Abstract
Emerging evidence underlined the crucial roles played by long non-coding RNAs (lncRNAs) in glioma. MINCR has been reported in multiple malignancies. Here, we studied its function and potential mechanism in glioma, which remain unclear. Gene expressions were analyzed by qRT-PCR assay. Both in vitro and in vivo assays were conducted to evaluate the cellular function of MINCR in glioma. The subcellular situation of MINCR was detected by subcellular fractionation and FISH assays. Luciferase reporter, RNA pull-down and RNA immunoprecipitation (RIP) assays were combined to investigate potential mechanisms of relevant genes. MINCR was up-regulated in glioma. MINCR depletion markedly refrained glioma cell proliferation, migration and invasion via sponging miR-876-5p. MiR-876-5p suppressed the malignant behaviors of glioma via binding to GSPT1. MINCR shared the binding sites with the 3’-untranslated region of GSPT1 and prevented the binding of miR-876-5p to GSPT1 mRNA, thus up-regulating the level of GSPT1. Moreover, miR-876 inhibition and GSPT1 up-regulation counteracted the functional effect induced by silencing MINCR on glioma progression. Our findings uncovered that MINCR might aggravated glioma cell proliferation and migration via acting as competing endogenous RNA (ceRNA), indicating prospective novel therapeutic target for glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is one primary intracranial malignant tumor that stems from neutral glial cells. As a kind of aggressive and lethal malignant tumor, it has become the leading cause of central nervous system cancer-related mortality in China [1]. Over the last decade, despite encouraging improvements have been achieved in the multimodal therapeutic methods for glioma, the general prognosis of patients suffering from glioma remains dim, with overall survival ranging from 12 to 14 month after diagnosis [2, 3]. The rising incidence and fatality of glioma necessitate the exploration of its underlying pathological mechanisms.
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts containing over 200 nucleotides in length, yet incapable of encoding proteins [4]. LncRNAs have emerged as important regulators in a set of biological processes, such as chromatin modification, histone modification, transcriptional control, and post-transcriptional mediation [5, 6]. Increasing lncRNAs have been reported act as miRNA sponges to regulate cellular activities, including proliferation [7], apoptosis [8], migration [9] and radio-resistance [10]. Aberrant expression of lncRNAs is implicated in the carcinogenesis process. For example, lncRNA CASC11 has been found overexpressed in osteosarcoma and positively correlated with cell migration, invasion and epithelial-mesenchymal transition (EMT) in osteosarcoma [11]. MINCR has been reported to act as a carcinogenic gene in promoting gallbladder cancer [12]. Overexpression of MINCR also leads to poor prognosis of hepatocellular carcinoma (HCC) and it modulates cellular activities [13, 14]. Besides, MINCR was an upregulated lncRNA in non-small cell lung cancer (NSCLC) and its overexpression promoted cell proliferation [15]. Nevertheless, the role and potential mechanism of MINCR in glioma have never been explored.
In recent years, the competing endogenous RNA (ceRNA) was revealed to be an essential post-transcriptional regulatory mechanism of lncRNAs. In ceRNA network, lncRNAs could indirectly regulate target genes via interacting with specific microRNA (miRNA) [16]. This lncRNA-miRNA-mRNA regulatory network has been discovered to be closely associated with the onset, development and advancement of cancers, including glioma. For example, lncRNA ZEB1-AS1 has been found to accelerate the tumorigenesis of glioma by targeting miR-200c/141-ZEB1 axis [17]. MINCR has been revealed to act as a ceRNA to up-regulate SLC7A5 through miR-126 in non-small cell of lung cancer [18]. Therefore, we intended to explore the potential ceRNA role of MINCR in the regulation of glioma development.
Numerous reports have indicated that miRNA is widely involved in multifarious biological behavior and pathological process of cancers, including glioma [19, 20]. MiR-876-5p is a relatively well-investigated gene. It has been reported to be a tumor suppressor in hepatocellular carcinoma [21], gastric cancer [22], head and neck squamous cell carcinoma [23] and esophageal squamous cell carcinoma [24]. Of note, its tumor inhibitor role in glioma has been verified. HOXC-AS2/miR-876-5p/ZEB1 constitutes a positive feedback loop to regulate EMT in glioma [25]. However, its relationship with MINCR in glioma has never been elucidated.
Further, miRNA induces mRNA degradation or translational repression by binding with target messenger RNA (mRNA), [26,27,28]. GSPT1 served as the target gene of miR-144 or miR-27b-3p in gastric cancer [29, 30]. Besides, GSPT1 was identified as an upregulated mRNA in NSCLC [31] and promoted the proliferation, invasion and migration of NSCLC cells [32]. Nonetheless, the role of GSPT1 in glioma hasn’t been explored yet.
In present study, we aimed to elucidate the function and the potential molecular mechanism of MINCR in glioma, confirming whether it impacted the progression of glioma via engaging in the hypothesized ceRNA network.
Materials and Methods
Cell Culture
Human glioma cell lines (A172, U87, U251, and LN229) and normal glial cell line (HA1800) from ATCC (Manassas, VA) were all maintained at 37°C with 5% CO2 in DMEM (Invitrogen, Carlsbad, ca.). 1% antibiotics and 10% FBS (Invitrogen) served as the supplements for DMEM.
RNA Extraction and qRT-PCR
Total cellular RNAs were extracted with the Trizol method (Invitrogen), and then used for cDNA synthesis with Reverse Transcription Kit (Takara, Shiga, Japan). Gene expression was estimated by qRT-PCR with SYBR Green PCR Master Mix (Invitrogen) and calculated by 2−ΔΔCT method, with GAPDH or U6 used as the internal reference.
Transfection
Specific shRNAs to MINCR and NC-shRNAs (sh-NC), miR-876-5p mimics and NC mimics, as well as miR-876-5p inhibitor and NC inhibitor were all available from Genechem (Shanghai, China). Besides, pcDNA3.1/MINCR, pcDNA3.1/GSPT1 and pcDNA3.1 vectors were acquired from Genepharma Company (Shanghai, China). Transfection kit Lipofectamine 2000 (Invitrogen) was obtained for 49 h of plasmid transfection in U87 and LN229 cells.
EdU Assay
5 × 104 U87 and LN229 cells were seeded to each well of the 96-well plates adding the EdU-medium for 2 h, following washing in PBS and fixed in 4% paraformaldehyde. Based on the protocol of EdU assay kit (Ribobio, Guangzhou, China), proliferative cells were measured. Cells stained with DAPI solution were seen as the total samples.
Colony Formation
Clonogenic cells of U87 and LN229 were collected after transfection for 14-day of seeding in 6-well plates at 37°C. After culturing with 0.5% crystal violet, clones were counted.
TUNEL Assay
Transfected cells of U87 and LN229 were fixed at room temperature for 1 h, then permeated on ice in 0.1% Triton X-100. Cells were washed in PBS and cultured in the dark with 50 µl of TUNEL reaction solution for 1 h as per guidebook of TUNEL assay kit (Beyotime, Shanghai, China). DAPI dye was finally used for nuclear detection.
Flow Cytometry Assay
Transfected cells were reaped for suspending in the Binding buffer adding the 5 µL of Annexin V-PE and 7AAD according to user manuals (BD Biosciences, San Jose, ca.). After 15 min of double-staining, apoptotic cells were examined by flow cytometry (BD Biosciences).
Transwell Assay
Invasion assay was conducted using Matrigel-coated transwell chamber (Corning Co, Corning, NY) in line with user guide. Conditioned medium was added into the lower chamber, while upper chamber was added with 5 × 103 glioma cells. 24 h later, invasive cells on the bottom were fixed for crystal violet staining. Five random fields were selected in each well and counted under microscope. Migration assay was conducted without Matrigel coating.
Subcellular Fractionation
Cytoplasmic and nuclear RNAs from U87 and LN229 cells were extracted severally using PARIS™ Kit method (Ambion, Austin, TX). Isolated RNAs were detected by qRT-PCR with GAPDH and U6 as fractionation indictors.
Fluorescence In Situ Hybridization (FISH)
Fixed glioma cell samples were cultured with MINCR-FISH probe (Ribobio) for RNA FISH assay. After hybridization, samples were washed for Hoechst staining and observed by Olympus fluorescence microscope (Tokyo, Japan).
RNA Pull Down
With Pierce Magnetic RNA-Protein Pull-Down Kit, RNA pull down assay was conducted as per manual (Thermo Fisher Scientific, Waltham, MA). Protein extracts from U87 and LN229 cells were mixed with biotin-labeled MINCR or miR-876-5p probes, and then streptavidin agarose magnetic beads were added. The collected pull-downs were assayed using qRT-PCR.
Luciferase Reporter Assay
Luciferase reporter gene vectors were acquired by inserting the wild-type or mutated MINCR or GSPT1 fragments covering miR-876-5p binding sites into pmirGLO (Promega, Madison, WI). Recombinant vectors, named as MINCR WT/Mut and GSPT1 WT/Mut, were co-transfected with miR-876-5p mimics or NC mimics in HEK-293T cells (ATCC). After 48 h, samples were subjected to Luciferase Reporter Assay System (Promega).
RNA Immunoprecipitation (RIP)
With Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit, RIP assay was conducted in U87 and LN229 cells in accordance with instruction (Millipore, Bedford, MA). Cell lysates were cultivated with the beads-bound specific antibody (anti-Ago2) in the RIP buffer. Normal control anti-IgG was used. qRT-PCR was performed for the relative RNA enrichment in immunoprecipitates.
In Vivo Tumor Growth Assay
6 week-old male nude mice (weight:16–20 g) were available from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) for the purposes of study, with the approval from the Animal Research Ethics Committee of Deyang People’s Hospital. 1 × 106 transfected U87 cells were used to inoculate subcutaneously to mice, with tumor growth recorded every 4 days. 28 days later, xenograft tumors were excised carefully and weighed.
Statistical Analyses
All assays were performed in at least triplicates. Data were presented as the mean ± standard deviations (S.D.). PRISM 6 software (GraphPad, San Diego, ca.) was applied for all statistical analysis with t-test and one-way analysis of variance (ANOVA), with significant level at p < 0.05.
Results
MINCR is Overexpressed in Glioma and MINCR Knockdown Restrains Glioma Development
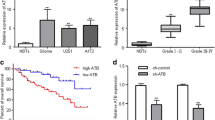
To identify the expression of MINCR in glioma, we implemented qRT-PCR and found the abnormal high expression of MINCR in glioma cell lines compared with normal glial cell (Fig. 1a). To understand the role of MINCR, we adopted loss-of-function assays in U87 and LN229 cells, where MINCR was observed significantly highly expressed. Firstly, the transfection efficiency of sh-MINCR#1, sh-MINCR#2 was validated by the results of qRT-PCR (Fig. 1b). EdU assay was utilized to evaluate cell proliferation, which showed that MINCR knockdown effectively suppressed proliferation in glioma cells (Fig. 1c). Besides, colony formation experiment confirmed the inhibitory effect on proliferation by MINCR knockdown (Fig. 1d). Moreover, TUNEL assay was conducted for the investigation of apoptosis after silencing MINCR. The higher ratio of apoptosis was found in U87 and LN229 cells transfected with sh-MINCR#1/2, indicating the accelerating effect of silencing MINCR on cell apoptosis (Fig. 1e). Flow cytometry also showed increased cells in apoptotic fraction by silencing MINCR (Fig. 1f). Later, transwell assays were performed and found that knockdown of MINCR could inhibit the migrating and invasive capacities in selected cell lines (Fig. 1g, h). Above observations elucidated the carcinogenic role of MINCR in glioma.
MINCR is overexpressed in glioma and MINCR knockdown restrains cell growth, migration and invasion, yet promoting apoptosis in glioma. a qRT-PCR analysis was conducted to detect MINCR expression in glioma cell lines (A172, U87, U251, and LN229) and normal glial cell line (HA1800). b The expression of MINCR was detected by qRT-PCR after transfecting sh-MINCR#1/2 in U87 and LN229. c, d EdU and colony formation assays were performed to study the effect of MINCR silence on glioma cell proliferation. e, f TUNEL and flow cytometry were performed to measure apoptotic rate after knockdown of MINCR. g, h Transwell assays were performed to evaluate migrating and invasive abilities of glioma cells after knockdown of MINCR. *P < 0.05, **P < 0.01
MINCR Serves as a Molecular Sponge of miR-876-5p
Growing number of lncRNAs have been reported to regulate malignancies via acting as miRNA sponges [33]. We presumed that MINCR facilitated glioma by serving as a molecular sponge for certain miRNA. Firstly, the location of MINCR in U87 and LN229 was identified via subcellular fractionation and FISH (Fig. 2a, b). Next, seven candidate miRNAs that might bind to MINCR were found in starBase (http://starbase.sysu.edu.cn/). We found that only miR-876-5p was successfully pulled down by Bio-MINCR (Fig. 2c). MiR-876-5p was also aberrantly down-regulated in glioma cell lines (Fig. 2d). The putative binding sites between miR-876-5p and MINCR were acquired by utilizing starBase bioinformatics analysis (Fig. 2e). To verify the interaction between miR-876-5p and MINCR, luciferase reporter assay was conducted in HEK-293T. It showed that the luciferase activity of MINCR WT was evidently impaired by miR-876-5p mimics, while no significant variation in that of MINCR Mut (Fig. 2f). Moreover, RNA pull down data suggested the enrichment of MINCR in Bio-miR-876-5p WT-precipitated complex (Fig. 2g). To certify that MINCR promoted glioma progression via acting as a sponge of miR-876-5p, we carried rescue assays. The expression of miR-876-5p was forced to down-regulate utilizing miR-876-5p inhibitor in selected glioma cells (Fig. 2h). MiR-876-5p inhibitor could significantly countervail the suppressive effect of sh-MINCR on proliferation (Fig. 2i, j). TUNEL assay and flow cytometry assay showed that the enhanced apoptosis ability by MINCR depletion was reversed by miR-876-5p inhibitor (Fig. 2k, l). Moreover, miR-876-5p inhibitor could offset the inhibiting effect of MINCR down-regulation on migration and invasion (Fig. 2Mm ,n). Thus, MINCR promoted glioma via sponging miR-876-5p.
MINCR serves as a molecular sponge of miR-876-5p
a, b Subcellular fractionation and FISH assays were adopted to determine the location of MINCR in glioma cells. c RNA pull down assay was performed to testify relative enrichment of candidate miRNAs. d qRT-PCR analysis was carried out to explore the expression level of miR-876-5p in cell lines. e Putative and mutate miR-876-5p binding site with MINCR. f Luciferase reporter assay was conducted to determine the relationship between miR-876-5p and MINCR. Luciferase activities were normalized to Renilla luciferase. g RNA pull down was performed to verify the interaction between miR-876-5p and MINCR. h MiR-876-5p inhibitor could decrease the expression of miR-876-5p efficiently. I-N. Rescue functional experiments were performed in U87 and LN229 cells to study the rescuing effect of miR-876-5p inhibitor on sh-MINCR mediated cellular activities. **P < 0.01
GSPT1 is the Target of miR-876-5p and is Regulated by MINCR
MiRNAs could bind to particular targets and realized mRNA degradation primarily in an Ago2-dependent way [34, 35]. We analyzed the ceRNA role of MINCR by searching downstream targets of miR-876-5p. By limiting binding conditions on starBase database and jointly utilizing microT, miRanda and miRmap bioinformatics tools afterwards, POLD3, GSPT1 and ARCN1 were found as potential targets of miR-876-5p (Fig. 3a). Subsequent qRT-PCR analysis showed that the expression of GSPT1 was obviously down-regulated by overexpressing miR-876-5p (Fig. 3b). Furthermore, GSPT1 was found to be up-regulated in glioma cells (Fig. 3c). Hence, GSPT1 was chosen for further study. The binding sites of miR-876-5p in the 3’ UTR of GSPT1 was detected and correspondingly mutated for conducting luciferase reporter assay (Fig. 3d). The luciferase activity of GSPT1 WT was diminished distinctly by overexpressing miR-876-5p. However, miR-876-5p overexpression exerted no effect on the luciferase activity of GSPT1 Mut (Fig. 3e). As shown in Fig. 3f, RIP assay manifested preferential enrichment of miR-876-5p, MINCR and GSPT1 in the Ago2-bound complexes. Furthermore, the expression of GSPT1 declined by overexpressing miR-876-5p, yet restored by co-transfection with MINCR (Fig. 3g), suggesting the ceRNA role of MINCR in glioma. To sum up, GSPT1 was the target gene of miR-876-5p, and MINCR could upregulate GSPT1 via sponging miR-876-5p.
GSPT1 is the target of miR-876-5p and is negatively regulated by MINCR. a The potential targets of miR-876-5p predicted by different databases. b The expression of candidate mRNAs was detected by qRT-PCR analysis after transfecting miR-876-5p mimics in U87 and LN229 cells. c qRT-PCR was carried out to evaluate the expression of GSPT1 in glioma cell lines and normal one. d Putative binding sites between miR-876-5p and GSPT1 predicted by starBase. e Luciferase reporter assay was performed to investigate the molecular relationship between miR-876-5p and GSPT1. f RIP assays manifested that miR-876-5p, MINCR and GSPT1 were co-immunoprecipitated by antibody targeting Ago2. g qRT-PCR was performed to study the expression of GSPT1 after co-transfecting MINCR into miR-876-5p-overexpressed U87 and LN229 cells. *P < 0.05, **P < 0.01
MINCR Regulates miR-876-5p/GSPT1 Axis to Aggravate Glioma Progression
To further validate whether MINCR exerted its carcinogenic effect in glioma via regulating miR-876-5p/GSPT1 axis, we overexpressed GSPT1 for the purpose of subsequent rescue assays (Fig. 4a). We firstly examined the effectiveness of MINCR/miR-876-5p/GSPT1 network on glioma cell proliferation. EdU and colony formation assays manifested that the declined proliferation ability caused by MINCR depletion was reversed by GSPT1 up-regulation (Fig. 4b, c). Additionally, the strengthened apoptotic capacity caused by MINCR silence was reversed by GSPT1 up-regulation (Fig. 4d, e). Then, transwell assays revealed that GSPT1 up-regulation rescued the suppressed migration and invasion induced by MINCR depletion in glioma (Fig. 4f, g). In vivo experiments further proved that GSPT 1 overexpression restore the inhibitory effect of sh-MINCR on tumor growth (Fig. 4h). The tumor volume and weight were dramatically decreased by sh-MINCR, yet increased in the presence of GSPT1 overexpression (Fig. 4i, j), which added strong evidence to the inhibitory impact of sh-MINCR on glioma development. Therefore, MINCR played a ceRNA role to regulate miR-876-5p/GSPT1 axis in glioma.
MINCR regulates miR-876-5p/GSPT1 axis to aggravate glioma progression. a qRT-PCR analysis was used to testify the expression of GSPT1 after transfecting pcDNA3.1/GSPT1. b, c EdU and colony formation assays were performed to elucidate the impact of GSPT1 up-regulation on sh-MINCR induced glioma cell proliferation. d, e TUNEL and flow cytometry were conducted to determine apoptosis after transfecting pcDNA3.1/GSPT1 in MINCR-silenced glioma cells. f, g Transwell assays were implemented to study the rescuing effect of GSPT1 overexpression on sh-MINCR mediated migration and invasion. h Xenograft tumor growth curve was illustrated. i, j Tumor volume and weight were measured and compared among sh-NC, sh-MINCR#2 and sh-MINCR#2 + pcDNA3.1/GSPT1 groups. **P < 0.01
Discussion
Occupying approximately 40–50% of all intracranial malignancies cases, glioma is a leading cause of intracranial tumor-associated death [36, 37]. Owning to the advancement achieved next-generation sequencing technology, lncRNAs have been revealed to play vital role in the development of human malignancies [38]. Similarly, the link between lncRNAs and glioma has been uncovered. For example, an array of lncRNAs have been identified to impact proliferation [39], autophagy [40], migration [41] and chemotherapy resistance [42] of glioma cells. MINCR has been discovered to positively regulate cell proliferation, migration, and invasion in hepatocellular carcinoma [14]. In current study, MINCR was abnormally highly expressed in glioma cells in contrast with normal one. Loss-of-function assays manifested that silencing MINCR curbed the proliferative, migrating and invasive activities of glioma cells, while stimulating apoptosis. This oncogenic property of MINCR accorded with the findings in oral squamous cell carcinoma (OSCC). According to Lyu et al., MINCR activated Wnt/β-catenin signaling pathway in promoting malignant biological behaviors in OSCC. Silencing MINCR dramatically decreased the activity of Wnt/β-catenin pathway, consequently leading to weakened proliferation and migration abilities of OSCC cells [43].
Accumulating lncRNAs have been reported to function as ceRNAs at post-transcription phase to regulate the mRNA and protein expressions of target genes via the competition for shared miRNAs. Regarding the regulatory system of MINCR in glioma, first and foremost, we investigated the distribution of MINCR in selected glioma cells and confirmed the cytoplasmic localization of MINCR. Utilizing starBase bioinformatics tool, miR-876-5p was identified to be a potential combinable miRNA for MINCR. Previous study demonstrated that miR-876-5p repressed cell viability, migration and invasion in colorectal cancer via targeting WNT5A and MITF [22]. Here, miR-876-5p was found to be abnormally down-regulated in glioma cell lines. Besides, miR-876-5p was validated to be sponged by LINC-ROR [44]. Here, we found that MINCR facilitated glioma process via sponging miR-876-5p. This study initially probed into the regulatory effect of MINCR and miR-876-5p in glioma.
GSPT1 was elucidated to function as critical regulator in the pathological progression of cancers. It was found to be involved in ceRNA network in non-small cell lung cancer cells via interacting with miR-27b-3p [31]. Besides, in gastric cancer, it was found to be targeted by miR-27b-3p and could advance the carcinoma progression, including proliferation, invasion and migration [30]. Hence, we supposed that GSPT1 might be targeted by miRNA and engaged in ceRNA network in glioma. GSPT1 was found as a downstream target of miR-876-5p via bioinformatics tool. Moreover, GSPT1 was found highly expressed in glioma cells. MINCR could up-regulate the expression of GSPT1 via sponging miR-876-5p. GSPT1 restoration could countervail the inhibitory effect of silencing MINCR on glioma. That is to say, MINCR facilitated the advancement of glioma via regulating miR-876-5p-medaited GSPT1.
All in all, our data initially revealed that MINCR regulates miR-876-5p/GSPT1 axis to aggravate glioma progression, indicating profound potential of MINCR as a novel therapeutic target for glioma patients.
References
Khasraw M, Ameratunga MS, Grant R, Wheeler H, Pavlakis N (2014) Antiangiogenic therapy for high-grade glioma. Cochrane Database Syst Rev 22:CD008218
Wang J, Su HK, Zhao HF, Chen ZP, To SS (2015) Progress in the application of molecular biomarkers in gliomas. Biochem Biophys Res Commun 465:1–4
Jaspan T, Morgan PS, Warmuth-Metz M, Sanchez Aliaga E, Warren D, Calmon R, Grill J, Hargrave D, Garcia J, Zahlmann G (2016) Response assessment in pediatric neuro-oncology: implementation and expansion of the RANO criteria in a randomized phase II trial of pediatric patients with newly diagnosed high-grade gliomas. AJNR Am J Neuroradiol 37:1581–1587
Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY (2013) Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol 425:3731–3746
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307
Wan G, Zhou W, Hu Y, Ma R, Jin S, Liu G, Jiang Q (2016) Transcriptional regulation of lncRNA genes by histone modification in Alzheimer’s disease. BioMed Res Int 2016:3164238
Liu B, Li J, Liu X, Zheng M, Yang Y, Lyu Q, Jin L (2017) Long non-coding RNA HOXA11-AS promotes the proliferation HCC cells by epigenetically silencing DUSP5. Oncotarget 8:109509–109521
Zhao X, Cheng Z, Wang J (2018) Long noncoding RNA FEZF1-AS1 promotes proliferation and inhibits apoptosis in ovarian cancer by activation of JAK-STAT3 pathway. Med Sci Monitor 24:8088–8095
Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X, Zai S (2017) LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep 37(6):BSR20171070
Han D, Wang J, Cheng G (2018) LncRNA NEAT1 enhances the radio-resistance of cervical cancer via miR-193b-3p/CCND1 axis. Oncotarget 9:2395–2409
Song K, Yuan X, Li G, Ma M, Sun J (2019) Long noncoding RNA CASC11 promotes osteosarcoma metastasis by suppressing degradation of snail mRNA. Am J Cancer Res 9:300–311
Wang SH, Yang Y, Wu XC, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW (2016) Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Lett 380:122–133
Jin XL, Lian JR, Guan YH (2018) Overexpression of long non-coding RNA MINCR contributes to progressive clinicopathological features and poor prognosis of human hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 22:8197–8202
Cao J, Zhang D, Zeng L, Liu F (2018) Long noncoding RNA MINCR regulates cellular proliferation, migration, and invasion in hepatocellular carcinoma. Biomed Pharmacother 102:102–106
Chen S, Gu T, Lu Z, Qiu L, Xiao G, Zhu X, Li F, Yu H, Li G, Liu H (2019) Roles of MYC-targeting long non-coding RNA MINCR in cell cycle regulation and apoptosis in non-small cell lung Cancer. Respir Res 20:202
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369
Meng L, Ma P, Cai R, Guan Q, Wang M, Jin B (2018) Long noncoding RNA ZEB1-AS1 promotes the tumorigenesis of glioma cancer cells by modulating the miR-200c/141-ZEB1 axis. Am J Transl Res 10:3395–3412
Wang J, Ding M, Zhu H, Cao Y, Zhao W (2019) Up-regulation of long noncoding RNA MINCR promotes non-small cell of lung cancer growth by negatively regulating miR-126/SLC7A5 axis. Biochem Biophys Res Commun 508:780–784
Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang Y, Li X, Xie X, Wang J, Huang M, Sun X, Ke Y (2019) circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res 38:398
Chen X, Yang F, Zhang T, Wang W, Xi W, Li Y, Zhang D, Huo Y, Zhang J, Yang A, Wang T (2019) MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res 38:99
Wang Y, Xie Y, Li X, Lin J, Zhang S, Li Z, Huo L, Gong R (2018) MiR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol Res Pract 214:1024–1030
Xu Z, Yu Z, Tan Q, Wei C, Tang Q, Wang L, Hong Y (2019) MiR-876-5p regulates gastric cancer cell proliferation, apoptosis and migration through targeting WNT5A and MITF. Biosci Rep 39:BSR20190066
Dong Y, Zheng Y, Wang C, Ding X, Du Y, Liu L, Zhang W, Zhang W, Zhong Y, Wu Y, Song X (2018) MiR-876-5p modulates head and neck squamous cell carcinoma metastasis and invasion by targeting vimentin. Cancer Cell Int 18:121
Sang M, Meng L, Sang Y, Liu S, Ding P, Ju Y, Liu F, Gu L, Lian Y, Li J, Wu Y, Zhang X, Shan B (2018) Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett 426:37–46
Dong N, Guo J, Han S, Bao L, Diao Y, Lin Z (2019) Positive feedback loop of lncRNA HOXC-AS2/miR-876-5p/ZEB1 to regulate EMT in glioma. OncoTargets Therapy 12:7601–7609
Meng L, Liu F, Ju Y, Ding P, Liu S, Chang S, Liu S, Zhang Y, Lian Y, Gu L, Zhang X, Sang M (2018) Tumor suppressive miR-6775-3p inhibits ESCC progression through forming a positive feedback loop with p53 via MAGE-A family proteins. Cell Death Dis 9:1057
Salem M, O’Brien JA, Bernaudo S, Shawer H, Ye G, Brkic J, Amleh A, Vanderhyden BC, Refky B, Yang BB, Krylov SN, Peng C (2018) miR-590-3p promotes ovarian cancer growth and metastasis via a Novel FOXA2-versican pathway. Cancer Res 78:4175–4190
Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J (2019) Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci 76:441–451
Tian QG, Tian RC, Liu Y, Niu AY, Zhang J, Gao WF (2018) The role of miR-144/GSPT1 axis in gastric cancer. Eur Rev Med Pharmacol Sci 22:4138–4145
Zhang C, Zou Y, Dai DQ (2019) Downregulation of microRNA-27b-3p via aberrant DNA methylation contributes to malignant behavior of gastric cancer cells by targeting GSPT1. Biomed Pharmacother 119:109417
Sun W, Zhang L, Yan R, Yang Y, Meng X (2019) LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. OncoTargets Therapy 12:3945–3954
Nair S, Bora-Singhal N, Perumal D, Chellappan S (2014) Nicotine-mediated invasion and migration of non-small cell lung carcinoma cells by modulating STMN3 and GSPT1 genes in an ID1-dependent manner. Mol Cancer 13:173
Chen Z, Hu X, Wu Y, Cong L, He X, Lu J, Feng J, Liu D (2019) Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed Pharmacother 109:2228–2236
Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553–563
Shukla GC, Singh J, Barik S (2011) MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 3:83–92
Seyithanoglu MH, Abdallah A, Kitis S, Guler EM, Kocyigit A, Dundar TT, Gundag Papaker M (2019) Investigation of cytotoxic, genotoxic, and apoptotic effects of curcumin on glioma cells. Cell Mol Biol 65:101–108
Chen H, Liu L, Li X, Shi Y, Liu N (2018) MicroRNA-1294 inhibits the proliferation and enhances the chemosensitivity of glioma to temozolomide via the direct targeting of TPX2. Am J Cancer Res 8:291–301
Hu X, Sood AK, Dang CV, Zhang L (2018) The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev 48:8–15
Hu Y, Jiao B, Chen L, Wang M, Han X (2019) Long non-coding RNA GASL1 may inhibit the proliferation of glioma cells by inactivating the TGF-beta signaling pathway. Oncol Lett 17:5754–5760
Jia L, Song Y, Mu L, Li Q, Tang J, Yang Z, Meng W (2019) Long noncoding RNA TPT1-AS1 downregulates the microRNA-770-5p expression to inhibit glioma cell autophagy and promote proliferation through STMN1 upregulation. J Cell Physiol 235(4):3679–3689
Yu M, Yu S, Gong W, Chen D, Guan J, Liu Y (2019) Knockdown of linc01023 restrains glioma proliferation, migration and invasion by regulating IGF-1R/AKT pathway. J Cancer 10:2961–2968
Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P, Lin K (2018) Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int 118:233–241
Lyu Q, Jin L, Yang X, Zhang F (2019) LncRNA MINCR activates Wnt/beta-catenin signals to promote cell proliferation and migration in oral squamous cell carcinoma. Pathol Res Pract 215:924–930
Zhi Y, Abudoureyimu M, Zhou H, Wang T, Feng B, Wang R, Chu X (2019) FOXM1-mediated LINC-ROR regulates the proliferation and sensitivity to sorafenib in hepatocellular carcinoma. Mol Therapy Nucleic Acids 16:576–588
Acknowledgements
The current study was supported of all laboratory staff.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Z., Xie, X., Fan, X. et al. Long Non-coding RNA MINCR Regulates miR-876-5p/GSPT1 Axis to Aggravate Glioma Progression. Neurochem Res 45, 1690–1699 (2020). https://doi.org/10.1007/s11064-020-03029-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-020-03029-8