Abstract
Medulloblastoma (MB) is the most common malignant brain tumor in childhood. The 5 year disease-free survival rate is rather low. There is a consensus that MB can be divided into at least four clinically, transcriptionally, and genetically distinct molecular variants, being designated as wingless (WNT), sonic hedgehog (SHH), Group 3 and Group 4. It poses a great challenge to the design of therapeutic strategy for MB patients. Intensive clinical intervention, including high dose radiotherapy, is commonly used in treatment of high risk MB, most of which are considered to be Group 3 patients. But such intensive therapy should be avoided to protect neurologic function of patients in the lower risk WNT group. In present study, MB subgroup assignment in formalin-fixed paraffin embedded (FFPE) specimens from 45 Chinese patients were performed by Nanostring platform using 22 well-known signature genes. Based on comparative expression profiles of miRNA real-time PCR microarray in MB cells with and without treatment of demethylation reagent, as well as MSP assay, miR-449a was demonstrated to be significantly silenced by aberrant DNA methylation in tumor cells. Real-time PCR showed that expression level of miR-449a in WNT group was significantly different from other subgroups, although it was down-regulated in most of the MB samples. In conclusion, current study demonstrates for the first time the feasibility of using the Nanostring assay for subgrouping of MBs in Chinese patients. In addition, MiR-449a, a candidate tumor suppressor regulated by hypermethylation, is a novel potential diagnostic marker for WNT group of MBs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Medulloblastoma (MB) is the most common malignant brain tumor in childhood and locates at patient’s cerebellum. Although the overall 5 year-survival rate is now higher than 70 % due to multimodal treatment strategy of surgical resection followed by cranio-spinal radiation (children older than 3 years) and chemotherapy, the 5 year disease-free survival rate is still rather low (36 %), and prognosis for patients with recurrent MB remains very poor [1, 2]. At the same time, neurocognitive impairment due to high-dose radiation therapy has a significant negative impact on the life quality of the children with this tumor.
There is a consensus based on current studies that MB is not a single entity but rather a heterogeneous group of tumors which differ not only in histologic appearance, but also in molecular biology. This poses a great challenge to the design of therapeutic strategy for this tumor. Integrated genomic studies have demonstrated MB can be divided into at least four clinically, transcriptionally, and genetically distinct molecular variants [3]. These four variants have been designated as wingless (WNT), sonic hedgehog (SHH), Group 3 and Group 4. Group 3 is known to have the poorest outcome, while the WNT group in which the Wnt signaling pathway is activated has a relatively good prognosis. Intensive clinical intervention, including high dose radiotherapy is commonly used in treatment of high risk MB [3]. Most of these high risk cases are considered to be Group 3, but such intensive therapy should be avoided to protect neurologic function of patients in the lower risk WNT group [3]. Thus, development of novel subgroup-specific markers to facilitate refinement of risk-stratification as well as individualization of therapy has become a critical area in next generation clinical trials for MB. Northcott et al. recently established an assay using a panel of 22 group-specific gene probes based on Nanostring nCounter Technology. Gene probe data allows assignment of MB into subgroups rapidly and economically according to transcriptional profiles [4, 5]. Being considered as the golden standard, this method is practical for subtyping MB from FFPE samples, and has great potential for use in clinical trials [5, 6].
Micro RNAs (miRNA) are 20–25-nucleotide RNAs [7]. Deregulation of miRNA expression and the genetic aberration within amplified or deleted regions of a few miRNA genes [8–10] are closely related to the occurrence of a variety of tumors [11–17] as they regulate the multistep processes of carcinogenesis as oncogenes or tumor suppressor genes (TSG) [18, 19]. Various studies show the value of miRNA alterations in molecular subtyping of MB, and their close relationship with tumor progression, invasiveness, metastasis and patient prognosis [17, 20–23]. Recent studies, including our own [24, 25], on DNA methylation-mediated downregulation of TSG in MB clearly demonstrate the concept of MB as an epigenetic disease [24–26]. However, to date there have been few reports on subtyping specific TS-miRNA related with DNA hypermethylation in MB.
In the present study, differing expression profiles of miRNA were evaluated by real-time PCR microarray in MB cells with and without treatment of DNA methyltransferase inhibitor 5-Aza. Several instances of re-expression of a given miRNA that was silenced by aberrant DNA methylation directly or indirectly were identified, including miR-449a. DNA hypermethylation of CpG island in the promoter region of miR-449a in MB cells was then confirmed. To further explore the specificity of miR-449a expression to MB tumor subtype, the expression status of miR-449a in 45 MB primary tumors compared with normal cerebellum, where the tumor was located, was analyzed. Correlation of expression levels of miR-449a among all molecular subtypes of MB was further evaluated in our cohort, based on subtyping data from Nanostring nCounter Technology assay.
Methods and materials
Clinical information of MB
Formalin-fixed paraffin embedded (FFPE) samples were obtained from 45 MBs in patients from Beijing Tian Tan Hospital (China). All these tumors samples were collected between September 2007 and December 2011. Each tumor was reviewed by two pathologists (Q.C, Y.Z) for histologic subtyping. MBs were classified as classic, desmoplastic nodular (DN), extensive nodular (EN) and anaplastic/large cell (A/LC) tumor variants according to the 2007 World Health Organization (WHO) classification [3]. This series of MB (n = 45), which all located in cerebellum, consisted of 24 classic MBs (53.3 %), 15 DN/EN MBs (33.33 %), and 6 A/LC MBs (13.33 %) tumor variants. The mean age of patients was 7.9 years old in 40 children and 31.6 years old in five adults. Clinico-pathologic information was summarized in Table 1. This study was approved by the Institutional Review Board of Peking University, Beijing, China (review reference number IRB00001052-14003).
Demethylation treatment
Three MB cell lines (Daoy, D283, and D341) obtained from American Type Culture Collection (Manassas, VA) were examined in this study. These cell lines were cultured in a-minimal essential medium (Sigma–Aldrich Corporation, St Louis, MO) supplemented with 10–20 % fetal bovine serum (FBS; Invitrogen Corporation, Carlsbad, Calif) and 2 mmol/L1-glutamine (Sigma–Aldrich Corporation). Cells growing at 80 % confluency were harvested and seeded on six-well plates (Corning, NY). At 30 % confluency, drug treatment was started by adding complete medium with 5-aza-2′-deoxycytidine (5-Aza) which is a demethylation reagent (Sigma–Aldrich Co., St. Louis, MO). The concentrations used were 0 μM (as a no reagent control) and 5 μM as previously reported [24]. Half of the medium was replaced every day with fresh complete medium containing 5-Aza for 3 days. On the fourth day, cells were harvested. Total RNA for mRNA analysis was extracted using the RNeasy Mini Kit (Qiagen, Basel, Switzerland) following the manufacturer’s instructions, and tested for restoration of gene expression with the MiRCURY LNA™ Universal RT miRNA PCR array.
MiRCURY LNA™ Universal RT miRNA PCR
According to the manufacturer’s protocol [27], a total of 25 ng RNA extracted from each MB cell line with or without 5-Aza treatment was reverse transcribed using the miRCURY Locked Nucleic Acid (LNA™) Universal Reverse Transcription (RT) microRNA PCR, Polyadenylation and cDNA synthesis kit (Exiqon miRNA qPCR panel, Vedbaek, Denmark). The products of reverse transcription were subjected to Exiqon miRCURY-Ready-to-Use PCR-Human-panel-I + II-V3.M (Exiqon miRNA qPCR panel, Vedbaek, Denmark) which can detect a total 766 different miRNAs in treated or untreated cells and allow identification of differently expressed miRNAs on a 7900HT real-time PCR system (Applied Biosystems, Foster City, CA, USA). An RNA spike-in (UniSp6) and a DNA spike-in (Sp3) were applied in the panel for quality control of technical performance for all samples. Appropriate melting curves and melting temperatures (Tm) were determined to be within known specifications in all assays. Normalization was further performed based on the average of the normalizer assays in panel-I + II, where normalized Ct (∆Ct) = average Ct (assay) − average Ct (normalizer assays). The relative different in expression of cells with and without drug treatment was assessed by the 2− ∆∆Ct method.
Sodium bisulfite treatment of DNA
The sodium bisulfite reaction converts unmethylated cytosine in DNA to uracil, while leaving methylcytosine intact, allowing amplification of methylated and unmethylated alleles with specific primers by polymerase chain reaction (PCR). Genomic DNA from MB cell lines was extracted using the QIAampR DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) and 0.1–1.0 µg DNA samples were treated with sodium bisulfite using the BIsulFlash DNA Modification Kit (Epigentek, NY, USA) according to the manufacturer’s recommendations [26].
Methylation specific PCR (MSP) for miR-449a methylation status in MB cells
The methylation status of miR-449a in MB cell lines was evaluated with MSP. MSP primers used for the unmethylated reaction were 5′-TTTGGTGTTTGGGTGTGTGT-3′ (sense) and 5′-CACCCCTACCAACCCTCTCT-3′ (antisense); for the methylated reaction, 5′-TTACGGTTCGCGGTAAAGTT-3′ (sense) and 5′-CCCCTACCGACCCTCTCTAA-3′ (antisense), with 71 bp and 87 bp amplification product, respectively. The annealing temperature for both sets of MSP primers was 59 °C. After thermocycling, PCR products were resolved with electrophoresis on 12 % polyacrylamide and stained with ethidium bromide, followed by examination with UV illumination. CpGenome universal methylated DNA (Millipore, MA, USA) was used as methylation control. Four normal cerebellum tissues were recruited from postmortem brain tissue of children. All experiments were repeated at least twice.
Nanostring analysis
Total RNA was extracted from FFPE material from 45 MB samples and 3 MB cell lines based on the Nanostring nCounter Analysis System (Nanostring Technologies) in the Department of Pathology, Peking University Health Science Center, and gene expression analysis was conducted using the classic MB subtyping gene CodeSet, including 22 genes with 5-6 signature genes for each subgroup: WNT (WIF1, TNC, GAD1, DKK2 and EMX2), SHH (PDLIM3, EYA1, HHIP, ATOH1 and SFRP1), Group 3 (IMPG2, GABRA5, EGFL11, NRL, MAB21L2 and NPR3) and Group 4 (KCNA1, EOMES, KHDRBS2, RBM24, UNC5D and OAS1) [5]. Each reaction contained 200 ng of total RNA in a 4 µl aliquot, together with the reporter and capture probes, and six pairs of positive controls and six pairs of negative control probes. Analysis and normalization of the raw Nanostring data was conducted using nSolver Analysis Software v2.5 (Nanostring Technologies). Raw counts were normalized to internal levels for three reference genes including CLTC, GAPDH and TUBB. A background count level was estimated using the average count of the six negative control probes in every reaction plus two standard deviations (SDs). All procedures related to mRNA quantification including sample preparation, hybridization, detection, and scanning were carried out as recommended by Nanostring Technologies.
Quantitative reverse transcriptase PCR
Reverse transcription was carried out with the miRcute miRNA First Strand cDNA Synthesis Kit (Tiangen, China) according to the manufacturer’s protocol. The miRcute miRNA qPCR Kit (Tiangen, China) was used for quantitative real time-PCR (qRT-PCR). Real-time PCR was performed on a Mx3000P Real-time PCR system (Agilent, USA). The reaction mixtures were incubated at 94 °C for 2 min, followed by 40 cycles at 94 °C for 20 s, and 60 °C for 34 s. The primer miR-449a (ID 000583), and the U6 small nuclear 2 (RNU6B) (ID 001093) used for an RNA control were designed and synthesized by Tiangen (Beijing, China). Human cerebellar total RNA pooled from three male Asian was obtained from Clontech Laboratories, Inc. (CA, US). All the real time-PCRs were performed in triplicate. Data are presented based on calculating 2− ∆∆Ct.
Statistical analysis
All statistical analysis was performed using the SPSS 20.0 statistical software package. Results were presented as mean ± SEM. The Student’s t test was used to determine statistical differences between groups (MB tissues and normal cerebellum tissue). ANOVA was used to evaluate the relationship of miRNA-449a expression with clinical-pathological characteristics in MBs. Other results were evaluated by independent samples T test. p values of <0.05 were considered to be statistically significant.
Result
Comparative analysis of miRNA expression profiles in MB cell lines with and without demethylation reagent treatment
In order to screen candidate miRNAs which may be regulated by DNA hypermethylation as candidate tumor suppressor in MB, the expression profile of miRNAs in three well established MB cell lines were detected by MiRCURY LNA™ Universal RT miRNA PCR array before and after treatment with 5-Aza, respectively. Comparative data analysis showed the expression level was significantly increased after demethylation drug treatment in 6 of 766 miRNAs (p < 0.05) (Fig. 1a). Among the 6 miRNAs, miR-449a expression has the relatively higher up-regulation fold change. CpG island has been found in its promoter region using bioinformatics tools (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/ and http://www.ncbi.nlm.nih.gov/gene/), demonstrating that miR-449a may be directly or indirectly regulated by DNA hypermethylation.
a Comparative expression profile of miRNAs in 3 well established MB cell lines detected by MiRCURY LNA™ Universal RT miRNA PCR array before and after treatment with 5-Aza. Expression level was significantly increased in 6 of 766 miRNAs after demethylation drug treatment (p < 0.05). Among the 6 miRNAs, miR-449a expression has the relatively higher up-regulation fold change. b The methylation status of CpG island in the promoter region of miR-449a in MB cell lines detected by methylation specific PCR. Hypermethylation was observed in D283 and D341. No hypermethylation was observed in Daoy. m methylation, u unmethylation, uni CpGenome universal methylated DNA as methylation control, a normal cerebellum tissue from as negative control, H 2 O blank control
Hypermethylation status of miR-449a in MB cell lines confirmed by MSP
To confirm the expression of miR-449a in MB cells was regulated by DNA hypermethylation directly, the methylation status of CpG island in the promoter region of miR-449a in MB cell lines were detected by methylation specific PCR. Hypermethylation was observed in D283 and D341. The methylation level was relatively higher in D283 than in D341 (Fig. 1b). Thus, hypermethylation of CpG island in the promoter region of miR-449a in these MB cell lines may induce the silence of miR-449a, which can be significantly restored by the demethylation reagent treatment. No methylation was detected in normal cerebellum tissues from 4 children, implying this epigenetic alteration in MB cells was tumor-specific. The results coming from MSP assay together with RT miRNA PCR gene profile demonstrated that DNA hypermethylation of miR-449a may be a potential epigenetic marker for MBs.
Subgroup assignment of medulloblastomas by Nanostring assay
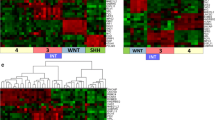
To further explore the specificity of miR-449a expression to MB subgroups, the first step is to demonstrate the utility of the Nanostring assay for MB subgrouping in Chinese patients using the well-known CodeSet [5]. Total 45 primary MBs were analyzed using 22 subgroup-specific signature genes validated from a large series of primary MBs [4, 5]. It was observed that the subgroup could be predicted in 97.8 % (44/45) tumors by non-hierarchical clustering analysis, with 4 tumors subgrouped as WNT, 8 as SHH, 16 as Group 3 and 16 as Group 4 (Table 1; Fig. 2). At the same time, the three well established MB cell lines were also analyzed for subtyping, showing Daoy cells belong to the SHH group, and both D283 and D341 cells subtyped as Group 3. Analysis and normalization of the raw Nanostring data using nSolver Analysis Software v2.5 (Nanostring Technologies) showed ‘normalization flag’ in 1/45 (2.2 %) case (M-T-45), which means our current Nanostring assay is unable to provide a high confidence of subgroup assignment to sub-group this case. Data in present study thus demonstrated that the Nanostring assay with CodeSet consisting of 22 well-established signature genes utilized at Peking University Health Science Center is capable of assignment of molecular subgroups in most cases of MB, which were all FFPE materials from Chinese patients.
MB subgroup predicted by nanostring assay using the 22 gene well-known CodeSet. With non-hierarchical clustering analysis, 97.8 % (44/45) of MBs can be assigned, with four tumors subgrouped as WNT, 8 as SHH, 16 as Group 3 and 16 as Group 4. Three well established MB cell lines were also analyzed for subtyping, showing Daoy cells belong to the SHH group, and both D283 and D341 cells subtyped as Group 3. Blue block WNT, green block SHH, orange block Group 3, Pink block Group 4
MiR-449a expression level and its relationship with molecular subgroups in primary MBs
After the Nanostring assay platform for MB subgrouping was established, the specificity of miR-449a expression to MB subgroups was explored. The expression of miR-449a in 45 primary MBs were detected by real-time PCR. 97.8 % (44/45) of the cases showed lower expression level compared with normal cerebellum, indicating miR-449a was down-regulated in most of the MB tumors (Fig. 3). Based on molecular subgroup assignment data coming from Nanostring assay, the relationship between miR-449a expression and MB subgroups was further analyzed. The result showed expression of miR-449a was significantly different within 4 subgroups (p = 0.0270, ANOVA) and was significantly higher in the WNT group than in other subgroups (p = 0.0026, t test) (Fig. 4), demonstrating it may be a potential diagnostic marker for subtyping of MB.
Expression of miR-449a in 45 primary MBs detected by real-time PCR. 97.8 % (44/45) of the cases showed lower expression level compared with normal cerebellum, indicating miR-449a was down-regulated in most of the MB tumors as a candidate tumor suppressor. NC normal cerebellum pooled from three male Asian
The relationship between miR-449a expression and MB subgroups. Based on molecular subgroup assignment data coming from nanostring assay, expression of miR-449a showed significantly difference in four subgroups (p = 0.0270, ANOVA) and was significantly higher in the WNT group than in other subgroups (p = 0.0026, t test)
Discussion
In the current study, the Nanostring gene expression assay with the 22 gene-Codeset showed forceful capacity for MB subgroup assignment in FFPE specimens from Chinese patients. Expression of miR-449a was down-regulated in most of the MB samples as compared with normal human cerebellum, but expression levels were relatively high in the WNT group in our cohort. The results of demethylation drug treatment accompanied by MSP in MB cells support understanding the loss of expression of miR-449a was due to regulation of DNA hypermethylation, indicating that this molecule is a potential epigenetic diagnostic marker for subtyping MBs.
MiRNAs regulate a variety of biological processes in human malignancy, and a given type of miRNA may show variable expression in different types of human cancer and function as either an oncogene or a tumor suppressor gene. MiR-449a functions as a tumor suppressor resulting in reduction of cell proliferation, migration and invasion, as well as inducing apoptosis in various human tumors, including GBM [28], neuroblastoma [14], prostate cancer [29], osteosarcoma [30], ovarian cancer [31] and hepatocellular carcinoma [32]. Conversely, it acts as an oncogene and drives malignant phenotypes in aggressive breast cancer [33] and colorectal carcinoma [34], which predicts poor prognosis in these tumors [34]. In the present study, based on comparative miRNA expression profiling assay, we found DNA methyltransferase inhibitor 5-Aza treatment allows for re-expression of miR-449a silenced by tumor-specific aberrant DNA methylation in MB cells. Further evaluation of miR-449a expression demonstrated that it is down-regulated in most MB primary tumors compared with normal cerebellum, indicating miR-449a may be a potential tumor suppressor being regulated by hypermethylation in this tumor. Epigenetic alterations of miR-449a in human disease have been observed in previous studies [29, 35]. It is reported that the expression of miR-449a is down-regulated by histone methylation through SUZ12 in human non-small cell lung cancer (NSCLC) [35]. Epigenetic inactivation of miR-449a/b expression by the oncogenic trimethylated histone H3 Lys27 (H3K27me3) can also be found in cancer cells, and this inactivation can be reversed by epigenetic drug treatment targeting histone modifications. This highlights the possibility of application of this newly characterized miRNA as an epigenetic biomarker and therapeutic target [29].
In order to develop novel subgroup-specific marker for refinement of risk-stratification to MB patients, we established the first Nanostring assay platform for MB molecular subtyping in China with the CodeSet as described in a previous study [5]. It is well known that RNA extracted from FFPE tissue is invariably degraded and fragmented, and therefore not suitable for most molecular profiling platforms [36]. The Nanostring assay is a powerful technology using unique digital color-coded barcodes that hybridize directly to specific nucleic acid targets. It is highly reproducible and is as sensitive as real-time PCR [37]. Northcott et al. applied the Nanostring assay to a series of FFPE cases and found tumors could be assigned to tumor subgroups with high confidence in 87.5 % of cases [5]. The accuracy of this assay was greater than 95 % as compared with Affymetrix expression array data from the same cohort of frozen tumor tissues [38, 39], suggesting that use of the Nanostring assay for MB classification based on FFPE material is comparable to the expression profiling data obtained with RNA from fresh-frozen tissue [5, 37]. In the present study, we used the same panel of gene signatures as described in the previous study [5] to evaluate paraffin-embedded samples from our MB patients of recent years, and this method allowed subgrouping of 97.8 % (44/45) of cases by non-hierarchical clustering analysis. The percentage of cases in subgroups is 9 % (4/44), 18.2 % (8/44), 36.4 % (16/44)and 36.4 % (16/44) for WNT, SHH, Group 3 and Group 4, respectively. This subgroup ratio is quite similar to reports from western groups [40]. In addition, we also firstly demonstrated the molecular subtype of three well established MB cell lines through Nanostring assay, showing Daoy cells belong to the SHH group, and both D283 and D341 cells subtyped as Group 3.
Based on the subtyping result through Nanostring assay, we further analyzed the specificity of miR-449a expression to MB subgroups. The result shows that although miR-449a was downregulated in most primary tumors in our cohort, indicating it is likely a tumor suppressor, its expression in the WNT group was significantly higher than other subgroups (p = 0.0026). This result was supported by Gokhale et al. who applied microRNA and protein-coding gene arrays in parallel on 31 MBs and 4 normal cerebellar tissues and found that miR-449 was overexpressed in WNT signaling associated MBs [41]. Although their result was not confirmed in their real time RT-PCR study, we performed q-RT-PCR detection assay for miR-449a in our cohort of 45 MBs and demonstrated the significance of miR-449a expression status in WNT group. A number of other miRNAs with potential tumor suppression roles were found to be overexpressed in the WNT group of MBs in Gokhale’s study, including miR-193a and miR-224. These miRNAs inhibit proliferation, increase radiation sensitivity and reduce invasion of MB cells [41], which may be related with relatively good prognosis in WNT group patients. It has been shown that miR-449 is up-regulated and targets WNT1-inducible signaling pathway protein 2 (WISP2) directly in primary pigmented nodular adrenocortical disease (PPNAD) [42]. Pharmacologic inhibition of (PKA)affects the Wnt signaling pathway, leading to up-regulation of miR-449 and suppression of WISP2, indicating that the Wnt signaling pathway can be affected by PKA via miR-449a regulation [42].
All four WNT tumors in present study were from female patients over 3 years old (100 %), while 16 of the remaining 40 (40 %) subgrouped patients in our data set were females. This larger proportion of female patients in the WNT group as compared with other groups was also found in Kool et al. and Gokhale et al.’s data sets [39, 40]. Prevalence of MBs resulting from deregulated WNT pathway activation in females over 3 years of age may explain the better survival of this subgroup as reported from the retrospective analysis of a large cohort of MB cases [43].
In summary, our results demonstrated for the first time the feasibility of using the Nanostring assay with the well known CodeSet for subgrouping of MBs in Chinese patients. In addition, MiR-449a, a candidate tumor suppressor regulated by hypermethylation, is a novel potential diagnostic marker for WNT group of MBs. Detailed functional studies on miR-449a differentially expressed in WNT signaling associated MBs and correlation of its expression with clinical outcome in a study of larger sample size would help support use of this miRNA as biomarker for risk stratification, and may have potential for use in personalized therapy of this neoplasm.
References
Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE et al (2006) Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol 7:813–820
Packer RJ, Gajjar A, Vezina G, Rorke-Adams L, Burger PC, Robertson PL et al (2006) Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol 24:4202–4208
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A et al (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol (Berl) 114:97–109
Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S et al (2011) Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 29:1408–1414
Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C et al (2012) Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol (Berl) 123:615–626
Beard RE, Abate-Daga D, Rosati SF, Zheng Z, Wunderlich JR, Rosenberg SA et al (2013) Gene expression profiling using nanostring digital RNA counting to identify potential target antigens for melanoma immunotherapy. Clin Cancer Res 19:4941–4950
Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6:376–385
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99:15524–15529
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866
Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM (2006) MicroRNA expression and function in cancer. Trends Mol Med 12:580–587
Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S et al (2013) Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res 3:465–477
Li H, Yu G, Shi R, Lang B, Chen X, Xia D et al (2014) Cisplatin-induced epigenetic activation of miR-34a sensitizes bladder cancer cells to chemotherapy. Mol Cancer 13:8
Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E et al (2014) miR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene 33:4307–4315
Zhao Z, Ma X, Sung D, Li M, Kosti A, Lin G et al (2015) microRNA-449a functions as a tumor suppressor in neuroblastoma through inducing cell differentiation and cell cycle arrest. RNA Biol 12:538–554
Genovesi LA, Carter KW, Gottardo NG, Giles KM, Dallas PB (2011) Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. PloS One 6:e23935
Venkataraman S, Birks DK, Balakrishnan I, Alimova I, Harris PS, Patel PR et al (2013) MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem 288:1918–1928
Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS et al (2009) MicroRNA profiling in human medulloblastoma. Int J Cancer 124:568–577
Miska EA (2005) How microRNAs control cell division, differentiation and death. Current Opin Genet Dev 15:563–568
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J et al (2012) Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 279:3159–3165
Pierson J, Hostager B, Fan R, Vibhakar R (2008) Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 90:1–7
Shalaby T, Fiaschetti G, Baumgartner M, Grotzer MA (2014) MicroRNA signatures as biomarkers and therapeutic target for CNS embryonal tumors: the pros and the cons. Int J Mol Sci 15:21554–21586
Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y et al (2009) The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res 69:3249–3255
Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ et al (2012) Pleiotropic effects of miR-183 ~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol (Berl) 123:539–552
Chang Q, Pang JC, Li KK, Poon WS, Zhou L, Ng HK (2005) Promoter hypermethylation profile of RASSF1A, FHIT, and sFRP1 in intracranial primitive neuroectodermal tumors. Hum Pathol 36:1265–1272
Pang JC, Chang Q, Chung YF, Teo JG, Poon WS, Zhou LF et al (2005) Epigenetic inactivation of DLC-1 in supratentorial primitive neuroectodermal tumor. Hum Pathol 36:36–43
Shao LW, Pan Y, Qi XL, Li YX, Ma XL, Yi WN et al (2016) ATRX loss in adult supratentorial diffuse astrocytomas correlates with p53 over expression and IDH1 mutation and predicts better outcome in p53 accumulated patients. Histol Histopathol 31:103–114
Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F et al (2015) Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep 5:11251
Yao Y, Ma J, Xue Y, Wang P, Li Z, Li Z et al (2015) MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol Oncol 9:640–656
Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M et al (2009) miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev 23:2388–2393
Chen J, Zhou J, Chen X, Yang B, Wang D, Yang P et al (2015) miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biol 36:8221–8229
Yuan JM, Shi XJ, Sun P, Liu JX, Wang W, Li M et al (2015) Downregulation of cell cycle-related proteins in ovarian cancer line and cell cycle arrest induced by microRNA. Int J Clin Exp Med 8:18476–18481
Chen SP, Liu BX, Xu J, Pei XF, Liao YJ, Yuan F et al (2015) MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer 15:706
Shi W, Bruce J, Lee M, Yue S, Rowe M, Pintilie M et al (2016) MiR-449a promotes breast cancer progression by targeting CRIP2. Oncotarget.doi:10.18632/oncotarget.7753
Chen S, Dai Y, Zhang X, Jin D, Li X, Zhang Y (2014) Increased miR-449a expression in colorectal carcinoma tissues is inversely correlated with serum carcinoembryonic antigen. Oncol Lett 7:568–572
You J, Zhang Y, Li Y, Fang N, Liu B, Zu L et al (2015) MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res 5:2730–2744
Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B et al (2011) mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol 11:46
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL et al (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325
Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H et al (2011) Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 29:1424–1430
Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P et al (2008) Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PloS One 3:e3088
Gupta T, Shirsat N, Jalali R (2015) Molecular subgrouping of medulloblastoma: impact upon research and clinical practice. Curr Pediatr Rev 11:106–119
Gokhale A, Kunder R, Goel A, Sarin R, Moiyadi A, Shenoy A et al (2010) Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J Cancer Res Ther 6:521–529
Iliopoulos D, Bimpaki EI, Nesterova M, Stratakis CA (2009) MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Res 69:3278–3282
Curran EK, Sainani KL, Le GM, Propp JM, Fisher PG (2009) Gender affects survival for medulloblastoma only in older children and adults: a study from the surveillance epidemiology and end results registry. Pediatr Blood Cancer 52:60–64
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81101900) and the Clinicopathologic Innovation Fund from Peking University Third Hospital, Peking University Health Science Center to Qing Chang, MD., Ph.D.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
Li, Y., Jiang, T., Shao, L. et al. Mir-449a, a potential diagnostic biomarker for WNT group of medulloblastoma. J Neurooncol 129, 423–431 (2016). https://doi.org/10.1007/s11060-016-2213-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-016-2213-y