intron, Pooideae members lost the second intron and Panicoideae members maize and sorghum show a common loss of the second and the fifth intron and species specific third and the sixth introns respectively. The structure of foxtail millet SiRca2 resembled closely the structure of maize ZmRca2 by losing the second, third and fifth introns. These precise intron exclusions appeared to have occurred after the divergence of the grass subfamilies as different introns were lost in different members without many changes in the coding sequences (Supplementary Fig. S7). Unlike the ancestral Rca gene, the OsRca2 gained an additional intron near the end of the fifth exon, which splits the exon, and created an additional exon with stop codon for the transcripts encoding OsRCA2-β. The CDS of the first exon of all studied grass Rca genes were 36 bp, except in the SiRca1 of Setaria italica, which has evolved to be an intron-less gene (Fig. 3). There is a deletion of nine nucleotides in the first exon of the grass Rca genes as compared with the other higher plants (Supplementary Fig. S5), and probably occurred before the tandem duplication event, as both the genes contain the deletion.
Expression and protein prediction analyses using the cDNA and EST sequences showed evidence for the divergent evolution of the duplicated Rca genes in grass families. The Rca1 gene of all three studied Pooideae members code for RCA-β isoform, whereas the Rca1 of all three Panicoideae species express only the RCA-α isoform (Table 2). The transcripts of Rca1 from both Pooideae and Panicoideae did not show any alternate splicing, except the 83 bp intron spliced from the 3′ UTR of TaRca1. The OsRca1 of rice did not contain any EST or cDNA sequences in the database, suggesting the possible loss of gene expression along with its function. The molecular weight of the mature Panicoideae RCA1-α was around 46 kDa, and the RCA1-β of Pooideae grasses was 42 kDa (Table 2). Although the alignment of RCA1 amino acid sequences from Pooideae and Panicoideae members showed overall sequence conservation between the isoforms, some domains accumulated more changes than others, as the cTP and Rubisco recognition domains had more changes than the ATPase domain (Supplementary Fig. S6).
The Rca 2 gene of rice, Brachypodium, barley, and wheat expresses both RCA2-α and RCA2-β isoforms via alternate splicing of the pre-mRNA. The splicing site at the end of the fifth exon determines whether the transcript will code RCA2-α or RCA2-β isoform as alternate splicing at the end of the fifth exon results in either a stop codon (RCA2-β) or splice five bases before (after 282 bp of the fifth exon), extending the intron, and allowing translation through the sixth exon (RCA2-α). The transcripts encoding RCA2-α splice an intron after the 282 bp of the fifth exon in Pooideae and extend the translation into the sixth exon to produce 46 kDa isoforms (Fig. 3). The smaller 42 kDa mature RCA-β isoform coding transcripts alternatively splice an intron at the end of the fifth exon, which results in a stop codon in the case of Rca2-β of Pooideae grasses; whereas in rice, alternate splicing of two introns produced an additional 85 bp exon, which contains a stop codon for RCA-β isoform (Fig. 3).
Unlike the Rca2 of Pooideae members and rice, the Rca2 gene of maize, sorghum, and foxtail millet have evolved differently by coding only the RCA-β from Rca2. Comparative analysis of Rca2 genomic sequences from Pooideae and Panicoideae revealed the loss of an evolutionarily conserved alternate splicing mechanism, which splices the stop codon at the end of sixth exon (ancestral Rca exon numbers) and extends to the seventh exon to code for RCA-α isoform (Fig. 3). The loss of the 5′ splice site, by point mutations before the divergence of Panicoideae members, abolished the alternate splicing mechanism and produced the transcripts to code for RCA-β in all three studied species (Supplementary Fig. S7). Interestingly, the angiosperm specific alternate splicing of an intron at 3′ UTR was still conserved in all the RCA2-β coding transcripts of all the grass species studied, except SbRca2 and ZmRca2, in which all or some transcripts retain the intron. Thus, the predicted molecular weight of mature RCA2-β in the Panicoideae species is 42 kDa (Table 2). Similar to the RCA1 protein alignment, the RCA2 alignment of all the studied grass species showed few mismatches in the ATPase region compared to the cTP, N-terminal and Rubisco recognition region (Supplementary Fig. S8).
Structure and expression pattern of Rca genes in Solanaceae
Comparative analysis of RCA coding genes from the whole genome sequenced plant species revealed the loss of the seventh exon, which codes for the redox regulated CTE domain in some Solanaceae species (Fig. 4). The RCA coding genes of three Solanaceae species viz., tomato, potato, and capsicum showed an exon–intron structure similar to the evolutionarily conserved higher plant Rca gene structure. However, mutations near the end of sixth exon created novel stop codons as with SlRca1-β, StRca1-β, StRca2-β, CaRca2-β, and CaRca3-β, or abolished the alternate splice site for producing larger isoform in SlRca2-β and CaRca1-β, resulting in only smaller isoform expression (Supplementary Fig. S9). The cultivated tetraploid tobacco (Nicotiana tabacum), contains six Rca genes (NtRca1- NtRca6) in the genome. Interestingly, the NtRca1 has seven exons as found in the other higher plants and the other five genes had similar mutations near the end of the sixth exon as identified in the other Solanaceae species (Supplementary Fig. S9). The first exon of SlRca2-β and CaRca1-β showed a six-nucleotide deletion, whereas all other genes contained the normal 45 bp exon. The second exon was highly variable among the Solanaceae species with the size range of 314 bp in tomato to 335 bp in CaRca3-β of capsicum. Sizes of the other three exons (third, fourth and fifth) were the same as those in the conserved Rca gene except for the mutations near the end of the sixth exon but before the alternate splicing site, which created a premature stop codon (Supplementary Fig. S9).
Unlike exons, the intron lengths varied significantly in size. The lengths of the first, second and fourth introns of all Solanaceae Rca showed more variation than the third and fifth introns (Table 3). The fifth intron of CaRca3-β was exceptionally large (5110 bp), compared to introns of other Rca genes of Solanaceae family. We suspected a transposable element (TE) insertion in the intron because of the change in size. To identify the element, we used the intronic sequence as a query in the NCBI nucleotide database of Solanaceae family, to find similarities with known TEs. We found a ~ 3.7 Kb putative retrotransposon insertion showing 83% similarity (E value 0.0) with Long terminal repeat (LTR) CopiaSL 36 element (GenBank ID: LC012662). The gene appears to be functional and expressing, as two ESTs (GD126554.1, and GD126641.1) were found in the NCBI’s EST database. The splicing of the flowering plant specific sixth intron in the 3′ UTR of SlRca1-β, StRca1-β, CaRca1-β, CaRca2-β, and in all the tobacco Rca genes were identified. However, the EST sequences of other gene copies SlRca2-β, StRca2-β, and CaRca3-β showed no evidence for intron splicing at the 3′ UTR sequences (Fig. 4).
Expression analysis and protein prediction using EST and cDNA sequences revealed that the Rca genes of Solanaceae species tomato, potato and capsicum showed expression of only RCA-β isoforms; whereas one of the six Rca genes of cultivated tobacco expresses both RCA-α and RCA-β isoform via alternate splicing, and the other five genes express RCA-β isoform. The transcripts coding NtRCA1-α isoform were less abundant in the tobacco EST database than of NtRCA1-β as only two ESTs (FG198824.1 and EB435319.1) out of 46 showed alternate splicing to code for NtRCA1-α isoform. The peptide sequence analysis of NtRCA1-α showed that both evolutionarily conserved redox sensitive Cys residues at the CTE domain were mutated to Ser (Serine) residues. It is not clear whether the NtRCA1-α is redox regulated or not in tobacco. In the Solanaceae species, the molecular weight of mature RCA-β isoforms (after removing the 55–58 aa long cTP sequence) was ~ 42 kDa, whereas the SlRCA2-β, and StRCA2-β showed molecular weights of 44.23 and 44.43 kDa respectively, because of the prediction of a shorter (43aa long) cTP domain (Table 3).
Evolution of RCA domains: integration of chloroplast transit and redox sensitive C-terminal extension domains
Domain and motif analyses showed that additional structural and functional domains were gained during the evolution of higher plant RCA from its prokaryotic form (Fig. 5a). For this analysis, we aligned the RCA protein sequences of cyanobacterium, chlorophyte, charophyte, bryophyte, and a higher plant, and compared the domain structures (Supplementary Fig. S10). Compared to the higher plants, RCA from cyanobacterium Anabaena sp. strain CA lacks the cTP and the N terminal domains. The ATPase and C terminal domains are conserved among all species, and appear to be critical for basic RCA function. The cyanobacterium RCA has a ~ 115aa CTE region that is similar to the Rubisco small like superfamily (NCBI conserved domain accession cd00307; Interval 328–411 and E-value 3.06e−44), but not to the redox sensitive CTE found in the RCA of higher plants (Fig. 5a). However, there were two Cys residues, separated by 17 amino acids in the cyanobacterial CTE region, which may form disulfide bridges at oxidative conditions. To further confirm the absence of cTP and N terminal domain, and presence of cyanobacterial CTE region, we aligned the RCA sequences of ten more cyanobacterial species (Nostoc sp., Cylindrospermum stagnale, Anabaena variabilis, Calothrix sp., Nodularia spumigena, Anabaena cylindrical, Nostoc azollae, Nostoc punctiforme, Fischerella sp., Trichodesmium erythraeum) with chlorophyte and higher plant RCA sequences (Supplementary Fig. S11). The RCA protein from Anabaena sp. strain CA is very similar to that from other cyanobacterial strains, and all the ten analyzed RCA proteins lacked the cTP or N terminal domains found in higher plants, and had different types of CTE domains.
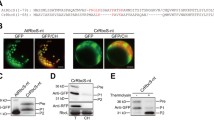
Structural and functional evolution of RCA in the photosynthetic organisms. A Detailed RCA domains of different group of organisms. The color bars and its length represent the presence of different RCA domains in each group, white box at the CTE region of cyanobacteria shows a Rubisco small like superfamily, shaded region of charophytes represent the CRISPR-associated RAMP protein region. The black downward arrows show redox regulated Cys residues in the region. B Protein BLAST analysis showing the light sensitive CTE like domains found in the genes of different chlorophytes. The black downward arrow mark the conserved cysteine residues involved in redox regulation. C Schematic diagram showing important changes occurred in plant RCA during the course of evolution (not on scale)
The RCA amino acid sequences of all 11 analyzed chlorophyte species (Chlamydomonas reinhardtii, Coccomyxa subellipsoidea C-169, Micromonas sp. RCC299, Ostreococcus lucimarinus CCE9901, Micromonas pusilla CCMP1545, Volvox carteri, Ostreococcus tauri, Bathycoccus prasinos, Micromonas sp. RCC299, Chlorella variabilis, and Auxenochlorella protothecoides 0710) showed some variations at amino acid levels, but the domain structure and sizes were highly similar (Supplementary Fig. S3). Compared to the cyanobacterial RCA, these Chlorophytic RCA proteins showed two additional domains, a short-cTP (33aa long in case of CrRCA1) domain, and an N-terminal domain along with the conserved ATPase, Rubisco recognition, and C-terminal domains. The chlorophytes’ RCA also showed loss of Rubisco small subunit like domain at the C terminal, which was found in the cyanobacterial RCA and resembled more more closely RCA-β of higher plants (Fig. 5a).
The RCA domains and structure of charophyte, bryophyte, and higher plants RCA, look similar and appeared to be evolved before the divergence of charophyte species and conserved during the evolution. One of the major evolutionary leaps occurred as the charophyte RCA gained a redox sensitive CTE domain at the end of C terminal of the protein (Fig. 5a). The KfRca1 gene of charophytes model organism K. flaccidum expresses two larger RCA isoforms, KfRCA1-α1 and KfRCA1-α2, with a size of 594 aa and 494 aa through mRNA alternate splicing. The KfRCA1-α1 contain 142 aa long CTE domains; whereas the KfRCA1-α2 contain 42aa long CTE domain, which is highly similar to higher plants CTE domain in terms of length, sequence identities and number of aa residues between conserved ‘Cys’ residues (Supplementary Fig. S4). In the CTE domain of KfRCA1-α1, the first 38 amino acids have high similarity with the CTE domain of early land and higher plant’s RCA, whereas the remaining 104 amino acids at the C-terminal end have some similarity with the CRISPR-associated RAMP protein (E value 1.74e−03). Although KfRCA-α1-CTE has high similarity with that from higher plants, out of two, only one of the Cys residues, which forms disulfide bridge at low oxidative conditions, is conserved (Supplementary Fig. S1). There are two more Cys residues towards the C-terminal end. However, the role of these residues in disulfide bridge formation is not known. Other charophytes, such as Penium margaritaceum, Spirogyra pratensis, and Chaetosphaeridium globosum, express RCA-α isoforms with CTE regions similar to higher plants. We did not find any ESTs for smaller RCA-β isoform expression in K. flaccidum; however, we found EST sequences supporting the expression of smaller RCA-β isoform in other charophyte species (Supplementary Fig. S4).
Bryophytes, known as early land plants, have RCA domains and structures similar to those present in higher plants. Unlike other lower organisms containing only a single RCA coding gene, these plants have multiple genes, along with conservation of redox sensitive CTE domain in at least one of these genes. The PpRca1 gene of P. patens expresses both larger and smaller RCA isoforms, as found in higher plants, and the other all four express RCA-β isoforms without redox regulated CTE domain. In higher plants, including gymnosperm, except Solanaceae species, which express RCA-β isoform all studied species express both RCA-α and RCA-β, and we did not find any change in the domain structure and order of these isoforms.
The presence of redox sensitive CTE domain in charophyte RCA, but not in the closely related and evolutionarily diverged green algae lineage chlorophytes, raises an intriguing question about its source and origin. Previous studies on the evolution of protein domains showed that gene fusion, or exon recombination mechanisms, majorly contributes to the emergence of novel domains in the existing proteins (Marsh and Teichmann 2010). We hypothesized that the insertion of CTE domain in the RCA of charophytes could have come from some other genes or genomic regions before the divergence of charophyte families and in the ancestral green algae species. To find the putative CTE domain coding DNA sequences in the genomic sequences of chlorophytes, we first analyzed the CTE domain of evolutionarily diverged plant RCA-α isoforms to get a consensus sequence of redox sensitive CTE domain, and use it as a query. The consensus sequence PVPEGCTDPXAXNFDPTARSDDGT/SCXYX, containing the two Cys residues that form disulfide bridges under oxidative conditions was conserved in most species. The length of the aa residues, which form a loop because of the Cys disulphide bridge, was 18, and this number is conserved in all RCA-α isoform (Supplementary Fig. S12).
To determine the source sequence of redox sensitive CTE domain in the land plant RCA, we performed a TBLASTN and BLASTP search using the consensus CTE sequences (28 aa long) in the NCBI genomic and protein sequences of chlorophytes. The results showed up to 69% amino acid sequence similarity among various proteins of different chlorophyte species. Interestingly, N-terminal region of Micromonas. sp. RCC299 IPT transcription factor protein (XP_002507277.1) showed 92% query coverage, with 50% sequence similarity with consensus CTE sequence. The predicted gene (MICPUN_55384) encoding the above-mentioned protein has three exons, and the first exon codes the aa sequence similar to the CTE region of RCA (Supplementary Fig. S13a). This gene could be a potential source of RCA-CTE domain, mainly because (1) it has two Cys residues separated by 18 aa loop, as found in consensus CTE sequence. (2) the peptide is coded from the 5′ end of the first exon, which has a high chance to fuse with a 3′ end of the Rca gene and create a C-terminal extension (Supplementary Fig. S13b). (3) most of the conserved residues of CTE domain are present in its position. The other sources could be the COCUSDRAFT_49151 gene of Coccomyxa subellipsoidea C-169, which codes for a hypothetical protein (GenBank ID: EIE18627.1) and has 82% query coverage and 57% sequence similarity. The gene contains 14 exons, and the putatively translated protein part of the fourth exon matched with the consensus sequence. Similarly, the ABO93836 of Ostreococcus lucimarinus CCE9901 has 62% sequence similarity with the consensus CTE sequence (Fig. 5b).
Evolution of the alternate splicing site in the 3′ end of the RCA coding mRNAs
Many plant species contain at least one Rca gene, which is capable of expressing both the RCA-α and RCA-β isoforms via alternate splicing of the pre-mRNA. In higher plants, the expression levels of RCA-α and RCA-β isoforms, regulated through alternate splicing, change during circadian rhythm, leaf development (Rundle and Zielinski 1991a, b) and stress conditions such as heat (Wang et al. 2010; DeRidder et al. 2012). We identified two types of alternate splicing of introns occuring at the 3′ end of the Rca mRNAs: (1) an intron is spliced before the RCA-β isoform stop codon, and translation is extended into the next exon to produce the larger RCA-α isoform. (2) an intron is spliced after the RCA-β isoform stop codon (i.e. in the 3′ UTR of RCA-β isoform coding transcripts) resulting in the production of essentially the same smaller RCA-β isoform. In both the cases, the end site of the splicing is the same, whereas the start site determines the type of isoform coding from the transcripts (Fig. 2). Comparative analysis of RCA coding genes of evolutionarily significant species showed that two types of alternate splicing mechanisms evolved at different time periods, the alternate splicing results in the larger RCA-α isoform originated in charophytes and the alternate splicing at 3′ UTR appeared after the divergence of flowering plants.
The alternate splicing mechanism, which determines the retention of the CTE region, probably evolved in charophytes as a part of CTE domain addition. We found that the RCA coding gene (KfRca1) of charophyte K. flaccidum contains sequences to code for the CTE domain in the 10th and 11th exons; however, no EST or cDNA sequence capable of expressing RCA-β isoforms was identified in the database. In other charophyte species, transcripts expressing both the isoforms via alternate splicing were identified (Supplementary Fig. S14 a). The EST sequences of charophytes Chaetosphaeridium globosum and Chlorokybus atmophyticus show alternate splicing of RCA coding transcripts similar to that of higher plants, in which the putative larger RCA-α isoform encoding transcripts splice additional intron sizes of 50 bp and nine bp respectively, with an in-frame stop codon, and extend the translation to sequences coding the CTE domain (Supplementary Fig. S14b, c). The transcripts encoding putative RCA-β did not splice the intron with in-frame stop codon, resulting in longer 3′ UTRs and smaller isoform translation. Similarly, the mRNA transcripts expressed from MpRca1 (Mapoly0022s0132) of bryophytic liverwort (Marchantia polymorpha) showed a splicing after 196th nucleotide position at the 3′ side of the fifth intron, resulting in an in-frame stop codon after 20 nucleotides of the sixth exon and translation of a smaller 445 aa RCA protein. The putative larger 479 aa isoform is translated from alternatively spliced transcripts, in which the fifth intron splices at 337th nucleotide, resulting in a 125 bp sixth exon with a stop codon at the end (Fig. 2). In the EST database analysis, out of 11 Marchantia polymorpha (Taxid: 3197) ESTs showing splicing of the fifth intron, only two (BJ857792 and BJ867397) showed 337 bp intron splicing, and the rest of the seven ESTs (BJ864269, BJ867834, C96375, BJ864479, BJ85774, BJ857034, and BJ868265) spliced 196 bp intron, suggesting the transcripts coding the smaller isoforms are relatively more abundant than the transcripts coding larger isoforms, as found in many higher plants. Similarly, one of five Rca genes of bryophytic model P. patens showed alternate splicing as found in charophyte’s Rca and M. polymorpha Rca1; the other four did not have any evidence to support alternate splicing. The PpRca1-α transcript splices the 158 bp fifth intron, extends the translation into the sixth exon, and produces a 477aa isoform with CTE domain; whereas the PpRca1-β transcript retains the fifth intron with an in-frame stop codon and transcribes the sixth exon as a UTR (Fig. 2). The transcripts of single Rca gene PaRca1 of gymnosperm species Picea abis splice an intron of 133 bp at the 3′ side to translate PaRca1-α, or retain to translate the PaRca1-β.
Discussion
Plants adapt to changing environmental conditions via modifications in basic biological processes. The early land plants (embryophytes) were exposed to extreme temperatures, high light intensities and prolonged drought conditions (Spicer et al. 1989; Waters 2003). Numerous molecular, cellular, and physiological changes allowed early plant species to colonize land areas, adapt slowly to harsh growing conditions, and radiate into entirely different ecosystems during evolution. Most of these adaptive changes, acquired by gene duplications and mutations in the genomes, played a vital role during the evolution by creating novel traits in the organisms for better survival (reviewed by Pires and Dolan 2012). Genes involved in important processes such as photosynthesis, cell division, and development, are believed to be highly conserved. However, without losing original functions, some genes gained new functions, improved activity or stability, via beneficial mutations and alternate or differential expressions, providing additional adaptive advantages to the organisms. Understanding these evolutionary changes is important for engineering future plants with better and wider adaptability. This study shows the changes that occurred at structural and functional levels in one of the important photosynthetic genes, Rubisco activase (Rca), during the course of evolution, and their implications on gene copy number, exon–intron evolution, expression, and function, in different plants.
Polyploidy and gene duplications increased Rca copy number in plant genomes
Although the number RCA coding genes from cyanobacteria to flowering plants showed variation in number from one to six genes in their genome, our analysis shows that most of the time, a single Rca gene was conserved in the genome until the evolution of monocots and eudicots. Unlike the genomes of cyanobacteria and chlorophytes, which have a single Rca gene, those early terrestrial plants bryophytes and tracheophyte showed rapid increase in Rca genes, and that could be because of the large scale genome and gene duplication events that occurred during the evolution (Rensing et al. 2008; Banks et al. 2011). The genome of another bryophyte M. polymorpha contains only one Rca gene, suggesting that not all the species of bryophytes underwent the same level of genome or gene duplication events as of P. patens. Polyploidy and gene duplication played a major role in copy number increase of Rca genes in plant genomes. Most of these copy number increases were observed after the divergence of families and subfamilies, and some were observed even at genus levels. For example, the tandem Rca gene duplication occurred before the divergence of grass families doubled the basic copy number to two and the hexaploid wheat contained up to six copies. Other monocot species, such as banana and pineapple have three Rca copies located on different chromosomes (Supplementary Table S1). Similarly, the Solanaceae family specific polyploidy event (Bombarely et al. 2016) could have changed the Rca gene copy number in the genome of Solanaceae species. These duplications could have relaxed the purifying selection pressure and probably allowed significant changes to occur with in the Rca genes.
Structure and expression pattern of Rca gene evolved before the divergence of charophytes
The exon–intron structure of higher plant Rca appears to have evolved in charophytes, as the bryophytic Rca gene structures were more similar to the Rca genes of higher plants. In most cases, species-specific intron loss was observed, and it was very frequent in the tandemly duplicated Rca genes of grasses (Fig. 3). This rapid loss of intron was not identified in some species with multiple Rca genes, present in different genomic locations. In most cases, the intron loss with very precise removal from the gene had little or no effect on the translated protein sequences. Irrespective of intron size, the intron phase, which is defined as the insertion location of an intron in a codon sequence of a gene, was essentially the same for the Rca gene exons of the same size within different species.
Tandem duplication of Rca genes in cereals was first identified in barley (Rundle and Zielinski 1991b), and then in wheat (Carmo-Silva et al. 2015). The tandem Rca duplication seems to have occurred twice during evolution, once in the bryophytes, or at least in the mosses (PpRca4 and PpRca5), and the second time in the common ancestor of the cereals (Fig. 3). The tandemly duplicated genes of cereals indicate that the changes occurred after the divergence from the common monocot ancestor. The capacity to express RCA-α by Rca1 in maize and sorghum, and Rca2 in cold cereals and rice, suggests that the ancestral copy of the tandemly duplicated genes was capable of coding both isoforms. After the divergence from a common ancestor species, random mutations occurred in the Rca2 of maize and sorghum, and the Rca1 of cold cereals have abolished the alternate splicing of mRNA and caused RCA-β expression (Fig. 3). Additionally, the alternate splicing at the 3′ UTR of maize ZmRca2 further strengthens the above-mentioned hypothesis. These observations show that the ancestral Rca gene involved in the duplication event of cereals should have coded both RCA-α and RCA-β isoforms, via the conserved alternate splicing mechanism found in other higher plants.
Identification of tandem Rca duplication in grass species sheds light on the heat stress induced expression patterns of different isoforms. During heat stress, photosynthetic capacity of a plant is affected by damages that occurred in electron transport, and reduced RUBP regeneration rate. These are important factors, but decreased Rubisco activity levels, caused by the failure of heat sensitive RCA to keep pace with the rate of Rubisco deactivation during heat stress, is widely recognized as the bottle neck (Salvucci and Crafts-Brandner 2004; Kim and Portis 2005; Sage et al. 2008). RCA expression and activity in plants acclimates to prolonged heat stress via various mechanisms viz., post transcriptional modifications of Rca-mRNA, as shown in Arabidopsis and cotton (DeRidder and Salvucci 2007; DeRidder et al. 2012); novel isoform expression as found in cotton, wheat and maize (Law et al. 2001; Ristic et al. 2009); and changes in the RCA-α: RCA-β ratio as seen in Rice (Wang et al. 2010). It is also shown that multiple mechanisms may activate within a single species to facilitate the acclimation of RCA to heat stress. Cross analysis between the previously published results and our findings revealed that the tandemly duplicated grass Rca genes evolved differently for heat induced gene expression. Previous studies showed that wheat expressed a novel smaller RCA isoform (41–42 kDa) during prolonged heat stress (Law and Crafts-Brandner 2001; Ristic et al. 2009). Later, Kumar et al. (Kumar et al. 2016) showed that heat stress increases the expression of TaRca1 in wheat cultivars, and its transcript levels correlated positively with TaRCA1 activity and rubisco activation. Similarly, heat stress experiments on maize showed expression of new 45 kDa larger isoform in the heat treated leaves, and its disappearance after recovery at normal temperatures (Sánchez de Jiménez et al. 1995; Ristic et al. 2009). These results suggest that the Rca1 genes of wheat and maize respond to high temperate stress conditions to express isoforms that are not expressed at high levels during normal conditions. It could be possible that the Rca1 gene of grasses, except the OsRca1 of rice, may respond differently to high-temperature stress conditions, and contain thermal tolerant amino acid variants in the isoform; however, we need more information on each isoform to identify their roles in Rubisco activation under heat stress conditions.
The protein crystallization studies of green type tobacco RCA-β suggest that this enzyme may function as a hexamer (Stotz et al. 2011). In Arabidopsis, a single RCA gene codes both RCA-α and RCA-β isoforms, which are the same at sequence level, except for the CTE domain of RCA-α, and these isoforms appear to form hetero-hexamers. These hetero RCA holoenzymes have a functional significance of conferring the redox regulated properties of RCA-α to the RCA-β isoform when present in multimeric state. In the case of plants with multiple Rca genes, it is not known whether the isoforms expressed from different genes with sequence changes could form hexamers or not. For example, amino acid level sequence similarity of TaRCA1 and TaRCA2 is ~ 88%, and the TaRCA2 is composed of RCA-α and RCA-β isoforms. Further research is needed to determine the conditions, capacity, rate, and ratio of different isoforms to form hexamers, and their effect on Rubisco activation.
Evolution higher plant RCA domains
All major changes that occurred in the Rca gene during evolution are summarized in Fig. 5c. Comparative analysis of RCA domains from prokaryotic algae to higher plants showed the addition of cTP and N-terminal domains to prokaryotic cyanobacterial type RCA before the divergence of eukaryotic green algae (chlorophytes and charophytes); the gaining of redox sensitive Cys residues containing CTE domain at the end of C-terminal and associated alternate splicing mechanism, which splices RCA-α and RCA-β isoform coding transcripts in some lineages of charophytes; and the loss of CTE domain, caused by mutations in the RCA coding sequences of some Solanaceae species. Structural and expressional level changes were also observed in the tandemly duplicated Rca gene of grass family members. In other words, the Rca gene underwent two major evolutionary changes: (1) probably during the primary endosymbiosis event in which the Rca gene migrated to the nuclear genome, along with many other genes, the Rca gene gained sequences coding cTP and N-terminal domains, and lost the sequences coding Rubisco smaller subunit like domain at 3′ end; (2) during the divergence of charophytes and land colonization, the Rca gene gained additional sequences coding redox sensitive CTE domain, possibly from another gene and alternate splicing of mRNA transcripts, producing both RCA-α and RCA-β isoforms.
Unlike plants, not all cyanobacteria code for RCA (Zarzycki et al. 2013). Species without RCA predominately code for another AAA+ protein called CbbX, which functions similarly to RCA, as shown in the α-proteobacterium Rhodobacter sphaeroides. Both cyanobacterial RCA and CbbX differed significantly for sequence identity but appeared to function the same (Mueller-Cajar et al. 2011). One of the major leaps in the RCA evolution is the migration of the RCA coding sequences to the nuclear genome and gaining of short cTP and N-terminal to the core RCA structure before the divergence of chlorophytes. This period also showed other major evolutionary changes at the organism level, including endosymbiosis, organelle DNA migration to nuclear genome, and cellular compartmentalization which marked the introduction of oxygenic photosynthetic eukaryotes (Archibald 2009; Keeling 2010). The Rca gene of cyanobacterial species is located adjacent to the Rubisco larger subunit (rbcL) and smaller subunit (rbcS) coding genes, and in the same transcription direction (Li et al. 1993); whereas in chlorophytes, as with other higher plants, the rbcL is in the chloroplast genome and rbcS and Rca genes are located in the nuclear genome (Khrebtukova and Spreitzer 1996). Thus, gaining of cTP becomes necessary for RCA, as with eukaryotes, it is transcribed in the nucleus, translated in the cytoplasm, and it must be transported to the pyrenoid of chlorophytes and chloroplasts of higher plants for activating Rubisco (Salvucci et al. 1985; McKay et al. 1991). Similarly, addition of the N-terminal domain to RCA is important for eukaryote Rubisco-RCA interaction, as the residues present in the N-terminal domain were necessary for Rubisco activation and the domain appears to be interacting directly with Rubisco (Van de Loo and Salvucci 1996; Esau et al. 1996). However, it is not clear how the cyanobacterial RCA, which lacks the N-terminal domain, interacts with its Rubisco. Interestingly, the crystallization studies on Arabidopsis (Hasse et al. 2015) and tobacco (Stotz et al. 2011) RCA showed flexible attachment of the N-terminal domain, which could be the result of later addition to the core RCA domains.
In most of the RCA-α coding genes of higher plants, the last exon contains CTE coding sequences, and alternate splicing of this exon determines the redox sensitivity of the isoform. Our analyses suggest that the plant RCA appeared to have acquired the CTE coding sequences from a gene coding IPT transcription factor of chlorophytes through exon fusion during the divergence of green algae and land plants (Supplementary Fig. S13). A similar example is the Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of higher plants, which underwent a gene duplication (Brinkmann et al. 1989), and gained the redox regulated CTE coding region from the CP12 gene (Pohlmeyer et al. 1996). The addition of C-terminal extension with redox regulated Cys residues in RCA-α for light sensitivity could be an evolutionary advantage for land plants to regulate overall photosynthetic flow. Besides RCA, many other Calvin-Benson cycle enzymes, including Phosphoribulokinase (PRK), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Fructose-1,6-bisphosphatase (FBPase), and Sedoheptulose-1,7-bisphosphatase (SBPase), are also regulated by the chloroplast redox state, and directly activated by thioredoxin (for review see Michelet et al. 2013; Gütle et al. 2017). Redox regulated Cys residues of these above-mentioned enzymes appear to have evolved at different time points of plant evolution (Gütle et al. 2017). During sub optimal light conditions, activity of these above-mentioned enzymes is reduced, which may affect metabolite generation or utilization. In these limiting conditions, the redox sensitive RCA-α down regulate Rubisco activation and carbon fixation (Zhang et al. 2002). The light regulation mechanism of Solanaceae RCA, which lost the CTE domain during evolution, is not known. We propose that the additions of these novel domains to RCA at different points of evolution were necessary for plant photosynthetic system development, and probably for adaptation to the new environment.
Although this study provides some aspects of structural and functional evolution of Rca genes in plants, still many important features are not covered. For example, a similar systematic analysis could be performed on the promoter sequences of Rca genes to see how and when different elements, such light and circadian controls (Rundle and Zielinski 1991a, b; To et al. 1999; Ayala-Ochoa et al. 2004), were integrated during evolution, and their influence on the gene expression. Additionally, an in-silico expression analysis for Rca gene copies using the available RNA-seq and microarray data of evolutionarily important plant species would provide information about the expression profile of individual Rca genes. By combining promoter and expression profile analyses, we can correlate the expression variations to different promoter elements and their evolution. Similarly, further investigation is needed on the factors regulating post transcriptional modifications of RCA coding pre-mRNAs, which determine the alternate polyadenylation sites, stability and translational efficiency.
In conclusion, Rca gene is one of the plant genes evolutionarily conserved for its structure and function. Copy number analysis of this gene showed that a single gene copy was maintained until the flowering plant evolution and multiple genome or gene duplication events increased the numbers in some lineages. The structure of higher plant Rca genes evolved in charophytes, and in most cases, intron sizes and sequences showed more variations than those of exons. In grasses, a tandem duplication of Rca gene occurred before the divergence of Poaceae family, and the duplicated genes evolved differently for intron loss and expression pattern. The Rca genes of Solanaceae species lost the last exon coding redox regulated CTE domains due to mutations in the coding region. At the protein level, the RCA gained cTP and N-terminal domains before the divergence of chlorophytes and redox sensitive CTE domain in the charophytes. Taken together, the RCA of higher plants evolved in a stepwise manner by incorporating different changes to adapt to the evolving photosynthetic machinery. Further understanding on these changes will help us to design a better RCA enzyme with increased Rubisco activation activity and thermal stability.
Abbreviations
- RCA:
-
Rubisco activase
- cTP:
-
Chloroplast transit peptide
- CTE:
-
C-terminal extension
- AAA+:
-
ATPases associated with diverse cellular activities
References
Amborella Genome Project. (2013) The Amborella genome and the evolution of flowering plants. Science 342, 1241089
Andersson I (2008) Catalysis and regulation in Rubisco. J Exp Bot 59:1555–1568
Archibald JM (2009) The puzzle of plastid evolution. Curr Biol 19:R81–R88
Ayala-Ochoa A, Vargas-Suárez M, Loza-Tavera H, León P, Jiménez-García LF, Sánchez-de-Jiménez E (2004) In maize, two distinct ribulose 1,5-bisphosphate carboxylase/oxygenase activase transcripts have different day/night patterns of expression. Biochimie 86:439–449
Badger MR, Lorimer GH (1981) Interaction of sugar phosphates with the catalytic site of ribulose-1,5- bisphosphate carboxylase. Biochemistry 20:2219–2225
Banks JA, Nishiyama T, Hasebe M et al (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332:960–963
Beuf L, Kurano N, Miyachi S (1999) Rubisco activase transcript (rca) abundance increases when the marine unicellular green alga Chlorococcum littorale is grown under high-CO2 stress. Plant Mol Biol 41:627–635
Bombarely A, Moser M, Amrad A et al (2016) Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nature Plants 2:16074
Brinkmann H, Cerff R, Salomon M, Soll J (1989) Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol 13:81–94
Brooks A, Portis AR (1988) Protein-bound ribulose bisphosphate correlates with deactivation of ribulose bisphosphate carboxylase in leaves. Plant Physiol 87:244–249
Carmo-Silva AE, Salvucci ME (2013) The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol 161:1645–1655
Carmo-Silva E, Scales JC, Madgwick PJ, Parry MA (2015) Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ 38:1817–1832
DeRidder BP, Salvucci ME (2007) Modulation of Rubisco activase gene expression during heat stress in cotton (Gossypium hirsutum L.) involves post-transcriptional mechanisms. Plant Sci 172:246–254
DeRidder BP, Shybut ME, Dyle MC, Kremling K, Shapiro G MB (2012) Changes at the 3′-untranslated region stabilize Rubisco activase transcript levels during heat stress in Arabidopsis. Planta 236:463–476
Dhaliwal AK, Mohan A, Gill KS (2014) Comparative analysis of ABCB1 reveals novel structural and functional conservation between monocots and dicots. Front Plant Sci 5:657
Emanuelsson O, Nielsen H, Heijne GV (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Esau BD, Snyder GW, Portis AR (1996) Differential effects of N- and C-terminal deletions on the two activities of rubisco activase. Arch Biochem Biophys 326:100–105
Feller U, Crafts-Brandner SJ, Salvucci ME (1998) Moderately high temperatures inhibit Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase-mediated activation of Rubisco. Plant Physiol 116:539–546
Gütle DD, Roret T, Hecker A, Reski R, Jacquot J-P (2017) Dithiol disulphide exchange in redox regulation of chloroplast enzymes in response to evolutionary and structural constraints. Plant Sci 255:1–11
Hasse D, Larsson AM, Andersson I (2015) Structure of Arabidopsis thaliana Rubisco activase. Acta Crystallogr Sect D: Biol Crystallogr 71:800–808
Jordan DB, Chollet R (1983) Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5-bisphosphate. J Biol Chem 258:13752–13758
Keeling PJ (2010) The endosymbiotic origin, diversification and fate of plastids. Philoso Trans R Soc London Series B Biol Sci 365:729–748
Khrebtukova I, Spreitzer RJ. (1996) Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA 93:13689–13693
Kim K, Portis AR (2005) Temperature dependence of photosynthesis in Arabidopsis plants with modifications in Rubisco activase and membrane fluidity. Plant Cell Physiol 46:522–530
Kramer EM (2009) Aquilegia: a new model for plant development, ecology, and evolution. Annual Rev Plant Biol 60:261–277
Kumar A, Li C, Portis AR (2009) Arabidopsis thaliana expressing a thermostable chimeric Rubisco activase exhibits enhanced growth and higher rates of photosynthesis at moderately high temperatures. Photosyn Res 100:143–153
Kumar RR, Goswami S, Singh K et al (2016) Identification of putative RuBisCo Activase (TaRca1)—the catalytic chaperone regulating carbon assimilatory pathway in wheat (Triticum aestivum) under the heat stress. Front Plant Sci 7:1–18
Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu G (2007) Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19:3230–3241
Law RD, Crafts-Brandner SJ (2001) High temperature stress increases the expression of wheat leaf ribulose-1,5-bisphosphate carboxylase/oxygenase activase protein. Arch Biochem Biophys 386:261–267
Law RD, Crafts-Brandner SJ, Salvucci ME (2001) Heat stress induces the synthesis of a new form of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in cotton leaves. Planta 214:117–125
Li L-A, Janet L, Tabita FR (1993) The Rubisco activase (rca) gene is located downstream from rbcS in Anabaena sp. strain CA and is detected in other Anabaena/Nostoc strains. Plant Mol Biol 21:753–764
Lorimer GH (1981) The carboxylation and oxygenation of Ribulose 1,5-biphoshate: the primary events in photosynthesis and photorespiration. Annu Rev Plant Physiol 32:349–383
Lorimer GH, Miziorko HM (1980) Carbamate formation on the epsilon-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by CO2 and Mg2+. Biochemistry 19:5321–5328
Marsh JA, Teichmann SA (2010) How do proteins gain new domains? Genome Biol 11:126
McKay RML, Gibbs SP, Vaughn KC (1991) RuBisCo activase is present in the pyrenoid of green algae. Protoplasma 162:38–45
Michelet L, Zaffagnini M, Morisse S et al (2013) Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci 4:470
Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M (2011) Structure and function of the AAA + protein CbbX, a red-type Rubisco activase. Nature 479:194–199
Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9:27–43
Pires ND, Dolan L (2012) Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc B: Biol Sci 367:508–518
Pohlmeyer K, Paap BK, Soll J, Wedel N (1996) CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol Biol 32:969–978
Pollock SV, Colombo SL, Prout DL, Godfrey AC, Moroney JV (2003) Rubisco activase is required for optimal photosynthesis in the green alga chlamydomonas reinhardtii in a Low-CO2 atmosphere1. Plant Physiol 133:1854–1861
Portis AR, Li C, Wang D, Salvucci ME (2008) Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot 59:1597–1604
Rensing SA, Lang D, Zimmer AD et al (2008) The physcomitrella genome reveals evolutionary insights into the of land by plants conquest. Science 319:64–69
Ristic Z, Momcilovic I, Bukovnik U et al (2009) Rubisco activase and wheat productivity under heat-stress conditions. J Exp Bot 60:4003–4014
Roesler KR, Ogren WL (1990) Primary structure of chlamydomonas-reinhardtii Ribulose 1,5-bisphosphate carboxylase oxygenase activase and evidence for a single polypeptide. Plant Physiol 94:1837–1841
Rundle S, Zielinski R (1991a) Alterations in barley Ribulose-1,5-bisphosphate carboxylase/oxygenase activase gene expression during development and in response to illumination. J Biol Chem 266:14802–14807
Rundle S, Zielinski R (1991b) Organization and expression of two tandemly oriented genes encoding ribulosebisphosphate carboxylase/oxygenase activase in barley. J Biol Chem 266, 4677–4685
Sage RF, Way DA, Kubien DS (2008) Rubisco, Rubisco activase, and global climate change. J Exp Bot 59:1581–1595
Salvucci ME (2004) Potential for interactions between the carboxy- and amino-termini of Rubisco activase subunits. FEBS Lett 560:205–209
Salvucci ME, Crafts-Brandner SJ (2004) Relationship between the heat tolerance of photosynthesis and the thermal stability of rubisco activase in plants from contrasting thermal environments. Plant Physiol 134:1460–1470
Salvucci ME, Portis AR, Ogren WL (1985) A soluble chloroplast protein catalyzes ribulosebisphosphate carboxylase/oxygenase activation in vivo. Photosyn Res 7:193–201
Salvucci ME, Werneke JM, Ogren WL, Portis AR (1987) Purification and species distribution of rubisco activase. Plant Physiol 84:930–936
Salvucci ME, van de Loo FJ, Stecher D (2003) Two isoforms of Rubisco activase in cotton, the products of separate genes not alternative splicing. Planta 216:736–744
Sánchez de Jiménez E, Medrano L, Martínez-Barajas E (1995) Rubisco activase, a possible new member of the molecular chaperone family. Biochemistry 34:2826–2831
Shen JB, Orozco EM, Ogren WL (1991) Expression of the two isoforms of spinach ribulose 1,5-bisphosphate carboxylase activase and essentiality of the conserved lysine in the consensus nucleotide-binding domain. J Biol Chem 266:8963–8968
Spicer RA, Alvin KL, Baas P et al (1989) Physiological characteristics of land plants in relation to environment through time. Trans R Soc Edinburgh: Earth Sci 80:321–329
Stotz M, Mueller-Cajar O, Ciniawsky S, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M (2011) Structure of green-type Rubisco activase from tobacco. Nat Struct Mol Biol 18:1366–1370
To KY, Suen DF, Chen SC (1999) Molecular characterization of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in rice leaves. Planta 209:66–76
Van de Loo FJ, Salvucci ME (1996) Activation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) involves Rubisco activase Trp16. Biochemistry 35:8143–8148
Wang D, Li X-F, Zhou Z-J, Feng X-P, Yang W-J, Jiang D-A (2010) Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiologia Plant 139:55–67
Waters ER (2003) Molecular adaptation and the origin of land plants. Mol Phylogenet Evol 29:456–463
Watillon B, Kettmann R, Boxus P, Burny A (1993) Developmental and circadian pattern of Rubisco activase mRNA accumulation in apple plants. Plant Mol Biol 23:501–509
Weston DJ, Bauerle WL, Swire-Clark GA, Moore BD, Baird WV (2007) Characterization of Rubisco activase from thermally contrasting genotypes of Acer rubrum (Aceraceae). Am J Bot 94:926–934
Xu K, He B, Zhou S, He B, Zhou S, Li Y, Zhang Y (2010) Cloning and characterization of the Rubisco activase gene from Ipomoea batatas (L.) Lam. Mol Biol Rep 37:661–668
Yin Z, Meng F, Song H, Wang X, Xu X, Yu D (2010) Expression quantitative trait loci analysis of two genes encoding rubisco activase in soybean. Plant Physiol 152:1625–1637
Yin Z, Zhang Z, Deng D, Chao M, Gao Q, Wang Y, Yang Z, Bian Y, Hao D, Xu C (2014) Characterization of Rubisco activase genes in maize: an α-isoform gene functions alongside a β-Isoform gene. Plant Physiol 164:2096–2106
Zarzycki J, Axen SD, Kinney JN, Kerfeld CA (2013) Cyanobacterial-based approaches to improving photosynthesis in plants. J Exp Bot 64:787–798
Zhang Z, Komatsu S (2000) Molecular cloning and characterization of cDNAs encoding two isoforms of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in rice (Oryza sativa L.). J Biochem 128:383–389
Zhang N, Portis AR (1999) Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Scie USA 96:9438–9443
Zhang N, Kallis RP, Ewy RG, Portis AR (2002) Light modulation of Rubisco in Arabidopsis requires a capacity for redox regulation of the larger Rubisco activase isoform. Proc Natl Acad Scie USA 99:3330–3334
Acknowledgements
This work was supported by the USDA National Institute of Food and Agriculture, Hatch (project number WNP00449) and United States Agency for International Development Feed the Future Innovation Lab-Climate Resilient Wheat (Grant Number AID-OAA-A-13-00008).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: RN KSG. Performed the experiments: RN. Analyzed the data: RN. Contributed reagents/materials/analysis tools: KSG. Wrote the paper: RN KSG.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nagarajan, R., Gill, K.S. Evolution of Rubisco activase gene in plants. Plant Mol Biol 96, 69–87 (2018). https://doi.org/10.1007/s11103-017-0680-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-017-0680-y