The antibacterial resistance (ABR) is a growing phenomenon and global threat to mankind. To circumvent the ABR, many approaches have been put forth, but none of them meet the pre-requisites associated with the resistance mechanisms. In this review, we focused on the importance of unexploited enzyme, ParE, a topoisomerase responsible for the bacterial survival. The bacterial topoisomerases maintain the topological state of DNA. The gyrases and topoisomerases IV are validated targets for the antibacterial activity. Both these enzymes are structurally similar and possess high degree conservation in the catalytic domain of the N-terminal region, which make them appealing targets for broad spectrum antibacterial activity. Despite being an attractive target for the development of new antibacterials, there are currently no antibiotics targeting gyrases and topoisomerase (topo) IV in the market. Availability of the high-resolution crystal structure data for ParE made it possible to design new classes of antibacterials. Here, we discuss the importance of targeting topo IV enzyme as it is less prone to bacterial resistance which has been disclosed in the literature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1. INTRODUCTION
The rapid evolution of emerging antibacterial resistance (ABR) have pushed the need to explore new and alternative antibacterial agents less prone to ABR. However, progressive ages of alternative antibiotics have met with advanced influxes of resistance as the demonstration of using new agents chosen for resistant organisms. In the twentieth century, the revelation of novel antibacterial agents depended on alteration of the synthetic structure of existing anti-infection agents or antibiotics [1]. Another strategy includes the development of structurally innovative classes of antibiotics acting on clinically approved targets to reduce the prevalence of antibiotic resistance. Although there are several antibacterials on the market, it remains a fact that among most of the antibacterials targets only few refer to bacterial cellular functions: cell wall synthesis, nucleic acid synthesis, protein synthesis, and folate synthesis. The reason for emergence of bacterial resistance is evolutionary adaptation of the receptor proteins which are subject to antibacterial attack. Repeated strike of antibacterials on the same active sites results in genetic mutations, which is a primary cause of ABR prevalence.
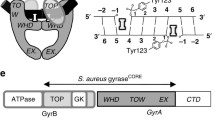
Bacterial DNAgyrase B (GyrB) and ParE enzyme are related to bacterial topoisomerases. These are clinically validated targets that utilize energy through ATP hydrolysis [2]. GyrB and ParE are considered to be exceedingly conserved topoisomerases that play essential roles in replication and transcription of DNA and are appealing targets for antibacterial drug discovery [3, 4]. DNA gyrase is a heterotetramer consisting of two GyrA and GyrB subunits. It utilizes the energy through hydrolysis of ATP to introduce negative supercoils into DNA and stabilizes the super-helical state of the bacterial chromosome [2].
On the other hand, topoisomerase (topo) IV, a related homolog of gyrase, is a heterotetramer consisting of two ParC and two ParE subunits. It is involved in the process of decatenation required for the separation of daughter chromatids behind the replication fork and after replication of DNA. The N-terminal domain (64 kDa) of GyrA and ParC subunits of topoisomerase provides the active-site tyrosine residue that irreversibly attaches to the cleaved DNA gate [2]. The C-terminal domain (33 kDa) of both GyrA and ParC differ in the similarity of genomic sequence and also in how they bind to the bacterial genome [5]. GyrB and ParE have similarities in their conserved active sites and subunit organization [6, 7]. The N-terminal region (43 kDa) of both these enzymes forms a functional dimer with adenylyl-imidodiphosphate (ADPNP) as supported by biochemical shreds of evidence [8]. The first 220 amino acid (domain 1) residues generate contacts which play a role in dimer stabilization and form a part of the ATP binding site. The remaining residues (domain 2) form the sides of a hole (20 Å) in the crystal structure of protein dimer that is supposed to serve as the cavity in which the T-segment seized for presentation to the DNA gate [9]. Both enzymes are essential for bacterial cell growth and have been successfully approved targets across a broad spectrum of bacterial species because of being structurally distinct from eukaryotic topoisomerases. Inhibition of these enzymes in bacteria results in the interruption of bacterial DNA synthesis and eventually cell death [2]. DNA gyrases are more susceptible to drug resistance because of mutations caused in the genome of gyrase subunits [10]. It is stated that GyrA subunit confers mutations associated with quinolone resistance, whereas the GyrB subunit confers mutations associated with coumarin resistance.
Studies revealed that quinolone resistance in species of E. coli k-12 strain is mainly due to the replacement of serine, aspartate, and alanine residues to tryptophan, glutamate, and serine at positions 83, 678 and 828, respectively [11]. Further studies convinced the presence of ciprofloxacin resistance in S. aureus GyrB-GyrA locus isolated from ciprofloxacin-susceptible clinical isolate 81231 [12]. The GyrA nucleotide sequence of S. aureus at N-terminus is highly homologous to that of E. coli counterpart. In particular, the substitutions of amino acid residues, responsible for the quinolone resistance in E. coli, Ala67, Ser83, and Gln106, are all conserved in the GyrA subunit at 68, 84 and 107 positions in S. aureus [13]. The review also demonstrated that GyrA of C. jejuni showed the mutations in quinolone resistance determining region (QRDR) at positions 86 and 90 [14]. Studies indicate that nalidixic acid-resistant mutants can emerge as the consequence of modified DNA gyrase. It has been suggested that MUG116 is resistant to nalidixic acid because of G-to-A transition, resulting in a substitution of the residue of aspartic acid by asparagine at position 419 [15]. Bacteria N. gonorrhoeae possess a variety of loci, and probable mutations at three loci are involved in low-level resistance to some antibiotics such as penicillin and tetracyclines [16]. The same structural modification was observed in GyrB protein as the nal-24 mutant of E. coli, which contained a G-to-A substitution at amino acid 426 [17]. Both enzymes exhibit high structural similarity and, hence, the scaffolds targeting GyrB have some affinity with that of ParE enzyme [18]. It was stated that excessive use of fluoroquinolones resulted in the emergence of resistance to these antibiotics mainly due to the mutations conferred in QRDR of ParC and GyrB and less frequently in ParE [19,20, – 21]. Because of the mutations occurring with DNA gyrase, ParE is a potential target for antibacterial agents. Till now, no drugs were filed by FDA that only be used for targeting ParE. The ParE enzyme was considered as a significant target in the strains suffering from mutations such as RecA, SeqA and GyrB and it also proved that drug norfloxacin from quinolone class of drugs showed 1/10 MIC value in in-vivo studies when compared with in vitro data and this value was even less (i.e., 1/100) for gyrase inhibition in E. coli [6, 22].
The catalytic domain of ParE (S. pneumonia) shows higher sequence similarity with the 43 kDa ATP binding pocket of GyrB (E. coli, T. thermophilus). The ParE (S. pneumonia) exhibits higher similarity at G-loop (glycine-rich segment) of N-terminal catalytic domain and EGDSA, which is highly conserved (pdb.4MOT) [19]. The 43 kDa structure of the ParE (E. coli) contained 1 to 390 amino acid residues and ordered into two distinct subdomains: a C-terminal domain (218 – 390 residues) containing a four α-helices and fourstranded β-sheets and the subdomain of N-terminus (1-217 residues) contains five α-helices and an eight-stranded β-sheets (Fig. 1).
The catalytic domain of ParE lined with arginine residues which involve in the transport of the T-segment DNA to the DNA gate present at the heterotetrameric interface [23]. The N-terminal end is considered as a catalytic domain responsible for the stabilization of monomer and binding of ATP. Though the gyrases and ParE are structurally homologous, there is largest difference in the long α-helices that are present at the unconserved C-terminus of each monomer. Because of the truncated nature of full-length protein at C terminus, the interactions are less extensive, and the region is distinct from gyrases and mainly involved in the interaction of ParC or DNA substrate, although several papers demonstrated the importance of DNA GyrB and ParE. This paper mainly describes the role of ParE inhibition in the discovery of new antibacterial drugs.
2. INHIBITORS OF ParE ENZYME
Studies revealed that ParE was more sensitive than gyrases to inhibition by levofloxacin (2), ciprofloxacin (3), sparfloxacin (5), tosufloxacin (6), gatifloxacin (4) and sitafloxacin (1), which directly indicated that ParE could be a primary target for quinolones [24]. It was reported that sparfloxacin (5) (IC50 = 0.39 mg/mL) and tosufloxacin (6) (IC50 = 0.39 mg/mL) showed highest levels of inhibitory activity against ParE (Fig. 2). Some examples of antibacterial drugs targeting ParE enzyme are pyrrolopyrimidines [25,26,27,28,29,30, – 31], pyrimidinoindoles [31,32,33,34, – 35], and some pyrrolamides. The benzo-fused heterocycles either five-membered or six-membered, showed inhibitory activity against ParE. Studies also showed that the dissociation constant (ki) is minimum for the pyrimidinoindole (11), a fused tricyclic system which exhibits maximum inhibitory activity against ParE. Pyrimidoindole scaffold showed better activity against ParE (ki = 0.9 nM) in comparison to other synthetic scaffolds like benzimidazole ethyl urea (7) (ki = 9 nM), benzothiazole ethyl urea (8) (ki = 20 nM), pyrazolopyridine (9) (ki = 183 nM), pyrazolopyrimidine (10) (ki = 1.7 nM), and pyrrolamide (12) (ki = 72 nM) (Fig. 3).
Other ParE inhibitors (Fig. 4) exhibited activity against Gram-negative organisms of E. coli strain and Gram-positive bacteria of S. aureus strain [36]. Compound 13 showed good activity against S. aureus ParE (IC50 = 0.086 mM) and E. coli ParE (IC50 = 0.94 mM) but the half-maximum inhibitory concentration (IC50) of compound 14 for S. aureus ParE was not determined and showed good activity against Gram-negative bacteria (IC50 = 0.078 mM). Compound 15 showed good activity against S. aureus ParE (IC50 = 0.80 mM) as compared to E. coli ParE (IC50 = 42 mM). The synthetic scaffold of imidazopyridine (compounds 16, 17, 18, 19, Fig. 5) along with ethyl urea side-chains showed moderate activity against ParE with MIC values >256 μg/mL for various bacteria (S. aureus, E. coli, S. pneumonia, E. faecalis) [30].
Based on analysis of binding modes of the pyrrolopyrimidine scaffold in the catalytic domain, a series of derivatives were designed and optimised. Among these, compound 20 exhibited inhibitory activity against E. coli ParE (ki = 1.2 μM) and compound 21 (Fig. 6) showed maximum inhibitory activity against Francisella tularensis ParE (ki < 1 nM). Compounds 22 and 23 (Fig. 6) containing bicyclic heteroaryl groups showed maximum interactions in the salt-bridge pocket and exhibited good activity against ParE of F. tularensis (ki < 1 nM) [37].
Compounds 24 and 25 (Fig. 7) exhibited excellent activity against Gram-positive and Gram-negative organisms with a MIC90 values less than 0.1 μg/mL, and significant inhibitory activity against E. faecalis ParE (ki < 0.3 nM) and F. tularensis ParE (ki < 0.4 nM) of (38). CompanyAstraZeneca performed virtual screening to identify 5-phenyl azaindole as a prospective dual inhibitor of topoisomerases [29]. However, compound 26 (Fig. 8) showed potent inhibitory activity against ParE (IC50 = 10 μM) at maximum interactions with various amino acid residues Asp78 (hydrogen bonding interaction) and interaction with bound water molecules. Based on the literature data, trans-cinnamic acid derivative 27 was designed (Fig. 8) and these molecules showed activity against ParE (IC50 = 7.7 μM) of S. pneumonia [28]. Compound 27 forms a salt bridge with Arg140, which seems to be key interaction for inhibition of the target enzyme.
In conclusion, ParE alone could be a potential target for various new antibacterial agents to treat multidrug resistant bacteria.
References
C. Walsh and G. Wright, ACS Publ., 105 (2), 391 – 394 (2005).
A. Maxwell and D. M. Lawson, Curr. Topics Med. Chem., 3(3), 283 – 303 (2003).
K Drlica and X Zhao, Microbiol. Mol. Biol. Rev., 61(3), 377 – 392 (1997).
P. Heisig, Planta Med., 67(01), 3 – 12 (2001).
H. Peng and K. J. Marians, J. Biol. Chem., 270(42), 25286 – 25290 (1995).
A. B. Khodursky, E. L. Zechiedrich, and N. R. Cozzarelli, Proc. Natl. Acad. Sci. U. S. A., 92, 11801 – 11805 (1995).
M. Takei, H. Fukuda, R. Kishii, et al., Antimicrob. Agents Chemother., 45(12), 3544 – 3547 (2001).
L. Brino, A. Urzhumtsev, M. Mousli, et al., J. Biol. Chem., 275(13), 9468 – 9475 (2000).
L. Brino, C. Bronner, P. Oudet, et al., Biochimie, 81(10), 973 – 980 (1999).
M. Stieger, P. Angehrn, B. Wohlgensinger, et al., Antimicrob. Agents Chemother., 40(4), 1060 – 1062 (1996).
M. Cullen, A. Wyke, R. Kuroda, et al., Antimicrob. Agents Chemother., 33(6), 886 – 894 (1989).
S. Sreedharan, M. Oram, B. Jensen, et al., J. Bacteriol., 172(12), 7260 – 7262 (1990).
R. Hopewell, M. Oram, R. Briesewitz, et al., J. Bacteriol., 172(6), 3481 – 3484 (1990).
R. Changkwanyeun, T. Yamaguchi, S. Kongsoi, et al., Drug Test. Anal., 8(10), 1071 – 1076.
D. C. Stein, R. J. Danaher, and T. M. Cook, Antimicrob. Agents Chemother., 35(4), 622 – 626 (1991).
P. Sparling, F. Sarubbi, and E. Blackman, J. Bacteriol., 124(2), 740 – 749 (1975).
J.-I. Yamagishi, K. Yoshida, M, Yamayoshi, and S. Nakamura, Mol. Gen. Genet., 204, 367 – 373 (1986).
Z. Liao, L. Thibaut, A. Jobson, et al., Mol. Pharmacol., 70(1), 366 – 732 (2006).
M. A. Azam, J. Thathan, and N. S. Tripuraneni, Struct. Chem., 28(4), 1187 – 200 (2017).
F. Sifaoui, V. Lamour, E. Varon, et al., J. Bacteriol., 185(20), 6137 – 6146 (2003).
C. Janoir, V. Zeller, M-D. Kitzis, et al., Antimicrob. Agents Chemother., 40(12), 2760 – 2764 (1996).
A. B. Khodursky and N. R. Cozzarelli, J. Biol. Chem., 273(42), 27668 – 27677 (1998).
S. Bellon, J. D. Parsons, Y. Wei, et al., Antimicrob. Agents Chemother., 48(5), 1856 – 1864 (2004).
Y. Onodera, J. Okuda, M. Tanaka, et al., Antimicrob. Agents Chemother., 46(6), 1800 – 1804 (2002).
P. Angehrn, E. Goetschi, H. Gmuender, et al., J. Med. Chem., 54(7), 2207 – 2224 (2011).
A.-L. Grillot, A. L. Tiran, D. Shannon, et al., J. Med. Chem., 57(21), 8792 – 8816 (2014).
J. T. Starr, R. J. Sciotti, D. L. Hanna, et al., Bioorg. Med. Chem. Lett., 19(18), 5302 – 5306 (2009).
PS Charifson, A-L Grillot, TH Grossman, et al., J. Med. Chem., 51(17), 5243 – 5263 (2008).
G. S. Basarab, J. I. Manchester, S. Bist, et al., J. Med. Chem., 56(21), 8712 – 8735 (2013).
S. P. East, C. B. White, O. Barker, et al., Bioorg. Med. Chem. Lett., 19(3), 894 – 899 (2009).
L. C. Axford, P. K. Agarwal, K. H. Anderson, et al., Bioorg. Med. Chem. Lett., 23(24), 6598 – 6603 (2013).
J. T. Palmer, L. C. Axford, S. Barker, et al., Bioorg. Med. Chem. Lett., 24(17), 4215 – 4222 (2014).
X. Huang, J. Guo, Q. Liu, et al., Med. Chem. Commun., 9(10), 1619 – 1629 (2018).
B. A. Sherer, K. Hull, O. Green, et al., Bioorg. Med. Chem. Lett., 21(24), 7416 – 7420 (2011).
A. Kumar, I. A. Khan, S. Koul, et al., J. Antimicrob. Chemother., 61(6), 1270 – 1276 (2008).
I. A. Yule, L. G. Czaplewski, S. Pommier, et al., Eur. J. Med. Chem., 86, 31 – 38 (2014).
Y. Li, Y. L. Wong, M. Y. Lee, et al., Biomol. NMR Assign., 10(1), 135 – 138 (2016).
Tricyclic Gyrase Inhibitors, US Patent 9732083, August 15 (2017).
Acknowledgements
The authors much acknowledge the All India Council of Technical Education, National Doctoral Fellowship (NDF) Grant-56149 for providing the funding support.
Conflict of Interest
The authors declare that they have no conflict of interest in this study. The authors alone are responsible for the content and writing of the paper.
Ethical Statement
The present study involved neither animals nor human volunteers. Thus, no ethical approval was required.
Author information
Authors and Affiliations
Contributions
Vidyasrilekha Yele was the author and synthesized the literature, involved in drafting the paper; Dr. Afzal Azam Md provided conceptual inputs and critical revision of the manuscript. All authors read and approved the final document.
Corresponding author
Rights and permissions
About this article
Cite this article
Yele, V., Md, A.A. Exploitation of Pare Topoisomerase IV as Drug Target for the Treatment of Multidrug-Resistant Bacteria: A Review. Pharm Chem J 54, 462–468 (2020). https://doi.org/10.1007/s11094-020-02223-w
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11094-020-02223-w