Abstract
Purpose
Alterations in the promoter of the telomerase reverse transcriptase (TERT) gene are a major mechanism of upregulating telomerase, which plays a crucial role in tumor development. Mutations in the TERT promoter have been observed in a subset of brain tumors, including adult gliomas and high-grade meningiomas. In pituitary adenomas (PAs), however, abnormalities in TERT are not fully understood. The present study aimed to investigate not only mutational but also methylation changes in the TERT promoter in PAs and to analyze their correlations with clinical variables.
Methods
We retrospectively studied 70 PAs consisting of 53 primary and 17 recurrent samples. Clinical data, including age at surgery, sex, largest tumor dimension, tumor subtype, resection rate, and progression-free survival (PFS), were obtained from medical records. We investigated TERT promoter hotspot mutations via Sanger sequencing and quantified the methylation status of the TERT promoter using methylation-sensitive high-resolution melting analysis (MS-HRM). Additionally, we investigated TERT mRNA expression using real-time quantitative PCR.
Results
TERT promoter hotspot mutations were not observed in any PA sample, while 16% of PAs exhibited TERT promoter methylation. PAs with methylated TERT promoters were significantly more likely to show disease progression, shorter PFS, and higher TERT expression levels compared to those with unmethylated promoters.
Conclusions
This is the first study showing that TERT promoter methylation is associated with disease progression and shorter PFS as well as upregulated TERT expression in PAs. Our results suggest that TERT promoter methylation may be a potential biomarker for predicting tumor recurrence in PAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Telomere lengthening is a critical step in tumorigenesis because it provides a mechanism for overriding normal proliferative limitations [1,2,3]. This is maintained by telomerase activation, which is directly related to the expression of its catalytic subunit, telomerase reverse transcriptase (TERT). TERT upregulation has been reported in the majority of human cancers [1]. While the underlying regulation of TERT expression is not fully understood in cancer cells, two potent mechanisms have been proposed. As one of the main mechanisms of TERT upregulation, two somatic promoter mutations, C228T and C250T, confer enhanced TERT promoter activity by generating binding sites for an activating E26 transformation-specific (ETS) transcription factor within the TERT promoter region [3,4,5]. These specific mutational hotspots are frequently found in malignant melanomas (71%) [4, 5], glioblastomas (70–84%) [6, 7], and high-grade meningiomas (20–28%) [8, 9]. The other mechanism is DNA hypermethylation of the TERT promoter, which has been reported to upregulate TERT expression in malignant pediatric brain tumors [1], medulloblastomas [10], and meningiomas [11].
Pituitary adenomas (PAs) account for 10–15% of intracranial tumors [2, 12, 13]. Most PAs are benign in nature, but some grow rapidly and show early postoperative recurrence [12, 13]. However, there are currently no specific biomarkers for predicting PA recurrence and aggressiveness [12, 13]. Increased telomerase activity was previously observed in 13% of 30 PAs and might represent a marker of PA prognosis [14]. Moreover, TERT was found via immunohistochemical staining to be expressed in 28.6% of 49 PAs, and its expression correlated with those of cellular proliferation markers [13]. These reports suggest that activation of telomerase or TERT expression may be useful as a biomarker of the clinical aggressiveness of PAs. However, it is unknown whether TERT alterations are involved in PA recurrence, and the underlying mechanisms of TERT upregulation in PAs remain unclear.
While previous studies have revealed that TERT upregulation contributes to telomerase activity [15, 16], genetic or epigenetic alterations in the TERT promoter that cause TERT upregulation may differ depending on tumor type. There have been no previous studies that have analyzed both mutation and methylation in the TERT promoter in a set of PA cases with extended follow-up information. Therefore, in this study, we aimed to examine TERT promoter hotspot mutations and methylation along with TERT expression levels in 70 PAs and to evaluate their correlations with clinical variables.
Methods
Patients and methods
We studied 70 PAs, including 53 primary diagnosed and 17 recurrent tumors, which were obtained by surgical resection or biopsy at our institution. Clinical data, including age at surgery, sex, largest tumor dimension, PA subtype, Ki-67 labeling index, resection rate, and progression-free survival (PFS), were obtained from medical records. The largest tumor dimension was measured in the coronal or sagittal view from preoperative magnetic resonance imaging (MRI). Resection rate was calculated by comparison of the tumor square measure between the preoperative MRI and the postoperative MRI performed within 72 h after the surgery. PA subtypes were determined according to clinical symptoms, serum hormone levels, and pathological findings. PFS was calculated from the date of surgery until disease progression was confirmed by neuroimaging. PFS is more informative in PA patients than overall survival, since there were no cases of tumor-induced death in our cohort. Disease progression was defined as evidence of one of the following: (1) 30% increase in tumor volume, (2) 10% increase in any dimension following incomplete resection, or (3) any detectable disease following complete resection [17]. This retrospective study was approved by the institutional review board of Saitama Medical University International Medical Center.
DNA/RNA extraction and cDNA synthesis
Genomic DNA was extracted from fresh frozen (56/70 samples) or paraffin-embedded formalin-fixed (14/70 samples) specimens using the Maxwell® RSC Blood DNA Kit (Promega, Madison, WI, USA) or Maxwell® RSC DNA FFPE Kit (Promega), respectively. Total RNA from tumor samples was purified using the Maxwell® RSC Simply RNA Tissue Kit (Promega) or Maxwell® RSC RNA FFPE Kit (Promega). Concentrations of extracted DNA and RNA were measured using Qubit® (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized from 200 ng RNA using the ReverTra Ace® qPCR RT Kit (Toyobo, Osaka, Japan) according to the manufacturer’s recommendations.
Analysis of TERT promoter hotspot mutations
A Sanger sequencing assay was designed to examine the two reported mutational hotspots in the TERT promoter at positions 1,295,228 and 1,295,250 on the reverse strand of chromosome 5, leading to cytidine to thymidine transitions at these positions (C228T and C250T, respectively) [5]. The following primer pair was designed to amplify a region including both sites: forward primer, 5ʹ-TCCCTCGGGTTACCCCACAG-3ʹ; reverse primer, 5ʹ-AAAGGAAGGGGAGGGGCTG-3ʹ [18]. Each PCR run contained 2 µl genomic DNA in a total volume of 25 µl, including 10 µl Amplitaq Gold® (Applied Biosystems, Carlsbad, CA, USA), 1 µl dimethyl sulfoxide, 0.5 µl each primer (10 µM), and distilled water. Cycling conditions were 95 °C for 10 min for initial denaturation, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 40 s, followed by final elongation at 72 °C for 7 min. Amplification products were purified and cleaned by agarose gel electrophoresis, and then gel-extracted with the NucleoSpin® gel and PCR clean-up kit (Macherey–Nagel, Dören, Germany) according to the manufacturer’s protocol. Purified products were submitted to cycle sequencing using the BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems) with the forward PCR primer as a sequencing primer. Purification was then performed using the BigDye® Xterminator Purification Kit (Applied Biosystems). Finally, sequences were determined using a Genetic Analyzer 3130 (Applied Biosystems).
Analysis of TERT promoter methylation
We quantified the methylation status of TERT promoter CpG sites using methylation-sensitive high-resolution melting (MS-HRM) analysis. Briefly, 500 ng of genomic DNA was treated with sodium bisulfite using the Epitect Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. PCR amplification and MS-HRM analysis were carried out sequentially on a LightCycler 480 Real-time PCR system (Roche Diagnostics, Basel, Switzerland). Primer sets for amplifying the region upstream of the transcription start site (UTSS), the methylation of which is observed in malignant pediatric brain tumors, were designed according to previous reports [1, 19] as follows: forward primer, 5ʹ-CCCCGCGTCCGAACCT-3ʹ; reverse primer, 5ʹ-TTCGAGGGAGGGGTTATGATGTG-3ʹ. Each PCR run contained 2.5 µl bisulfite-converted DNA in a total volume of 10 µl, including 5 µl 2 × master mix containing high-resolution melting dye (Roche Diagnostics), 0.9 µl Mg2+, 0.2 µl each primer (10 µM), and distilled water. Cycling conditions were 5 min at 95 °C, followed by 55 cycles of denaturation at 95 °C for 10 s and annealing and extension at 60 °C for 30 s. The melting step was 95 °C for 5 s and 70 °C for 1 min, followed by continuous acquisition to 95 °C at 25 acquisitions/1 °C. As positive (100% methylated) and negative (0% methylated) controls, we used CpGenome™ Universal Methylated and Unmethylated DNA (Chemicon, Millipore, Billerica, MA, USA), respectively. All reactions were performed at least in triplicate.
Analysis of TERT expression
The mRNA expression levels of TERT were determined by real-time quantitative PCR (qPCR) using a LightCycler 480 Real-time PCR system (Roche Diagnostics). The following primer pair was used according to a previous report [18] to amplify TERT: forward primer (located in exon 5), 5ʹ-GCCTGAGCTGTACTTTGTC-3ʹ; reverse primer (located in exon 6), 5ʹ-CGTGTTCTGGGGTTTGATG-3ʹ. The expression level of H6PD (hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase), determined using the primer pair 5ʹ-GATCCTGCCTTTCCGAGAC-3ʹ and 5ʹ-GACCTCCGTCAGATGGTTC-3ʹ, was used as an internal control for normalization. The qPCR reaction volume of 10 µl contained 2 µl cDNA, 5 µl LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics), 0.5 µl each primer (10 µM), and distilled water. Cycling conditions were 5 min at 95 °C, followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 56 °C for 20 s, and extension at 72 °C for 10 s. The melting step was 95 °C for 5 s and 65 °C for 1 min, followed by continuous acquisition to 95 °C at 30 acquisitions/1 °C. Expression was measured relative to that of human total brain RNA (Takara Bio, Shiga, Japan). Relative quantification levels were analyzed using the LightCycler 480 software version 1.5.1 (Roche Diagnostics). All reactions were performed at least in triplicate.
Statistical analysis
We performed Fisher’s exact tests or Fisher-Freeman-Halton test to investigate associations between methylation status and clinical variables [sex, largest tumor dimension, tumor subtype, resection rate, tumor status (primary or recurrent tumor), and disease progression]. The Mann–Whitney U test was performed to compare TERT methylation status by age or Ki-67 labeling index. PFS was calculated using Kaplan–Meier estimates and compared between methylated with unmethylated PAs using the log-rank test. The Mann–Whitney U test was performed to compare TERT mRNA levels with TERT promoter methylation status or disease progression. PFS was evaluated using uni- and multivariate analyses with the Cox proportional hazards model, including the following variables: age, sex, largest tumor dimension, resection rate, tumor status and methylation status. A monotonic trend between the degree of TERT methylation and tumor progression was assessed by the Cochran-Armitage test for trend. A p-value below 0.05 was considered statistically significant. SPSS Statistics, version 25 (SPSS Inc., Chicago, IL, USA) was used for statistical calculations.
Results
Baseline clinical data
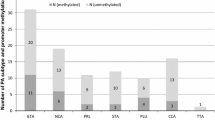
The clinicopathological characteristics of the investigated samples are summarized in Table 1. The median age at surgery was 59 years old, ranging from 22 to 81 years old, and cases were predominantly male (61%). The largest tumor dimension was more than 4 cm (giant adenoma) in 12 cases (17%) and 4 cm or less in 58 cases (83%). Among the 70 PAs, 59 were non-functioning adenomas (NFAs), eight were growth hormone-producing adenomas (GHomas), one was an adrenocorticotroph hormone-producing adenoma (ACTHoma), one was a thyrotroph stimulating hormone-producing adenoma (TSHoma), and one was a prolactin-producing adenoma (PRLoma). Disease progression, confirmed by follow-up neuroimaging, occurred in 18 cases among 70 PAs (26%). We also examined the Ki-67 labeling index in 38 out of the 70 samples. The median value was 1.0%, ranging from 0.1 to 5.1%.
No PA samples harbored TERT promoter hotspot mutations
All 70 samples were analyzed for the TERT promoter hotspot mutations C228T and C250T using Sanger sequencing. Neither of these mutations were found in any of the samples.
TERT promoter methylation was more common in recurrent PAs and correlated with shorter PFS
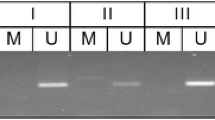
We performed MS-HRM analysis to quantitatively evaluate TERT promoter methylation status in PAs. Melting data collected using the LightCycler 480 was analyzed using the ‘‘Tm (Melting Temperature) calling’’ algorithm, which converts melting profiles into derivative plots, allowing methylated and unmethylated samples to be distinguished. Products amplified from methylated DNA have a higher Tm due to the presence of CpGs in the amplicon. In contrast, products amplified from unmethylated DNA have a lower Tm due to the conversion of unmethylated cytosine to uracil in the bisulfite-modified DNA sample, which results in thymine in the amplicon [20]. If the sample contains a mixture of methylated and unmethylated DNA, two peaks are displayed [20]. We generated standard curves using serial samples with known ratios of methylated to unmethylated template (Fig. 1). The TERT methylation level of an unknown sample could then be estimated from these standard curves. Samples were analyzed at least in triplicate, and the methylation level was calculated as the average value of the experiments. We defined a methylation level of more than 10% as methylated and a level of 10% or less as unmethylated. This cut-off was used because a low level of TERT promoter methylation (less than 10%) was observed in certain normal tissues [21], and a previous study that investigated TERT promoter methylation using MS-HRM used the same threshold [19].
Methylation-sensitive high-resolution melting (MS-HRM) analysis with 100% methylated and 0% methylated controls and methylation standards at 10%, 25%, and 50%. The TERT methylation level of an unknown sample can be estimated from these standard curves. Data were analyzed using the Tm calling software module
All 70 samples were successfully analyzed by MS-HRM. The TERT promoter was determined to be methylated in 11 samples (16%) and unmethylated in 59 samples (84%). Age, sex, largest tumor dimension, subtype, Ki-67 labeling index and resection rate did not differ significantly between methylated and unmethylated cases (Table 1). Recurrent PAs were significantly more likely to be methylated (7 out of 17 samples) than primary PAs (4 out of 53 samples) (p = 0.003, Table 1). Disease progression occurred significantly more frequently in methylated PAs (55%) than in unmethylated PAs (20%) (p = 0.03, Table 1). Next, the impact of TERT promoter methylation on PFS was assessed. Compared to PAs with unmethylated TERT promoters, those with methylated TERT promoters exhibited significantly shorter PFS (median: 34 vs. 109 months, p = 0.002, Fig. 2). Among primary PA samples, PFS was also significantly shorter in methylated PAs (median: 30 vs. 133 months, p = 0.002). Cox proportional hazards regression analysis confirmed that PFS was correlated with methylation status (hazard ratio: 5.804, 95% confidence interval: 1.407–23.940, p = 0.02, Table 2). Finally, a trend between the degree of TERT methylation and tumor progression was assessed. Tumor progression occurred in 7 out of 45 PAs with 0% methylation; 5 out of 14 PAs with 1–10% methylation; 2 out of 4 PAs with 10–25% methylation; all 3 PAs with 25–50% methylation; and 1 out of 4 PAs with 50–100% methylation. The Cochran-Armitage test for trend showed that there was a statistically significant trend for a monotonic increase in the proportions of tumor progression cases dependent on the degree of TERT methylation (p = 0.007).
TERT expression was higher in PAs with methylated TERT promoters
To assess the influence of TERT promoter methylation on TERT expression in PAs, relative quantification analysis was performed to determine TERT mRNA expression levels using real-time qPCR. Samples were analyzed at least in triplicate, and expression levels were based on the average value of the experiments. The mean expression levels of TERT were 1.5 times and 37 times higher in unmethylated and methylated samples, respectively, than in normal brain tissue (Fig. 3). The expression level was significantly higher in the methylated group than in the unmethylated group (p = 0.008). The Mann–Whitney U test revealed that higher TERT mRNA levels correlated with increased disease progression (p = 0.04).
TERT expression levels in unmethylated and methylated pituitary adenomas. Mean expression levels are 1.5 times and 37 times higher in unmethylated and methylated samples, respectively, than in human total brain tissue. The mean expression level is significantly higher in the methylated group than in the unmethylated group (p = 0.008)
Discussion
TERT upregulation due to TERT promoter alterations has been implicated in various human cancers [1, 10, 11, 18]. Recent studies have identified TERT promoter hotspot mutations at C228T or C250T in various brain tumors [3, 6, 7, 9, 16, 22, 23]. These mutations increase the transcriptional activity of the TERT promoter [3,4,5]. However, TERT promoter mutations are primarily seen in malignant tumors [3], and therefore, TERT promoter mutations in PAs, which are mainly classified as benign tumors, have not been thoroughly investigated. To our knowledge, there have been only two reports of TERT promoter mutations in PAs [23, 24], which revealed the absence of TERT promoter hotspot mutations in 11 and 15 PA cases. None of our 70 PAs exhibited mutations in the TERT promoter, confirming these previous results. Therefore, it appears that TERT promoter mutations are uncommon in PAs.
Accumulating evidence has indicated a positive association between TERT promoter methylation and TERT upregulation in several tumors, such as malignant pediatric brain tumors [1], medulloblastomas [10], and high-grade meningiomas [11]. We quantitatively assessed TERT promoter methylation status using MS-HRM, and our results showed that 16% of PAs exhibited methylation of the TERT promoter, which was predominantly found in recurrent samples. There are several methods for analyzing TERT promoter methylation [25]. Köchling et al. [2] reported that 27% of primary PAs demonstrated TERT promoter methylation according to methylation-specific PCR (MSP), and the frequency of TERT promoter methylation was similar between primary and recurrent PAs. While MSP is sensitive, a qualitative method is needed to examine epigenetic gene alterations, so it cannot distinguish between high and low levels of methylation [25]. In contrast, the assay used in the present study, MS-HRM, can detect 1.0% methylated DNA over a background of unmethylated DNA and is capable of better distinguishing between high and low levels of TERT promoter methylation [20, 25]. This is likely why the TERT promoter methylation frequencies in our primary PAs were lower than those found in the previous study using MSP [2]. Moreover, because a low level of TERT promoter methylation (less than 10%) has been observed in certain normal tissues [19, 21], the MSP method may overestimate the tumor-related frequency of TERT promoter methylation.
We first showed that disease progression occurred significantly more frequently in PAs with methylated TERT promoters than in PAs with unmethylated ones. Disease progression occurred in 55% of PAs with methylated TERT promoter. Moreover, PFS was significantly shorter in methylated PAs than in unmethylated PAs, and multivariate analyses confirmed that PFS was correlated with methylation status alone. These results were obtained using a quantitative method that more accurately evaluates TERT promoter methylation status. While most PAs are benign in nature, some cases exhibit clinically aggressive behavior and display early postoperative recurrence [12, 26]. Although proliferation potential or angiogenesis may reflect the aggressiveness of PAs, no molecular marker has yet been identified as a reliable predictor, including Ki-67, CD34, epidermal growth factors, and vascular endothelial growth factors [12, 13, 26]. Although we investigated the Ki-67 labeling index in 38 out of the 70 samples, all samples exhibited a staining index of less than approximately 5%, and this was not associated with recurrence or TERT methylation status (Table 1). Therefore, based on our results, TERT promoter methylation may have potential for development into a sensitive predictive marker.
The effect of promoter methylation on gene expression differs depending on the type of tumor and the gene involved. Promoter hypermethylation of O6-methylguanine-DNA methyltransferase (MGMT), a DNA repairing enzyme, induces loss of its expression [6]. Cyclin-dependent kinase inhibitor 2A/p16 (CDKN2A) shows decreased expression following methylation, and its methylation status is reported to be a poor prognostic marker in gliomas [27]. Conversely, TERT promoter methylation and TERT expression is reported to be positively correlated in malignant brain tumors [1, 10, 11], although TERT expression is not related to TERT promoter methylation in some tumors, such as adult gliomas [18, 21]. This study is the first to investigate and identify a correlation between TERT promoter methylation and TERT expression in PAs. TERT promoter methylation may prevent the binding of transcriptional repressors that normally prevent TERT expression, promoting TERT upregulation [21]. TERT methylation thus subsequently elevates telomerase activity and may lead to PA tumor recurrence. There are likely to be numerous genetic and epigenetic factors involved in the aggressiveness of PAs. However, to our knowledge, this is the first study showing that TERT promoter methylation upregulates TERT expression and is associated with disease progression in PAs. Further investigations with larger sample sizes are needed to clarify the mechanisms of PA pathogenesis.
Conclusions
In this study, we found that no PA samples exhibited TERT promoter hotspot mutations, whereas TERT promoter methylation was found in 16% out of 70 PAs. PAs with methylated TERT promoters were significantly more likely to show disease progression and shorter PFS compared to those with unmethylated promoters. Moreover, we found that TERT promoter methylation upregulated TERT expression and was associated with disease progression in PAs. Our results suggest that TERT promoter methylation may be a potential biomarker for predicting disease progression in a subset of PAs.
References
Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, Zhukova N, Walker EJ, Martin D, Merino D, Wasserman JD, Elizabeth C, Alon N, Zhang L, Hovestadt V, Kool M, Jones DT, Zadeh G, Croul S, Hawkins C, Hitzler J, Wang JC, Baruchel S, Dirks PB, Malkin D, Pfister S, Taylor MD, Weksberg R, Tabori U (2013) Methylation of the TERT promoter and risk stratifi cation of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol 14:534–542. https://doi.org/10.1016/S1470-2045(13)70110-4
Kochling M, Ewelt C, Furtjes G, Peetz-Dienhart S, Koos B, Hasselblatt M, Paulus W, Stummer W, Brokinkel B (2016) hTERT promoter methylation in pituitary adenomas. Brain Tumor Pathol 33:27–34. https://doi.org/10.1007/s10014-015-0230-8
Bell RJA, Rube HT, Xavier-Magalhaes A, Costa BM, Mancini A, Song JS, Costello JF (2016) Understanding TERT promoter mutations: a common path to immortality. Mol Cancer Res 14:315–323. https://doi.org/10.1158/1541-7786.MCR-16-0003
Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R (2013) TERT promoter mutations in familial and sporadic melanoma. Science 339:959–961
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339:957–959. https://doi.org/10.1126/science.1229259
Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, Shibui S, Ichimura K (2013) Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol 126:267–276. https://doi.org/10.1007/s00401-013-1141-6
Huang DS, Wang Z, He XJ, Diplas BH, Yang R, Killela PJ, Meng Q, Ye ZY, Wang W, Jiang XT, Xu L, He XL, Zhao ZS, Xu WJ, Wang HJ, Ma YY, Xia YJ, Li L, Zhang RX, Jin T, Zhao ZK, Xu J, Yu S, Wu F, Liang J, Wang S, Jiao Y, Yan H, Tao HQ (2015) Recurrent TERT promoter mutations identified in a large-scale study of multiple tumor types are associated with increased TERT expression and telomerase activation. Eur J Cancer 51:969–976. https://doi.org/10.1016/j.ejca.2015.03.010
Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M (2014) High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol 24:184–189. https://doi.org/10.1111/bpa.12110
Juratli TA, Thiede C, Koerner MVA, Tummala SS, Daubner D, Shankar GM, Williams EA, Martinez-Lage M, Soucek S, Robel K, Penson T, Krause M, Appold S, Meinhardt M, Pinzer T, Miller JJ, Krex D, Ely HA, Silverman IM, Christiansen J, Schackert G, Wakimoto H, Kirsch M, Brastianos PK, Cahill DP (2017) Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget 8:109228–109237. https://doi.org/10.18632/oncotarget.22650
Lindsey JC, Schwalbe EC, Potluri S, Bailey S, Williamson D, Clifford SC (2014) TERT promoter mutation and aberrant hypermethylation are associated with elevated expression in medulloblastoma and characterise the majority of non-infant SHH subgroup tumours. Acta Neuropathol 127:307–309. https://doi.org/10.1007/s00401-013-1225-3
Fürtjes G, Köchling M, Peetz-Dienhart S, Wagner A, Heß K, Hasselblatt M, Senner V, Stummer W, Paulus W, Brokinkel B (2016) hTERT promoter methylation in meningiomas and central nervous hemangiopericytomas. J Neurooncol 130:79–87. https://doi.org/10.1007/s11060-016-2226-6
Chatzellis E, Alexandraki KI, Androulakis II, Kaltsas G (2015) Aggressive pituitary tumors. Neuroendocrinology 101:87–104. https://doi.org/10.1159/000371806
Ortiz-plata A, Suck MLT, Lopez-Gomez M, Heras A, Sánchez García A (2007) Study of the telomerase hTERT fraction, PCNA and CD34 expression on pituitary adenomas. Association with clinical and demographic characteristics. J Neurooncol 84:159–166. https://doi.org/10.1007/s11060-007-9365-8
Yoshino A, Katayama Y, Fukushima T, Watanabe T, Komine C, Yokoyama T, Kusama K, Moro I (2003) Telomerase activity in pituitary adenomas: significance of telomerase expression in predicting pituitary adenoma recurrence. J Neurooncol 63:155–162
Boldrini L, Pistolesi S, Gisfredi S, Ursino S, Ali G, Pieracci N, Basolo F, Parenti G, Fontanini G (2006) Telomerase activity and hTERT mRNA expression in glial tumors. Int J Oncol 28:1555–1560
Dome JS, Chung S, Bergemann T, Umbricht CB, Saji M, Carey LA, Grundy PE, Perlman EJ, Breslow NE, Sukumar S (1999) High telomerase reverse transcriptase (hTERT) messenger RNA level correlates with tumor recurrence in patients with favorable histology Wilms’ tumor. Cancer Res 59:4301–4307
Hightower E, Cabanillas ME, Fuller GN, McCutcheon IE, Hess KR, Shah K, Waguespack SG, Corley LJ, Devin JK (2012) Phospho-histone H3 (pHH3) immuno-reactivity as a prognostic marker in non-functioning pituitary adenomas. Pituitary 15:556–561. https://doi.org/10.1109/TMI.2012.2196707
Arita H, Narita Y, Takami H, Fukushima S, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Shibui S, Ichimura K (2013) TERT promoter mutations rather than methylation are the main mechanism for TERT upregulation in adult gliomas. Acta Neuropathol 126:939–941. https://doi.org/10.1007/s00401-013-1203-9
Bougel S, Lhermitte B, Gallagher G, de Flaugergues JC, Janzer RC, Benhattar J (2013) Methylation of the hTERT promoter: a novel cancer biomarker for leptomeningeal metastasis detection in cerebrospinal fluids. Clin Cancer Res 19:2216–2223. https://doi.org/10.1158/1078-0432.CCR-12-1246
Adachi J, Mishima K, Wakiya K, Suzuki T, Fukuoka K, Yanagisawa T, Matsutani M, Sasaki A, Nishikawa R (2012) O6-methylguanine-DNA methyltransferase promoter methylation in 45 primary central nervous system lymphomas: quantitative assessment of methylation and response to temozolomide treatment. J Neurooncol 107:147–153. https://doi.org/10.1007/s11060-011-0721-3
Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J (2002) Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer 101:335–341. https://doi.org/10.1002/ijc.10593
Sahm F, Schrimpf D, Olar A, Koelsche C, Reuss D, Bissel J, Kratz A, Capper D, Schefzyk S, Hielscher T, Wang Q, Sulman EP, Adeberg S, Koch A, Okuducu AF, Brehmer S, Schittenhelm J, Becker A, Brokinkel B, Schmidt M, Ull T, Gousias K, Kessler AF, Lamszus K, Debus J, Mawrin C, Kim YJ, Simon M, Ketter R, Paulus W, Aldape KD, Herold-Mende C, von Deimling A (2016) TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst 108:1–4. https://doi.org/10.1093/jnci/djv377
Koelsche C, Sahm F, Capper D, Reuss D, Sturm D, Jones DT, Kool M, Northcott PA, Wiestler B, Böhmer K, Meyer J, Mawrin C, Hartmann C, Mittelbronn M, Platten M, Brokinkel B, Seiz M, Herold-Mende C, Unterberg A, Schittenhelm J, Weller M, Pfister S, Wick W, Korshunov A, von Deimling A (2013) Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol 126:907–915. https://doi.org/10.1007/s00401-013-1195-5
Martins C, Castro M, Calado R (2016) Absence of TERT promoter mutations in pituitary adenomas. J Endocrinol Invest 39:933–934
Wojdacz TK, Dobrovic A (2007) Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 35:e41. https://doi.org/10.1093/nar/gkm013
Di Ieva A, Rotondo F, Syro LV, Cusimano MD, Kovacs K (2014) Aggressive pituitary adenomas-diagnosis and emerging treatments. Nat Rev Endocrinol 10:423–435. https://doi.org/10.1038/nrendo.2014.64
Costello JF, Berger MS, Huang HS, Cavenee WK (1996) Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res 56:2405–2410
Acknowledgements
This work was supported by a grant from the Hidaka Research Project and a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS KAKENHI Grant Number 15K10369), 2015–2017. We would like to thank Ms. Kozue Watanabe and Ms. Ayako Kubota for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Miyake, Y., Adachi, Ji., Suzuki, T. et al. TERT promoter methylation is significantly associated with TERT upregulation and disease progression in pituitary adenomas. J Neurooncol 141, 131–138 (2019). https://doi.org/10.1007/s11060-018-03016-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-03016-8