Abstract
Brain-invasive growth of a subset of meningiomas is associated with less favorable prognosis. The molecular mechanisms causing invasiveness are only partially understood, however, the expression of matrix metalloproteinases (MMPs) has been identified as a contributing factor. We have previously found that loss of density enhanced phosphatase-1 (DEP-1, also designated PTPRJ), a transmembrane protein-tyrosine phosphatase, promotes meningioma cell motility and invasive growth in an orthotopic xenotransplantation model. We have now analyzed potential alterations of the expression of genes involved in motility control, caused by DEP-1 loss in meningioma cell lines. DEP-1 depleted cells exhibited increased expression of mRNA encoding MMP-9, and the growth factors EGF and FGF-2. The increase of MMP-9 expression in DEP-1 depleted cells was also readily detectable at the protein level by zymography. MMP-9 upregulation was sensitive to chemical inhibitors of growth factor signal transduction. Conversely, MMP-9 mRNA levels could be stimulated with growth factors (e.g. EGF) and inflammatory cytokines (e.g. TNFα). Increase of MMP-9 expression by DEP-1 depletion, or growth factor/cytokine stimulation qualitatively correlated with increased invasiveness in vitro scored as transmigration through matrigel-coated membranes. The studies suggest induction of MMP-9 expression promoted by DEP-1 deficiency, or potentially by growth factors and inflammatory cytokines, as a mechanism contributing to meningioma brain invasiveness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meningiomas, arising from arachnoidal cap cells, are the most frequently occurring intracranial tumors in adults. They are largely benign (>90 % WHO grade I), but a smaller fraction is classified as atypical or invasive (WHO grades II and III) [1]. Treatment of meningioma is surgical or through radiation. Thus far, no effective chemotherapy has been established [2]. Most meningiomas are sporadic, but inheritance of inactivating mutations in the NF2 gene (encoding the protein merlin) predispose for meningioma formation. Mutation of NF2 is also frequent in sporadic tumors, and genetic inactivation of Nf2 in mouse meningeal cells is sufficient for meningiomagenesis [1, 3, 4]. Among further genetic defects, the loss of CDK-inhibitor encoding loci CDKN2A/B in NF2-defective meningiomas has been associated with a more aggressive phenotype [5], which has been recapitulated in a mouse model [6]. Mutations in the genes encoding TRAF7, KLF4, AKT1, and SMO have recently been identified as genetic lesions in tumors with intact NF2 [7, 8].
Brain invasive growth of meningiomas, more common in WHO grade II/III tumors, is considered the major cause of recurrence. As of now, the molecular basis for brain invasion remains to be completely understood. Enhanced cell motility and alterations in cell–matrix interaction are likely contributors to an invasive phenotype. As important molecules in this process, the matrix metalloproteinases 2 and 9 (MMP-2 and 9) have been proposed. Their expression, notably that of MMP-9, has been shown to correlate with an invasive/recurrent meningioma phenotype [9–12]. The mechanisms leading to enhanced MMP-expression in invasive meningioma entities, are currently elusive.
We have previously identified the protein-tyrosine phosphatase (PTP) DEP-1/PTPRJ as a negative regulator of meningioma cell motility [13]. As we show in the present study, loss of DEP-1/PTPRJ caused a marked upregulation of MMP-9, and was associated with increased invasion in vitro. Moreover, inflammatory cytokines and growth factors enhanced MMP-9 expression and tended to also promote cell invasion. We propose that DEP-1 loss and cytokine/growth factor exposure may contribute to the invasiveness of meningioma through regulation of MMP expression.
Materials and methods
RT2 Profiler™ PCR Array analysis and qRT-PCR
To analyze the effect of DEP-1 and NF2 knockdown on migration-related genes, the RT2 Profiler™ PCR Array System (Qiagen, Hilden, Germany) was used. DEP-1-depleted, NF-2 depleted, DEP-1/NF2-depleted or control SF4068 cell lines were grown for 2 days until they reached confluence. RNA was extracted using RNeasy-Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. The optional on column-DNAse treatment was performed by incubating the column with 5 Units of DNase I, RNAse-free (Thermo Scientific, Schwerte, Germany) in 50 µl reaction buffer for 20 min at room temperature. Concentration of eluted RNA was measured photometrically, and 0.5 µg of isolated RNA was converted to cDNA using RT2 First Strand Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. 102 µl of cDNA synthesis reaction was mixed with 2× RT2 SYBR Green ROX qPCR Mastermix (Qiagen, Hilden, Germany) and H2O to a total volume of 2,700 µl. 25 µl of the reaction mixture were distributed into each well of RT2 Profiler™ PCR Array Human Cell Motility (PAHS-128Z; Qiagen, Hilden, Germany). The PCR was performed in an Eppendorf realplex4 Mastercycler under the following conditions: 1 min of initial denaturation at 95 °C followed by 40 cycles of [15 s at 95 °C and 1 min at 60 °C]. Melting curve analysis was subsequently performed. Data Analysis was performed using web-based RT2 Profiler PCR Array Data Analysis software version 3.5, provided by the array manufacturer, which calculates fold-changes based on the ΔΔCt method. The mean of five housekeeping genes served for normalization.
For normal qRT-PCR analysis RNA isolation was performed as described; including the column-DNAse treatment step. cDNA was synthesized using First Strand cDNA Synthesis Kit (Thermo Scientific, Schwerte, Germany) according to manufacturer’s protocol, with 1 µg RNA template and Oligo-dT-based priming. qRT-PCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (2×) (Thermo Scientific, Schwerte, Germany). The diluted master mixes containing 0.125 μM of each GAPDH primer or 1x QuantiTect primer assay mixture for MMP-9, MMP-2, EGF or FGF-2, respectively, were prepared according to the manufacturer’s protocol, with analysis performed in duplicates or triplicates using 1 μl of the cDNA preparation in a total reaction volume of 12 µl. The PCR was performed in an Eppendorf realplex4 Mastercycler under the following conditions: 10 min of initial denaturation at 95 °C followed by 45 cycles of [15 s at 95 °C, 30 s at 55 °C and 40 s at 72 °C]. Melting curve analysis was subsequently performed. Data analysis was performed with the aforementioned software, using GAPDH expression levels for normalization.
Invasion assays
BD Matrigel™ Basement Membrane Matrix, growth factor reduced (GFR) (BD Biosciences, Heidelberg, Germany; 356230) was thawed on ice and diluted with cold water to a concentration of 0.125 µg/ml. 12 well-migration inserts containing membranes with 8 µm-pores (Thin Certs; Greiner Bio-One, Frickenhausen, Germany) were coated with 135 µl diluted Matrigel by drying the solution overnight onto the membranes. Matrigel was rehydrated with 135 µl DMEM for 1 h at 37 °C before use.
Cells were trypsinized, resuspended with DMEM containing 10 % FCS, washed twice with DMEM containing 1 mg/ml BSA, counted and diluted to a concentration of 50,000 cells/ml DMEM + 1 mg/ml BSA. Bottom wells were filled with 1,400 µl of either DMEM + 1 mg/ml BSA alone, or with the addition of either 100 ng/ml TNFα, 10 ng/ml EGF, 10 ng/ml EGF + 100 ng/ml TNFα, or 10 % FCS (positive control). 600 µl of cell suspension corresponding to 30,000 cells were added to the inserts. After 24 h incubation, medium was removed, inserts were fixed in 5 % formaldehyde solution (Fischar, Saarbrücken, Germany) for 30 min at 37 °C, washed once with PBS and stained with 0.5 % crystal violet in 1× PBS/20 % methanol for 30 min at room temperature. Inserts were washed three times with water. Non-migrated cells on the upper side of the membrane were removed with a cotton swab. Migrated cells on the bottom side of the membrane were quantified by taking microscopic images of the whole membrane, then counting the cells on those images.
Statistics
Statistical analysis was performed using SigmaPlot 11.0 software. Data are shown as mean ± standard error of mean (SEM), and were analyzed for statistically significant differences by two-tailed t test, or one-way or two-way ANOVA with all pairwise multiple comparison procedure (Holm-Sidak method), as specified in the respective figure legends. Differences with p < 0.05 are indicated as significant.
Results
DEP-1 loss causes MMP-9 upregulation in meningioma cells
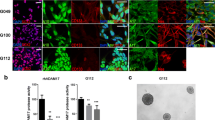
We have previously observed that loss of the PTP DEP-1/PTPRJ occurs in a fraction of human meningiomas, and its recapitulation by shRNA-mediated depletion in meningioma cell lines revealed an inhibitory role of DEP-1 for meningioma cell motility. Moreover, DEP-1 depleted cells exhibited an invasive potential in an orthotopic xenotransplantation model in mice [13]. The basis for enhanced invasion was not explored in the previous study. We considered the possibility that DEP-1/PTPRJ loss may cause alterations in the expression of genes, which are known to relate to invasive properties and motility of cells. We employed the meningioma cell line SF4068 [14] to screen for such genes. This cell line expresses DEP-1/PTPRJ and also has intact NF2. It was therefore of interest to explore also a possible combined effect of DEP-1 loss with NF2/merlin loss. A set of cell line pools with depletion of DEP-1, NF2, or both—and a control cell pool harboring non-targeting shRNA vectors only—was obtained by lentiviral transduction and appropriate selection (Fig. 1a). For RNA extraction, cell cultures with similar cell densities were used, since both DEP-1 and NF2 are reportedly regulated by cell density in several cell types. Expression profiling for 84 genes was done using the Human Cell Motility RT2 Profiler™ PCR Array (Qiagen). Interestingly, DEP-1 depletion led to the upregulation of only a few genes, most prominently MMP-9, and, to a lesser extent, MMP-2. Moreover, the genes encoding the growth factors EGF and FGF-2 were also increased in mRNA expression (Electronic Supplementary Material, Supplementary Fig. S1). These findings were confirmed by independent RT-qPCR assays (Fig. 1b). In contrast to DEP-1 depletion, NF2 knockdown had little effect on expression of the genes in the analyzed panel (Supplementary Fig. S1). To establish regulation of MMP-expression on protein level, immunoblotting and zymography with gelatine-containing SDS-PAGE gels was performed. MMP-9 protein levels in conditioned media were robustly increased by DEP-1 depletion, as seen in zymography (Fig. 1c). The available antibodies for MMP-9 were not sufficiently sensitive in immunoblotting (not shown). Consistent with the RNA data, MMP-2 levels detected by zymography and blotting were also increased, albeit only mildly (Fig. 1c–e). In addition to this, we detected increased MMP-9 (but not MMP-2) mRNA and protein expression in the cell line SF3061 (Fig. 2a, b) upon depletion of DEP-1.
DEP-1 knockdown enhances expression of MMP-9 and MMP-2 in SF4068 meningioma cells. a Generation of DEP-1- and/or NF2-depleted SF4068 cell lines. Stable knockdown of DEP-1 was achieved by lentiviral transduction of SF4068 cells with a DEP-1 shRNA or a non-targeting shRNA expression construct. In stably transduced cells, lentiviral transduction with a NF2 shRNA expressing or control construct was performed. For detection of DEP-1 by immunoblotting, equal protein amounts of the cell lysates were subjected to enrichment by wheat-germ agglutinin affinity precipitation. Vinculin and NF2 were analyzed in equal amounts of total cell lysate. b mRNA expression of MMP-9, MMP-2, EGF and FGF-2 in DEP-1-depleted and control SF4068 cell lines was measured by qRT-PCR. Expression levels were normalized to the expression of the housekeeping gene GAPDH. Expression levels in DEP-1 -depleted cells are plotted relative to levels in the control cells. Mean of 3 independent experiments ± SEM; *p < 0.05; **p < 0.01 by t test. c Detection of MMP-9 and MMP-2 in cell culture supernatants of DEP-1 -depleted or control SF4068 cell lines by zymography. One representative zymography out of 3 independent experiments is shown and mean ± SEM of quantification of MMP activities of all three experiments are also presented. *p < 0.05 by t test. d Detection of MMP-2 in cell culture supernatants. Serum-free culture supernatants of DEP-1 -depleted or control SF4068 cell lines collected after 24 h incubation were analyzed by immunoblotting. One representative experiment and quantification of 4 independent experiments are shown. Mean ± SEM; *p < 0.05 by t test. e Detection of MMP-2 in total cell lysates of DEP-1 -depleted or control SF4068 cell lines by immunoblotting. Mean of 4 independent experiments ± SEM; **p < 0.01 by t test
DEP-1 knockdown enhances expression of MMP-9, EGF and FGF-2 in SF3061 meningioma cells. a Expression of MMP-9, MMP-2, EGF and FGF-2 in SF3061 meningioma cells stably transduced with DEP-1 shRNA or control shRNA expression constructs was measured by qRT-PCR. Expression levels were normalized to the expression of the housekeeping gene GAPDH. Expression levels in DEP-1-depleted cells are plotted relative to levels in the control cells. Mean of 5 (for MMP-2, EGF, FGF-2) or 4 (for MMP-9) independent experiments ± SEM; *p < 0.05; **p < 0.01; ***p < 0.001 by t test. b Detection of MMP-9 in cell culture supernatants of DEP-1-depleted or control SF3061 cell lines by zymography. DEP-1-depleted and control SF3061 cells were starved for 16 h and subsequently treated with either 10 ng/ml EGF, 100 ng/ml TNFα or 10 ng/ml EGF + 100 ng/ml TNFα. Controls were left untreated. Supernatants were collected after 24 h, concentrated and analyzed by zymography
We assessed sections of mouse brain too—these sections were derived from xenotransplantation experiments of DEP-1 depleted MG-KT21 human meningioma cells [13]—for MMP-9 expression by IHC. Tumors harboring a non-targeting control shRNA did not reveal MMP-9 protein staining (Supplementary Fig. S2a). MMP-9 expression was, however, clearly detectable in tumors grown from DEP-1 depleted cells. It was confined to the cells in the invasion front, whereas the—partially necrotic—center of the invading tumor mass was negative (Supplementary Fig. S2b). This MMP-9 expression pattern was reminiscent of that in human invasive meningiomas (shown for comparison in Supplementary Fig. S2c).
These findings show that, depending on the cellular context, DEP-1 loss can upregulate MMP-9, and, depending on the cell context, MMP-2 in meningioma cells—which may relate to the invasive phenotype of DEP-1 deficient cells observed previously [13].
Growth factors and cytokines stimulate MMP-9 expression in meningioma cells
DEP-1 depletion in SF4068 and SF3061 cells also caused upregulation of EGF and FGF-2 mRNA expression (Figs. 1b, 2a). Production of EGF and FGF-2 by meningiomas has previously been reported [15, 16]. It appeared possible that these growth factors may play a role in MMP induction. Stimulation of MMP-9 expression by growth factors and cytokines has been described in fibroblasts, different brain tumor cells and epithelial cancer cells [17–19]. Activation of NFκB and ERK1/2 were implicated in the induction [17, 19]. Therefore, we tested the effect of EGF and FGF-2, as well as the inflammatory cytokines TNFα and IL1β, on MMP mRNA expression in SF4068 cells. As shown in Fig. 3a, all factors tended to increase the MMP-9 expression in control cells, while also further enhancing MMP-9 expression in DEP-1 depleted cells. Due to the large variation of the magnitude of responses in individual experiments, these effects were, however, only partially statistically significant when individually normalized experiments were analyzed (Supplementary Figure S3). These findings could qualitatively be recapitulated in zymography assays, although the combined enhancement effect of cytokines and growth factors was quite variable (Fig. 3b). A trend of cytokine-mediated stimulation of MMP-9 levels could also be detected by zymography in SF3061 cells (Fig. 2b). In contrast, little stimulation of MMP-2 protein levels could be detected by any of the growth factor- or cytokine-treatments in any of the tested cell lines (Supplementary Fig. S4 and data not shown). Small differences of signals and relatively high background unfortunately precluded a quantitative assessment of these experiments. Still, taken all findings together, growth factor and cytokine exposure tend to increase MMP-9 expression in meningioma cells, and may further enhance the effect of DEP-1 loss.
Growth factors, cytokines and DEP-1 knockdown enhance expression of MMP-9 in SF4068 meningioma cells. a DEP-1 -depleted (DEP-1/Ctrl) and control (Ctrl/Ctrl) SF4068 cells were starved for 16 h and subsequently treated with either 100 ng/ml TNFα, 100 ng/ml ΙL1β, 10 ng/ml EGF, 30 ng/ml FGF-2, 10 ng/ml EGF + 100 ng/ml TNFα, 10 ng/ml EGF + 100 ng/ml ΙL1β, 30 ng/ml FGF-2 + 100 ng/ml TNFα or 30 ng/ml FGF-2 + 100 ng/ml ΙL1β. Controls were left untreated. After 24 h incubation RNA was extracted and MMP-9 mRNA expression was measured by qRT-PCR. Expression levels were normalized to the expression of the housekeeping gene GAPDH. Expression levels in DEP-1 -depleted cells are plotted relative to levels in the control cells. Mean of three independent experiments ± SEM is shown. *p < 0.05; **p < 0.01 by two-way ANOVA. §§§ The difference in MMP-9 expression between the series of DEP-1 depleted cells versus control cells was highly significant (p < 0.001). (b) Zymographic detection of MMP-9 in supernatants of DEP-1 -depleted (D/C) and control (C/C) SF4068 cells. Cells were treated as described in (a), supernatants were collected after 24 h, concentrated and analyzed by zymography. One representative out of 3 independent experiments is shown
DEP-1 deficiency has been shown to stimulate growth factor signaling in meningioma and other cell types, including EGF and FGF signaling [13, 20, 21]. However, DEP-1 has not previously been implicated in NFκB signaling. Given that both signaling axes can contribute to MMP-9 induction, we intended to differentiate whether DEP-1 depletion would affect growth factor or cytokine signaling pathways, or both. Thus, we assessed the effects of well characterized signaling inhibitors on MMP-9 expression and its increase by DEP-1 depletion. As Fig. 4 illustrates, inhibitors of EGF receptor (Gefitinib), FGF receptor (PD173074), and the downstream mediators PI3 K (phosphoinositide 3-kinase; LY294002) and MEK (MAP/ERK-kinase; UO126) all tended to attenuate basal MMP-9 expression in SF4068 cells (although effects did not reach significance); whereas the IKK (inhibitor of NFκB-kinase) inhibitor BMS-345541, which blocks NFκB activation, apparently had less effect. MMP-9 induction by DEP-1 depletion was significantly affected by all inhibitors except Gefintinib. The strongest effect on MMP-9 induction by DEP-1 depletion was exerted by LY294002. Taken together, these experiments suggest that, primarily, growth factor/receptor-tyrosine kinase (RTK) signaling—notably PI3 K activation—contributes to both basal and DEP-1-loss mediated MMP-9 upregulation in this meningioma cell line. A (weaker) modulation of NFκB signaling may additionally contribute to MMP-9 regulation by DEP-1 deficiency.
MMP-9 expression in SF4068 meningioma cells is reduced by various signal transduction inhibitors. DEP-1 -depleted (DEP-1/Ctrl) and control (Ctrl/Ctrl) SF4068 cell lines were serum starved for 8 h, followed by a treatment with 0.1 % DMSO (solvent control), 2 µM BMS-34551, 2 µM Gefitinib, 25 µM LY294002, 1 µM PD173074 or 10 µM UO126 for 16 h. RNA was isolated and mRNA expression of MMP-9 was measured by qRT-PCR. Expression levels were normalized to the expression of the housekeeping gene GAPDH. Expression levels in DEP-1 depleted cells are plotted relative to levels in the control cells. Mean of 3 independent experiments ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001 by two-way ANOVA; #p < 0.05; ##p < 0.01; ###p < 0.001 for comparisons relative to DMSO-treated cells. §§§The difference in MMP-9 expression between the series of DEP-1 depleted cells versus control cells was highly significant (p < 0.001)
Effects of DEP-1 loss, growth factors and cytokines on meningioma cell invasion
It appeared reasonable that the increased MMP expression described above would translate into enhanced invasive properties of the meningioma cells. This was tested in invasion assays, using matrigel-coated membranes. We limited the analysis of growth factor and cytokine effects to EGF and the cytokine TNFα. As shown in Fig. 5a and b, DEP-1 loss led to a robust increase in cell migration through matrigel, consistent with the pronounced upregulation of MMPs under these conditions. EGF stimulated basal invasion and tended to further enhance invasion of DEP-1 depleted cells. TNFα was ineffective as a sole treatment. In some experiments, the combination of EGF and TNFα was more potent than the single factors. Overall, the invasive behavior of the meningioma cells largely correlated with the expression level of MMP-9 as seen through mRNA analyses and zymography; with the exception that exposure to TNFα alone was incapable of stimulating invasion.
DEP-1 knockdown and MMP-9-inducing factors enhance invasiveness of SF4068 meningioma cells. DEP-1 -depleted (DEP-1/Ctrl) and control (Ctrl/Ctrl) SF4068 cell lines were seeded in matrigel coated migration inserts in serum-free medium. Bottom chambers were filled with serum-free medium containing 10 ng/ml EGF, 100 ng/ml TNFα, 10 ng/ml EGF + 100 ng/ml TNFα or no stimulant. After 24 h incubation, cells were fixed and stained with crystal violet. Non-invasive cells on the upper side of the membrane were removed and invasive cells on the bottom side of the membrane were counted. For each condition, duplicates were measured. a Mean of three independent experiments ± SEM. *p < 0.05 by t test; #p < 0.05 by t test relative to unstimulated cells. §§The difference in invasion between the series of DEP-1 depleted cells versus control cells was significant as tested by two-way ANOVA (p < 0.015). b One representative microscopic image per condition of one representative experiment is shown
Discussion
MMP expression has been linked with the invasive behavior and recurrence of meningioma [11, 12]. This study expands knowledge of this phenomenon in two directions: 1. Loss of the PTP DEP-1, which occurs in a significant fraction of human meningioma, drives upregulation of MMP-9 (and to a lesser extent MMP-2). MMP-9 induction may underlie the invasive phenotype of DEP-1 depleted cells previously shown in an orthotopic xenotransplantation model [13]. 2. Inflammatory cytokines and the growth factors EGF and FGF-2 also tended to enhance MMP-9 expression in meningioma cells. Elevation of MMP-9 expression was correlated with promoting an invasive phenotype in vitro, and may indicate the importance of inflammatory conditions for meningioma invasion in vivo.
Of interest, depletion of NF2 in the meningioma cell lines did not increase expression of MMP-9 (Supplementary Fig. S1) or any other of the analyzed motility associated genes (not shown). This finding may correspond with the known lack of correlation of NF2 status and meningioma invasiveness [1].
An initial analysis of the signaling pathways involved in induction of MMP-9 expression by DEP-1 depletion with pharmacological inhibitors, revealed a role of typically growth factor-induced signaling. The best inhibitor of MMP-9 induction was LY294002, a PI3 K inhibitor. Also, interference with ERK/MAPK signaling by inhibition of the upstream kinase MEK, and direct inhibition of FGF receptor tyrosine kinases with PD173074, reduced MMP-9 induction by DEP-1 depletion. These observations are consistent with the possibility that DEP-1 depletion causes MMP-9 upregulation, primarily, via enhancement of RTK signaling. Indeed, DEP-1 has previously been shown to negatively regulate several RTKs [13, 20] and also specific downstream steps, including PI3 K activation [22] and ERK1/2 activation [23]. Assessment of the regulation of EGF and FGF receptor signaling by DEP-1 in meningioma cells will be of interest to better characterize the pathways for MMP-9 upregulation. Since expression of EGF and FGF-2 were also found upregulated in DEP-1 depleted cells, the possibility of an autocrine circuit in MMP-9 induction merits attention.
Several promoter elements, such as AP-1 binding sites, an AP-2 site, an Ets consensus binding site and an NFκB binding site, have been previously shown to be involved in MMP-9 transcriptional regulation [17, 24–28]. Interestingly, a synergistic upregulation of MMP-9 expression by growth factors and inflammatory cytokines has been observed in rabbit aortic smooth muscle cells [29] and dermal fibroblasts [17]. For this synergistic MMP-9 upregulation, transcription factor binding to the proximal AP-1 site and the NFκB site were involved; with, neither AP-1 nor NFκB activation alone being sufficient for synergistic regulation [17]. Our findings in meningioma cells are consistent with such a mechanism of MMP-9 expression control.
Elevated expression of MMP-9 by DEP-1 depletion, or cytokine and growth factor treatment qualitatively correlated with enhanced invasion through matrigel in vitro. TNFα stimulation alone was, however, insufficient to promote invasion in this assay, in contrast to EGF as single agent. It is conceivable that invasion may require both a motogenic stimulus and the promotion of MMP expression. TNFα tended to elevate MMP-9 induction, but may be ineffective in stimulating migration.
Few studies have addressed the possible role of inflammatory cytokines for meningioma recurrence and invasion. For example, early studies showed responses of meningioma cells in vitro to IL1 stimulation in terms of IL6 production [30]; also, the presence of CD70/CD27 ligand, a relative of TNFα, has been demonstrated in a meningioma cell line [31]. Recently, the cytokine MIF—a potent activator of NFκB—and upregulation of MMP-9 have been linked with meningioma recurrence [32]. Clearly, further investigation of this issue is highly warranted.
References
Mawrin C, Perry A (2010) Pathological classification and molecular genetics of meningiomas. J Neurooncol 99:379–391
Wilisch-Neumann A, Pachow D, Wallesch M, Petermann A, Böhmer FD, Kirches E, Mawrin C (2014) Re-evaluation of cytostatic therapies for meningiomas in vitro. J Cancer Res Clin Oncol 140:1343–1352
Kalamarides M, Niwa-Kawakita M, Leblois H, Abramowski V, Perricaudet M, Janin A, Thomas G, Gutmann DH, Giovannini M (2002) Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev 16:1060–1065
Kalamarides M, Stemmer-Rachamimov AO, Niwa-Kawakita M, Chareyre F, Taranchon E, Han ZY, Martinelli C, Lusis EA, Hegedus B, Gutmann DH, Giovannini M (2011) Identification of a progenitor cell of origin capable of generating diverse meningioma histological subtypes. Oncogene 30:2333–2344
Goutagny S, Yang HW, Zucman-Rossi J, Chan J, Dreyfuss JM, Park PJ, Black PM, Giovannini M, Carroll RS, Kalamarides M (2010) Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res 16:4155–4164
Peyre M, Stemmer-Rachamimov A, Clermont-Taranchon E, Quentin S, El-Taraya N, Walczak C, Volk A, Niwa-Kawakita M, Karboul N, Giovannini M, Kalamarides M (2013) Meningioma progression in mice triggered by Nf2 and Cdkn2ab inactivation. Oncogene 32:4264–4272
Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, Raza A, Sunkavalli A, Macconaill LE, Stemmer-Rachamimov AO, Louis DN, Hahn WC, Dunn IF, Beroukhim R (2013) Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 45:285–289
Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, Avsar T, Li J, Murray PB, Henegariu O, Yilmaz S, Gunel JM, Carrion-Grant G, Yilmaz B, Grady C, Tanrikulu B, Bakircioglu M, Kaymakcalan H, Caglayan AO, Sencar L, Ceyhun E, Atik AF, Bayri Y, Bai H, Kolb LE, Hebert RM, Omay SB, Mishra-Gorur K, Choi M, Overton JD, Holland EC, Mane S, State MW, Bilguvar K, Baehring JM, Gutin PH, Piepmeier JM, Vortmeyer A, Brennan CW, Pamir MN, Kilic T, Lifton RP, Noonan JP, Yasuno K, Gunel M (2013) Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339:1077–1080
Nordqvist AC, Smurawa H, Mathiesen T (2001) Expression of matrix metalloproteinases 2 and 9 in meningiomas associated with different degrees of brain invasiveness and edema. J Neurosurg 95:839–844
Okada M, Miyake K, Matsumoto Y, Kawai N, Kunishio K, Nagao S (2004) Matrix metalloproteinase-2 and matrix metalloproteinase-9 expressions correlate with the recurrence of intracranial meningiomas. J Neurooncol 66:29–37
Okuducu AF, Zils U, Michaelis SA, Mawrin C, von Deimling A (2006) Increased expression of avian erythroblastosis virus E26 oncogene homolog 1 in World Health Organization grade 1 meningiomas is associated with an elevated risk of recurrence and is correlated with the expression of its target genes matrix metalloproteinase-2 and MMP-9. Cancer 107:1365–1372
Barresi V, Vitarelli E, Tuccari G, Barresi G (2011) MMP-9 expression in meningiomas: a prognostic marker for recurrence risk? J Neurooncol 102:189–196
Petermann A, Haase D, Wetzel A, Balavenkatraman KK, Tenev T, Guhrs KH, Friedrich S, Nakamura M, Mawrin C, Böhmer FD (2011) Loss of the protein-tyrosine phosphatase DEP-1/PTPRJ drives meningioma cell motility. Brain Pathol 21:405–418
Cuevas IC, Slocum AL, Jun P, Costello JF, Bollen AW, Riggins GJ, McDermott MW, Lal A (2005) Meningioma transcript profiles reveal deregulated Notch signaling pathway. Cancer Res 65:5070–5075
Carroll RS, Black PM, Zhang J, Kirsch M, Percec I, Lau N, Guha A (1997) Expression and activation of epidermal growth factor receptors in meningiomas. J Neurosurg 87:315–323
Takahashi JA, Mori H, Fukumoto M, Igarashi K, Jaye M, Oda Y, Kikuchi H, Hatanaka M (1990) Gene expression of fibroblast growth factors in human gliomas and meningiomas: demonstration of cellular source of basic fibroblast growth factor mRNA and peptide in tumor tissues. Proc Natl Acad Sci U S A 87:5710–5714
Bond M, Fabunmi RP, Baker AH, Newby AC (1998) Synergistic upregulation of metalloproteinase-9 by growth factors and inflammatory cytokines: an absolute requirement for transcription factor NF-kappa B. FEBS Lett 435:29–34
Rooprai HK, Rucklidge GJ, Panou C, Pilkington GJ (2000) The effects of exogenous growth factors on matrix metalloproteinase secretion by human brain tumour cells. Br J Cancer 82:52–55
Kim S, Choi JH, Lim HI, Lee SK, Kim WW, Cho S, Kim JS, Kim JH, Choe JH, Nam SJ, Lee JE, Yang JH (2009) EGF-induced MMP-9 expression is mediated by the JAK3/ERK pathway, but not by the JAK3/STAT-3 pathway in a SKBR3 breast cancer cell line. Cell Signal 21:892–898
Tarcic G, Boguslavsky SK, Wakim J, Kiuchi T, Liu A, Reinitz F, Nathanson D, Takahashi T, Mischel PS, Ng T, Yarden Y (2009) An unbiased screen identifies DEP-1 tumor suppressor as a phosphatase controlling EGFR endocytosis. Curr Biol 19:1788–1798
Giampietro C, Taddei A, Corada M, Sarra-Ferraris GM, Alcalay M, Cavallaro U, Orsenigo F, Lampugnani MG, Dejana E (2012) Overlapping and divergent signaling pathways of N-cadherin and VE-cadherin in endothelial cells. Blood 119:2159–2170
Tsuboi N, Utsunomiya T, Roberts RL, Ito H, Takahashi K, Noda M, Takahashi T (2008) The tyrosine phosphatase CD148 interacts with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem J 413:193–200
Sacco F, Tinti M, Palma A, Ferrari E, Nardozza AP, Hooft van Huijsduijnen R, Takahashi T, Castagnoli L, Cesareni G (2009) Tumor suppressor density-enhanced phosphatase-1 (DEP-1) inhibits the RAS pathway by direct dephosphorylation of ERK1/2 kinases. J Biol Chem 284:22048–22058
Sato H, Kita M, Seiki M (1993) v-Src activates the expression of 92-kDa type IV collagenase gene through the AP-1 site and the GT box homologous to retinoblastoma control elements. A mechanism regulating gene expression independent of that by inflammatory cytokines. J Biol Chem 268:23460–23468
Sato H, Seiki M (1993) Regulatory mechanism of 92 kDa type IV collagenase gene expression which is associated with invasiveness of tumor cells. Oncogene 8:395–405
Bond M, Chase AJ, Baker AH, Newby AC (2001) Inhibition of transcription factor NF-kappaB reduces matrix metalloproteinase-1, -3 and -9 production by vascular smooth muscle cells. Cardiovasc Res 50:556–565
Himelstein BP, Lee EJ, Sato H, Seiki M, Muschel RJ (1997) Transcriptional activation of the matrix metalloproteinase-9 gene in an H-ras and v-myc transformed rat embryo cell line. Oncogene 14:1995–1998
Himelstein BP, Lee EJ, Sato H, Seiki M, Muschel RJ (1998) Tumor cell contact mediated transcriptional activation of the fibroblast matrix metalloproteinase-9 gene: involvement of multiple transcription factors including Ets and an alternating purine-pyrimidine repeat. Clin Exp Metastasis 16:169–177
Fabunmi RP, Baker AH, Murray EJ, Booth RF, Newby AC (1996) Divergent regulation by growth factors and cytokines of 95 kDa and 72 kDa gelatinases and tissue inhibitors or metalloproteinases-1, -2, and -3 in rabbit aortic smooth muscle cells. Biochem J 315(Pt 1):335–342
Boyle-Walsh E, Hashim IA, Speirs V, Fraser WD, White MC (1994) Interleukin-6 (IL-6) production and cell growth of cultured human ameningiomas:-interactions with interleukin-1 beta (IL-1 beta) and interleukin-4 (IL-4) in vitro. Neurosci Lett 170:129–132
Held-Feindt J, Mentlein R (2002) CD70/CD27 ligand, a member of the TNF family, is expressed in human brain tumors. Int J Cancer 98:352–356
Huang Q, Zhao SL, Tian XY, Li B, Li Z (2013) Increased co-expression of macrophage migration inhibitory factor and matrix metalloproteinase 9 is associated with tumor recurrence of meningioma. Int J Med Sci 10:276–285
Acknowledgments
We are grateful to Anita Lal for kind provision of meningioma cell lines. This work was supported by funding of the Deutsche Forschungsgemeinschaft (SFB604 A1 to F.D.B and C.M., BO 1043/9-1, and MA2530/6-1) and the Wilhelm Sander Stiftung (to C.M. 2014.092.1.).
Conflict of interest
The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Petermann, A., Stampnik, Y., Cui, Y. et al. Deficiency of the protein-tyrosine phosphatase DEP-1/PTPRJ promotes matrix metalloproteinase-9 expression in meningioma cells. J Neurooncol 122, 451–459 (2015). https://doi.org/10.1007/s11060-015-1740-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-015-1740-2