Abstract
N-Methyl-d-aspartate receptors are localized to synaptic and extrasynaptic sites of dendritic spines and shafts. Here, the ontogenic profiles of GluN3A and GluN3B subunits in the rat brain were determined. A developmental switch from GluN3A to GluN3B proteins was detected within the first two postnatal weeks of crude synaptosomes prepared from forebrain and midbrain. Further fractionation of crude synaptosomes revealed the preferential localization of GluN3B to synaptic regions from P7 onwards. Immunolabeling and biochemical fractionation of rat P7 cultured hippocampal neurons showed that GluN3B was predominantly at synaptic sites. Unlike GluN2A and GluN2B, both GluN3 subunits were mostly associated with peripheral components of the postsynaptic density (PSD) rather than its core. When considering the non-PSD fraction, the overall extrasynaptic/synaptic spatial profile of GluN3B differed from GluN3A. Heterologous expression of GluN3B with GluN1 in HEK293FT cells could not be co-immunoprecipitated with PSD-95 unless co-expressed with a PSD-95-interacting GluN2 subunit, suggesting that anchoring of GluN3B at synaptic sites may require co-assembly with another scaffold-interacting NMDAR subunit.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
N-Methyl-d-aspartate receptors (NMDARs) consist of GluN1, GluN2A-D and GluN3A-B subunits. These NMDAR subtypes, localized at postsynaptic sites in neurons, affect channel activity and downstream signalling [1]. Activation of synaptic NMDARs occurs during normal neurotransmission whereas activation of more remote receptors requires glutamate “spillover” during more intense stimulation [1]. These include receptors located at perisynaptic (defined as 100–200 nm beyond the PSD edge) and extrasynaptic (defined as beyond the perisynaptic regions) sites [2–4].
Postsynaptic density (PSD) defines a synaptic region in the dendrites whereby a high concentration of neurotransmitter receptors, scaffold proteins and signalling molecules are organised to facilitate neurotransmission. Biochemically, further subsynaptic regions can be isolated from the PSD with detergents. PSD structures are insoluble in the detergent Triton X-100, thus become enriched in the pellet after first centrifugation (resulting in PSD I pellet). Proteins with increasing association with the PSD core can be obtained from the PSD I pellet by a second round of extracting using Triton X-100 (resulting in PSD II pellet) or by using sarcosyl (resulting in PSD III pellet) followed by centrifugation with increasing speed. The PSD I, II and III pellets are later dissolved in buffer [5–7]. NMDARs are among the proteins which enrich differentially in these fractions [8].
NMDARs are anchored to the PSD on postsynaptic membrane juxtaposed to the active zone on pre-synaptic terminal. This NMDAR–PSD interaction is accomplished by C-terminal interactions of NMDAR subunits with family members of the membrane guanylate kinases (MAGUK), of which postsynaptic density protein 95 (PSD-95) is the major scaffolding protein essential for maintaining the molecular organization of the PSD [9, 10]. Association of NMDAR with PSD-95 has been shown to be responsible for regulating synaptic retention and synaptic localization [11].
GluN1 and GluN2 subunits have been found in synaptic and extrasynaptic regions [2]. Using electron microscopy, populations of perisynaptic NMDARs have been reported [12]. Studies on GluN3A protein have revealed its preferential localization at perisynaptic and extrasynaptic regions [7]. Much less is known about the GluN3B subunit synaptic localization. This study investigated the developmental expression and the subcellular distribution of GluN3B protein at the synapse, and examined the ability of different configurations of GluN3B-containing NMDAR to be anchored to PSD-95.
Materials and Methods
Crude Synaptosomal Membranes Preparation
The Sprague-Dawley rats used in the experiments were supplied by Centre for Animal Resources. Experiments were approved and performed according to Institutional Animal Care and Use Committee guidelines of the National University of Singapore (NUS). Crude synaptosomes were prepared from combined regions of forebrain and midbrain from rats of defined ages obtained from different litters according to the protocol stated in Huttner et al. [13]. In brief, brain tissues were dounced twenty times in ice-cold buffered sucrose (320 mM sucrose, 10 mM Tris HCl, 5 mM EDTA, pH 7.4). Homogenates were centrifuged at 800g for 10 min. The resultant supernatant was centrifuged at 9200g for 15 min to obtain the pellet which constituted a crude synaptosomal fraction (P2). The P2 pellet was rinsed in buffered sucrose once, and solubilized in 50 mM Tris–HCl, pH 9.0 containing 1 % sodium deoxycholate at 37 °C for 30 min [14, 15].
Subcellular Fractionation
Biochemical fractionations of the PSD fractions were performed on combined regions of rat forebrain and midbrain of 6-week-old male Sprague-Dawley rats [6, 13]. The brain tissues were processed as per processing for crude synaptosome except that the P2 pellet was rinsed with buffered sucrose and pellet resuspended in the buffered sucrose and centrifuged at 10,200g for 15 min to obtain a P2’ pellet. P2’ pellet was resuspended using 50 mM Tris pH 7.4 containing 0.5 % Triton X-100, incubated for 15 min and centrifuged at 32,000g for 20 min to obtain PSD I pellet. This pellet was resuspended and incubated for 10 min with a second round of buffer containing 0.5 % (v/v) Triton X-100 then centrifuged at 201,800g for 1 h to obtain the PSD II pellet. In a separate tube, the PSD I pellet was resuspended and incubated for 10 min in ice-cold 50 mM Tris, pH 7.4 containing 3 % (w/v) sacrosyl followed by centrifugation at 201,800g for 1 h to obtain the PSD III pellet. The supernatant obtained from the PSD I pellet extraction is termed the Triton-soluble supernatant (TxS), which is the non-PSD component. All pellets were solubilized in 50 mM Tris, pH 7.4 containing 1 % (w/v) SDS and centrifuged at 14,000g for 15 min. The clarified supernatant was collected and stored at −80 °C for subsequent analyses.
Cell Culture and Transfection
HEK293FT cells (R700-07; Invitrogen, Carlsbad, CA) were cultured and transfected using Lipofectamine 2000 according to manufacturer’s recommendation (#11668027; Invitrogen). cDNAs were transfected at 1:1 ratio for most combinations of NMDAR subunits and together with FLAG-tagged PSD-95 plasmid (#15463; Addgene, Cambridge, MA) [16]. Plasmid DNA ratios were adjusted, where necessary, to obtain equal protein expression between GluN1-1a and GluN1-4b (gifts from Dr. S. F. Heinemann, Salk Institute).
Immunoprecipitation
One hundred microgram of cell total lysates were pre-cleared for 1.5 h using 45 µL of Protein A Sepharose (1:3 slurry in lysis buffer) to remove the non-specific proteins and endogenous IgG that bind to the Protein A Sepharose. Anti-PSD-95 antibody was added to the pre-cleared cell lysates for binding overnight at 4 °C followed by incubation with 45 µL of Protein A Sepharose. The mixture was rotated for 1.5 h at 4 °C and washed with 500 µL lysis buffer four times. Sample buffer was added and slurry was boiled before resolving by SDS-PAGE. Two anti-PSD-95 antibodies were used (MA1-045 and MA1-046, 1 and 2.5 μg per 450 µL, respectively; Affinity Bioreagents/Thermo Scientific, Rockland, IL), with their specificities (clone 6G6-1C9 and clone 7E3-1B8, respectively) previously described by Kornau et al. [17].
Western Blot and Densitometry
Western blot was performed according to Wee et al. [18] using the following antibodies: anti-GluN2A (#07-632, 1:500; Upstate, Lake Placid, NY), anti-GluN2B (#06-600, 1:800; Upstate), anti-GluN3A (#07-356, 1:500; Upstate,), anti-GluN3B (#07-351, 1:600, Upstate), anti-GluN1 (#556308, 1:1500; BD Pharmingen, San Jose, CA)), anti-PSD-95 (MA1-046, 1:2000; Affinity Bioreagents), rabbit anti-calnexin (1:1000; Stressgen, British Columbia, CA) and anti-synaptophysin (sc-9116; 1:500; Santa Cruz, Tx, USA). Densitometry, the intensity of the protein band on the exposed X-ray film,was performed on scanned images using UN-SCAN-IT gel™ Version 6.1 (Silk Scientific Corp., USA). For each data set (TxS, PSD I, PSD II and PSD III), all other values were expressed as a percentage of the fraction with the highest protein expression level for most ages.
Primary Hippocampal Cultures
Hippocampi from P7 Sprague-Dawley pups were dissected in ice-cold Hanks Balanced Salt solution (HBSS) containing 10 mM HEPES, pH 7.35 (HBSS-HEPES), cut into pieces and incubated flat at 37 °C for 30 min in papain enzyme mix: 15U/mL papain (P4762; Sigma Aldrich, St Louis, MO, USA), 0.5 mM EDTA, 10 mg/µL DNase I (Sigma Aldrich), 1.5 mM CaCl2, 0.2 mg/mL l-cysteine (Sigma Aldrich) in HBSS-HEPES. Glial culture medium (Basal Eagle’s Medium containing 10 % FBS, 0.297 % d-(+)glucose, 0.1 % mito serum extender, 1 mM sodium pyruvate, 1 % penicillin/streptomycin, 10 mM HEPES, pH 7.35) was added immediately. The supernatant was discarded after settling of tissue.
Tissue was triturated in glial-conditioned neuronal medium consisting of Neurobasal-A, 3 % (v/v) B27 supplement (specific lot numbers used due to batch variation on cell viability), 2 mM GlutaMAX, 0.594 % (w/v) d-(+) glucose, 1 % (v/v) penicillin/streptomycin. Tissue was triturated through a 1 mL pipette tip, and left to stand for 3 min before collecting the supernatant. Cells were plated onto glass coverslips pre-coated with 0.5 mg/mL polyornithine in 10 mM Na2B4O7, pH 8.4, and cultured in conditioned neuronal medium. 4 µM of cytosine arabinofuranoside was added at 2 days in vitro (DIV). Full medium change was performed on 7DIV, and every subsequent week.
Immunocytochemistry, Confocal Imaging and Correlation Analyses
Immunocytochemistry and acquisition of fluorescence images were performed according to Wee et al. [18]. Antibodies used were: anti-GluN3B (sc-55727, 1:100; Santa Cruz), anti-synaptophysin (sc-9116, 1:400; Santa Cruz), Alexa Fluor488 donkey anti-rabbit and Alexa Fluor555 donkey anti-goat (Molecular Probes, Eugene, OR, USA). Pearson’s correlation coefficient of the amount of colocalization between GluN3B and synaptophysin at different stages of development of cultured hippocampal neurons was determined using NIS Elements™ version 3.10 (Nikon, Melville, NY, USA). Images were calibrated according to actual dimensions. For each neuron, four neurite regions of 30 µm length were used to define four polygonal regions-of-interests (ROI) from three independent cultures. Cell somas were excluded from analysis. Average values per neuron was computed and entered into GraphPad Prism™ version 5.0 (Graphpad Software Inc., La Jolla, CA) for graph plotting and one-way ANOVA with Tukey’s post hoc statistical analyses were performed.
Results
Ontogenic Profiles of GluN3B is Reciprocal to GluN3A
To investigate the developmental expression profile of GluN3A and GluN3B, we extracted crude synaptosomes of the pooled forebrain and midbrain from the rat to conduct western blotting. In crude synaptosomes, GluN3B protein levels were the lowest during P0–P1 but rapidly increased to two-thirds of its maximal expression between P7 and P14 (Fig. 1a). Maximal protein expression was observed in 6-weeks-old adults. Reciprocally, GluN3A protein levels were the highest at birth but progressively declined, reaching its lowest levels in rats >6 months old (Fig. 1a). The expression levels of GluN3B (P0–P1 to 6 weeks) were plotted relative to its own protein expression at >6 months. The GluN3A expression levels (P7 to >6 months) were plotted relatively to its own protein expression at P0–P1. An inverse protein expression pattern between GluN3B and GluN3A was revealed (Fig. 1b). As compared to the GluN3A and GluN3B, GluN1 showed the most stable expression among all four subunits (see supplementary Fig. 1).
Protein expression profile of NMDAR subunits during rat ontogenesis from combined regions of forebrain and midbrain. Protein expression level (%) for GluN3A and GluN3B in crude synaptosomes across six developmental ages. Data represent mean ± SEM of protein expression (%) of three independent experiments of different animals obtained from different litters. Representative blots are shown
GluN3B is Localized at the Postsynaptic Regions of Synapses
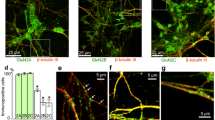
To determine the localization of the GluN3B proteins in the neurons, hippocampal neurons from P7 rat pups were cultured in vitro and GluN3B proteins were immunostained with the axonal (Tau) and dendritic (MAP2) markers on 35DIV (Fig. 2a). This in vitro paradigm was set up in an attempt to mimic the established synapses and peak GluN3B protein expression as observed in Fig. 1 (>6 weeks). The GluN3B proteins were found to colocalize with MAP2 with little or no detectable signals of colocalization with Tau suggesting that the GluN3B may be mostly expressed at the soma and the dendrites. To address whether GluN3B is localized at synaptic sites, a synaptic marker (synaptophysin) was co-immunolabeled with Tau. Synaptophysin-positive synaptic sites are present both at the axons as well as the dendrites in our hippocampal neuronal culture (Fig. 2b). Next, in the co-immunolabeling of 35DIV neurons for PSD-95 and synaptophysin, it was also shown that synaptophysin localized to most of the PSD-95 puncta (but not completely). Collectively, the preceding suggests that synaptophysin can be used in our system to label the active synaptic site (Fig. 2c) as suggested by other studies [19–21]. Immunostaining of GluN3B together with the synaptophysin shows that GluN3B is located at the synaptic sites (Fig. 2d).
GluN3B is expressed in the synaptic contacts at the postsynaptic density at the dendrites. a GluN3B (red) is expressed at the dendrites (MAP2 immunopositive; blue) but rarely on the axons (Tau immunopositive; green). b Synaptophysin (red) puncta which indicates synaptic sites (arrowheads) are interacting with the axons labeled by Tau. c Synaptophysin puncta are colocalized with PSD-95 (red) expression. d GluN3B is colocalized with synaptophysin puncta at the synaptic sites (insert; arrowheads). Non-colocalized synaptophysin and GluN3B sites were present too (insert; arrow). All cultured hippocampal neurons are 35DIV. Scale bars in (a) and (b) represent 10 µm while (d) represents 20 µm (Color figure online)
GluN3B Expression at the Synaptic Sites Increases with Neuronal Maturation
To investigate the localization of GluN3B proteins upon neuronal maturation, hippocampal neurons from P7 rat pups were cultured for 8, 19 and 35DIV, and immunostained for GluN3B and synaptophysin to examine its presence at the active synaptic sites [19–21]. With increasing time in culture, the correlation coefficient between GluN3B and synaptophysin increases significantly from 8DIV to 19DIV and 8DIV to 35DIV (Fig. 3; *p < 0.001, Turkey’s post hoc tests). Indeed, it was observed that the majority of GluN3B-immunoreactive puncta was localized with synaptophysin (Fig. 3, upper panel). GluN3B protein in these cultures was distributed in a punctate fashion throughout the dendrites.
Quantitative analysis of the amount of colocalization between immunopositive GluN3B and synaptophysin at the different stages of development of cultured hippocampal neurons. Average of Person’s correlation coefficients were calculated from four 30 μm segments of neurites per neuron collected from different neuronal cultures to obtain a mean for each developmental time point in culture. Bars represent mean ± SEM of Pearson’s correlation coefficient for n = 10 (8DIV), n = 18 (19DIV) and n = 16 (35DIV) neurons from three experiments (***p < 0.001, one-way ANOVA with Tukey’s post hoc tests)
Developmental Changes in Intrasynaptic Distribution Profile for GluN3B
When crude synaptosomes from P0, P7, P14, P21, 6 week and adult (>6 months) rats were further fractionated into PSD fractions (PSD I, PSD II, PSD III) and non-PSD fraction (TxS), the fractionation profile revealed a shift in peak GluN3B protein expression from PSD II to PSD I at the very early developmental stage of P0–P7 (Fig. 4). The GluN3B protein distribution profile between TxS to PSD III remained unchanged post P7 (Fig. 4).
Expression profile of GluN3B subunit (98 kD) across synaptic regions over six development stages. Percentage of protein expression of GluN3B in four fractions (TxS, PSD I, PSD II, PSD III) were plotted against different developmental stages of SD rats. Data represent mean ± SEM of protein expression (%) of three to five independent experiments
GluN3B Co-immunoprecipitates with PSD-95 Only When Co-expressed with GluN1-4, GluN2A or GluN2B
PSD-95 has been shown to bind to all GluN2 subunits and C2’-cassette containing GluN1-3 and GluN1-4 splice variants [17, 22]. C2’-cassette containing GluN1 are splice variants eliminated a stop codon and resulting in an additional 22 amino acid C-terminal domain. Examining GluN3B protein sequence did not reveal putative PSD-95-binding motif within its C-terminus [10]. To confirm it, FLAG-tagged PSD-95 (PSD-95-FLAG) was co-expressed with HA-tagged GluN3B (GluN3B-HA) (1) in the absence of other NMDAR subunit, (2) in the presence of GluN1 (GluN1-1a or GluN1-4b), or (3) in the presence of GluN1-1a and another subunit (GluN2A, GluN2B or GluN3A). PSD-95-FLAG pulldown did not co-immunoprecipitate GluN3B-HA when the following combinations were co-expressed with PSD-95 (GluN3B; GluN1-1a/GluN3B; GluN1-1a/GluN3A/GluN3B) (Fig. 5, right panel). However, in the presence of GluN1-4b or GluN2A/B, GluN3B-HA could be co-immunoprecipitated with PSD-95-FLAG (Fig. 5, right panel).
Co-immunoprecipitation of PSD-95 with various combinations of GluN3B-containing subunits. Pulldown by anti-PSD-95 was performed. Immunoprecipitates were probed with anti-HA to detect GluN3B-HA (top panels) and anti-PSD-95 (bottom panels) (n = 3). 10 % of input was loaded to show comparable total protein expression between samples (input). IB immunoblot, IP immunoprecipitation
NMDAR Intrasynaptic Distribution in 6 Weeks Old Rat
To further substantiate the requirement for the co-assembly of GluN3B with the other NMDAR subunits for surface transport and interaction with PSD-95, individual NMDAR subunits intrasynaptic distribution in the PSD and non-PSD fractionation was examined. The intrasynaptic distributions of GluN1, GluN2A and GluN2B suggest that in all instances, these subunits were present in the rat brain to support the surface transport and insertion of the GluN3B subunit via interacting with PSD-95 (Fig. 6).
Expression profiles of five NMDAR subunits across synaptic regions at 6 weeks old adult SD rats. Percentage of protein expression of the NMDAR subunits in four fractions (TxS, PSD I, PSD II, PSD III) were plotted. Data represent mean ± SEM of protein expression (%) of three to five independent experiments
When densitometric values of each individual PSD fraction were plotted alongside the non-PSD fractions, GluN2A and GluN2B expression showed the highest in the PSD III fraction—the fraction representing the PSD core (Fig. 6). The PSD core was first described by Kennedy et al. [23, 24]. It consists of highly insoluble proteins which are associated with the “core” of the PSD after removal of weakly associated proteins by N-lauroyl-sacrosinate (sarcosyl) treatment. In contrast to GluN2A and GluN2B, the expression of GluN3A and GluN3B were both low in PSD III. However, what distinguishes GluN3A spatial profile from that of GluN3B is the high level of GluN3A compared to the low levels of GluN3B in the TxS fraction (Fig. 6). On the other hand, GluN3A shows a descending profile—least expressed in PSD III and most expressed in TxS (Fig. 6). This is in contrast to the ascending profile of GluN1, GluN2A and GluN2B where these proteins were progressively enriched from the peripheral region towards the PSD core (Fig. 6).
Discussion
This study showed that GluN3B synaptic/extrasynaptic distribution profile differs from GluN1, GluN2A, GluN2B and GluN3A. Here, four main findings are: firstly, the development profiles of GluN3A and GluN3B subunits from the crude rat brain synaptosomes were found to be diametrically opposed (Fig. 1). The level of GluN3A is high in expression from P0 to P14, followed by a gradual decrease as the rat matures. However, the level of GluN3B is low at birth and steadily rises after first week to reach near adult levels by 14 days. This ontogenic trend of GluN3B and GluN3A mirrors that of GluN2A and GluN2B, respectively [25–27].
Secondly, GluN3B protein is enriched in PSD fractions regardless of developmental age (Fig. 4). This enrichment is supported by the high frequency of GluN3B puncta localized with synaptophysin, a synaptic marker in primary cultured neurons (Figs. 2, 3).
Thirdly, the predominant expression of GluN3B in the PSD fraction is in sharp contrast to GluN3A, where a rather large proportion of the GluN3A was mostly expressed outside the PSD (Fig. 6). This corroborates with a previous study that reported that GluN3A is concentrated in extrasynaptic and perisynaptic regions of the PSD [8]. Interestingly, the decreasing level of GluN3 proteins towards the PSD core was also observed for GluN3B, suggesting that a lack of enrichment at the PSD core is a common characteristic for GluN3 subunits (Fig. 4).
Fourthly, despite GluN3B sharing this common intrasynaptic distribution with GluN3A, the low expression of GluN3B protein in the TxS fraction is distinct from GluN3A where there are decreasing levels of GluN3A from TxS to PSD III (Fig. 6). Further differences are that GluN3B levels peaked at PSD II in neonates but peaked at PSD I from P7 onwards. The enrichment of GluN3B in the PSD I fraction most likely reflects the matured distribution pattern for GluN3B in rats since GluN3B levels remain the highest in PSD I from P7 up to 6 months-old (Fig. 4). The fact that GluN3B becomes progressively less enriched as the fraction is extracted with the strong detergent, sarcosyl, is indicative that GluN3B is a peripherally bound protein in the synaptic region. However this study is unable to rule out the possibility that GluN3B might be associated with both pre- and post-synaptic membranes of the synaptic junction.
While the ontogenic profiles (Fig. 1) and synaptic localization (Figs. 4, 6) data discussed above should be interpreted cautiously, they suggest that the developmental switch and preferential localization of GluN3 subunits present significant capacity for synaptic plasticity and dynamic regulation with respect to NMDARs. Such developmental switch in subunit levels have already been reported for GluN2A/GluN2B as GluN2B is expressed significantly earlier than GluN2A [25–27]. GluN3A has been shown to undergo activity-dependent endocytic removal from the synapse, thus altering the subunit composition of NMDARs within the PSD [8]. Others have shown that activation of perisynaptic receptors produces a functional response distinct from that of synaptic or extrasynaptic receptors [11, 28]. Similar to the perisynaptic region, peripherally bound proteins at the PSD edge may represent a region where receptors undergo synaptic removal or incorporation. Whether such temporal and spatial shifts would have important functional consequences on GluN3 subunit-containing NMDARs remain to be elucidated [4, 29].
What might account for the preferential localization of GluN3B at the edge of the PSD? The apparent lack of a PSD-95/discs large/ZO-1 homologous (PDZ)-binding domain in the C-terminus of GluN3B suggests that GluN3B is not able to bind directly to the usual synaptic scaffolding proteins. The current results indeed supported this phenomenon (Fig. 5). By showing that GluN3B can only be co-immunoprecipitated with PSD-95 in the presence of another PSD-95-binding NMDAR subunit in this study, it provided a rationale to explain how GluN3B-containing NMDARs could be enriched in synaptic regions despite the inability of GluN3B to anchor to scaffolding proteins.
References
Kohr G (2006) NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res 326:439–446
Groc L, Choquet D (2006) AMPA and NMDA glutamate receptor trafficking: multiple roads for reaching and leaving the synapse. Cell Tissue Res 326:423–438
Newpher TM, Ehlers MD (2008) Glutamate receptor dynamics in dendritic microdomains. Neuron 58:472–497
Zhang J, Diamond JS (2006) Distinct perisynaptic and synaptic localization of NMDA and AMPA receptors on ganglion cells in rat retina. J Comp Neurol 498:810–820
Carlin RK, Grab DJ, Cohen RS, Siekevitz P (1980) Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol 86:831–845
Cho KO, Hunt CA, Kennedy MB (1992) The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9:929–942
Naisbitt S, Valtschanoff J, Allison DW, Sala C, Kim E, Craig AM, Weinberg RJ, Sheng M (2000) Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J Neurosci 20:4524–4534
Perez-Otano I, Lujan R, Tavalin SJ, Plomannm M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD (2006) Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci 9:611–621
Chen X, Nelson CD, Li X, Winters CA, Azzam R, Sousa AA, Leapman RD, Gainer H, Sheng M, Reese TS (2011) PSD-95 is required to sustain the molecular organization of the postsynaptic density. J Neurosci 31:6329–6338
Kornau HC, Seeburg PH, Kennedy MB (1997) Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol 7:368–373
Groc L, Bard L, Choquet D (2009) Surface trafficking of N-methyl-D-aspartate receptors: physiological and pathological perspectives. Neuroscience 158:4–18
Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge L, Stephenson FA, Wenthold RJ (2010) Organization of NMDA receptors at extrasynaptic locations. Neuroscience 167:68–87
Huttner WB, Schiebler W, Greengard P, De Camilli P (1983) Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol 96:1374–1388
Dunah AW, Standaert DG (2003) Subcellular segregation of distinct heteromeric NMDA glutamate receptors in the striatum. J Neurochem 85:935–943
Blahos J, Wenthold RJ (1996) Relationship between N-methyl-d-aspartate receptor NR1 splice variants and NR2 subunits. J Biol Chem 271:15669–15674
Zhang J, Vinuela A, Neely MH, Hallett PJ, Grant SG, Miller GM, Isacson O, Caron MG, Yao WD (2007) Inhibition of the dopamine D1 receptor signaling by PSD-95. J Biol Chem 282:15778–15789
Kornau HC, Schenker LT, Kennedy MB, Seeburg PH (1995) Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269:1737–1740
Wee KS, Zhang Y, Khanna S, Low CM (2008) Immunolocalization of NMDA receptor subunit NR3B in selected structures in the rat forebrain, cerebellum, and lumbar spinal cord. J Comp Neurol 509:118–135
Huang YS, Jung MY, Sarkissian M, Richter JD (2002) N-methyl-d-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J 21:2139–2148
Gerrow K, Romorini S, Nabi SM, Colicos MA, Sala C, El-Husseini A (2006) A preformed complex of postsynaptic proteins is involved in excitatory synapse development. Neuron 49:547–562
Hörster F, Schwab MA, Sauer SW, Pietz J, Hoffmann GF, Okun JG, Kölker S, Kins S (2006) Phenylalanine reduces synaptic density in mixed cortical cultures from mice. Pediatr Res 59:544–548
Cousins SL, Kenny AV, Stephenson FA (2009) Delineation of additional PSD-95 binding domains within NMDA receptor NR2 subunits reveals differences between NR2A/PSD-95 and NR2B/PSD-95 association. Neuroscience 158:89–95
Kennedy MB (1993) The postsynaptic density. Curr Opin Neurobiol 3:732–737
Kennedy MB (1997) The postsynaptic density at glutamatergic synapses. Trends Neurosci 20:264–268
Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH (1992) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12:529–540
Wang YH, Bosy TZ, Yusada RP, Grayson DR, Vicini S, Pizzorusso T, Wolfe BB (1995) Characterization of NMDA receptor subunit-specific antibodies: distribution of NR2A and NR2B receptor subunits in rat brain and ontogenic profile in the cerebellum. J Neurochem 65:176–183
Laurie DJ, Bartke I, Schepfer R, Naujoks K, Seeburg PH (1997) Regional, developmental and interspecies expression of the four NMDAR2 subunits, examined using monoclonal antibodies. Brain Res Mol Brain Res 51:23–32
Yang Y, Wang XB, Frerking M, Zhou Q (2008) Delivery of AMPA receptors to perisynaptic sites precedes the full expression of long-term potentiation. Proc Natl Acad Sci USA 105:11388–11393
Conti R, Lisman J (2003) The high variance of AMPA receptor- and NMDA receptor mediated responses at single hippocampal synapses: evidence for multiquantal release. Proc Natl Acad Sci USA 100:4885–4890
Acknowledgments
This research was funded by BMRC grants (09/1/21/19/617) to C-M.L. K.S-L.W was a recipient of the graduate research scholarship from the Yong Loo Lin School of Medicine, NUS. We thank Dr. Tang Bor Luen for his gift of rabbit polyclonal anti-synaptophysin (Santa Cruz, sc-9116) and Dr. Yao Wei-Dong for PSD-95-FLAG plasmid deposited in Addgene.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Protein expression profile of GluN1 subunit during rat ontogenesis from combined regions of forebrain and midbrain. Protein expression level (%) for GluN1 in crude synaptosomes across six developmental ages. Data represent mean ± SEM of protein expression (%) of three independent experiments of different animals obtained from different litters. Representative blots are shown
Rights and permissions
About this article
Cite this article
Wee, K.SL., Tan, F.C.K., Cheong, YP. et al. Ontogenic Profile and Synaptic Distribution of GluN3 Proteins in the Rat Brain and Hippocampal Neurons. Neurochem Res 41, 290–297 (2016). https://doi.org/10.1007/s11064-015-1794-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-015-1794-8