Abstract
Paracoccidioidomycosis (PCM) is a systemic mycosis caused by a group of cryptic species embedded in the Paracoccidioides brasiliensis complex and Paracoccidioides lutzii. Four species were recently inferred to belong to the P. brasiliensis complex, but the high genetic diversity found in both human and environmental samples have suggested that the number of lineages may be higher. This study aimed to assess the 43-kilodalton glycoprotein genotypes (PbGP43) in paraffin-embedded samples from PCM patients to infer the phylogenetic lineages of the P. brasiliensis complex responsible for causing the infection. Formalin-fixed, paraffin-embedded (FFPE) tissue samples from patients with histopathological diagnosis of PCM were analyzed. DNAs were extracted and amplified for a region of the second exon of the PbGP43 gene. Products were sequenced and aligned with other PbGP43 sequences available. A haplotype network and the phylogenetic relationships among sequences were inferred. Amino acid substitutions were investigated regarding the potential to modify physicochemical properties in the proteins. Six phylogenetic lineages were identified as belonging to the P. brasiliensis complex. Two lineages did not group with any of the four recognized species of the complex, and, interestingly, one of them comprised only FFPE samples. A coinfection involving two lineages was found. Five parsimony-informative sites were identified and three of them showed radical non-synonymous substitutions with the potential to promote changes in the protein. This study expands the knowledge regarding the genetic diversity existing in the P. brasiliensis complex and shows the potential of FFPE samples in species identification and in detecting coinfections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paracoccidioidomycosis (PCM) is a severe deep systemic mycosis, sometimes fatal, difficult to diagnose in its early stages, and can remain latent for up to 40 years. The illness sometimes manifests as a self-limited pulmonary infection, which may remain as a latent or lifelong infection; on the other hand, upper respiratory tract paracoccidioimycosis is a slowly progressive disease which do not heal spontaneously. but PCM can also result in a disseminated disease affecting lymph nodes, liver, spleen, gut, bones, joints, and meninges, and that may be refractory to treatment [1]. PCM is endemic in Central and South America areas and about 80% of the cases have been reported in Brazil [2, 3]. However, the fact that PCM takes years to develop and its notification is not mandatory by the Ministry of Health makes it challenging to track and determine the exact number of cases in Brazil. Studies reported that approximately 10 million people could be infected and that 2% in endemic areas could develop the disease [4, 5]. Among the systemic mycoses, PCM presents the highest mortality rate, reaching the ratio of 1.65 deaths per million inhabitants [6]. A recent study that analyzed available data on fungal infections, obtained from autopsy reports in Brazil between 1930 and 2015, corroborated that PCM was the most frequent infection in this period [7].
Paracoccidioides species are the etiological agents of PCM, comprising the Paracoccidioides brasiliensis complex and Paracoccidioides lutzii [8,9,10,11]. The infection occurs by inhalation of conidia from the environment. For the disease development, it is essential that conidia (infective form) develop as yeast cells (pathogenic form) in the pulmonary alveoli, an event that is dependent on the temperature increase once inside the host [12]. There is no evidence of human-to-human transmission [13]. Paracoccidioides brasiliensis has been rarely isolated from soil, but isolates have been successfully obtained from armadillos (Dasypus novemcinctus), which are considered important natural reservoirs [14, 15]. Some manifestations of the disease were also observed in dogs [16, 17]. Since the incidence in endemic areas is mainly associated with deforestation, armadillo hunting, and agriculture practices, PCM becomes an increasingly important public health subject insofar as deforestation to agricultural expansion affects more people [3, 18].
The P. brasiliensis complex comprises four currently recognized phylogenetic species based on multilocus analyses that included microsatellites, mitochondrial and nuclear loci [19]. More recent studies using whole genome typing also support the presence of those four species that are reciprocally monophyletic. Paracoccidioides brasiliensis sensu stricto (former S1 phylogenetic group, and recently divided into two cryptic populations, S1a and S1b) is spread in Brazil, Argentina, Bolivia, Paraguay, Peru and Venezuela; P. americana (former PS2) is found in Venezuela and Brazil; P. restrepiensis (former PS3) is found mainly in Colombia, being recently also identified in Argentina, Brazil, Venezuela, Peru; and P. venezuelensis (former PS4), described as occurring exclusively in Venezuela, but recently detected in Brazil [8, 19,20,21,22,23,24,25]. Nevertheless, the high genetic diversity that has been found in both human and environmental samples suggests that the number of lineages in the complex may be higher.
The main antigenic/diagnostic component described in P. brasiliensis is a glycoprotein of 43 kDa (gp43) [26]. This molecule is secreted into the exocellular medium and contains a single oligosaccharide chain. The gp43 gene was the first to be fully characterized in P. brasiliensis, being named PbGP43. The open reading frame is within a DNA fragment of 1,329 base pairs, with two exons interrupted by an intron of 78 nucleotides [27]. Early studies have revealed a high level of polymorphism in the PbGP43 sequence among P. brasiliensis isolates, most of them located at the second exon [20, 28]. It was the most informative gene compared with other nuclear genes studied in P. brasiliensis, showing the highest occurrence of non-synonymous substitutions and evidence of positive selection [8, 20, 29]. Besides its diagnostic relevance, PbGP43 is also a vaccine candidate against P. brasiliensis [30], specifically its P10 peptide holding a protective T-cell epitope, which can induce protective immune responses in vitro and in vivo, promising therapeutic alternatives to combat PCM [31].
One of the main obstacles to the reliable diagnosis of PCM, especially in its early symptoms, is that it can be misinterpreted with other mycoses. Clinical profile and characteristic features can be easily detected in the late stage based on serology, histopathology, and culture. According to Guarner and Brandt [32], histopathology is still considered a faster and inexpensive way of diagnosing fungal infections. However, biopsies are the only material available in some cases, so it is highly relevant to improve the molecular assays using this type of material. Specifically, polymerase chain reaction (PCR)-based strategies are additive to the histopathology findings because they can define the etiological agent of fungal infection at species or genus level [32]. Although the PCR is sensitive and specific for identification of P. brasiliensis in various types of samples, as far as we know, few research groups have carried out analyzes using formalin-fixed, paraffin-embedded (FFPE) tissue in human biopsies [17, 33,34,35,36,37], mainly because of the limitations of amplifying DNA from this type of sample. It is stated that factors such as the size of the fragment to be amplified, the fixation technique, the time of the material storage, and the use of different DNA extraction kits and protocols may affect the efficacy of the method when applied in paraffin-embedded biopsies [38].
Molecular studies considering the P. brasiliensis complex are extremely important for understanding the extent of genetic diversity, their geographic distribution, and different clinical manifestations. In this context, this study aimed to assess genotypes of the PbGP43 gene in FFPE tissue samples from PCM patients to infer the phylogenetic lineages embedded in the P. brasiliensis complex responsible for causing the infection.
Material and Methods
Samples
We analyzed 31 tissue samples stored as FFPE blocks from 20 patients with histopathological diagnosis of PCM. The period of sample collection was from 1998 to 2014. Tissue specimens were fixed in 10% buffered formalin and embedded in paraffin blocks. DNA extraction and purification procedures were performed using the QIAamp™ DNA Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions.
To assess the quality of the extracted DNA, a PCR was performed with primers for the human β-globulin housekeeping gene (β1: 5′-CAA CTT CAT CCA CGT TCA CC-3′; β2: 5′-GAA GAG CCA AGG ACA GGT AC-3′) [39]. The samples were subjected to PCR in a final volume of 25 µl, containing 40 ng of the genomic DNA, 2.0 μM of each primer, and 12.5 µl of PCR Master Mix 2X (Promega, Madison, USA), with the remaining volume made up of sterile distilled water. PCR amplifications were performed in a thermal cycler GeneAmp (PCR System 9700), in the following conditions: an initial 5-min denaturation step at 94 °C coupled to a repeating cycle at 94 °C (1 min), 55 °C (1 min), and 72 °C (1 min) for 40 cycles, followed by a 7-min completion step at 72 °C. PCR products were visualized with ethidium bromide using 1.5% agarose gel electrophoresis.
PbGP43 Amplification and DNA Sequencing
Each DNA sample was used as a template for PCR amplification of the PbGP43 gene, using a nested PCR method with the two primer pairs [40] listed in Table 1. The samples were subjected to a first PCR round in a final volume of 25 µl, containing 40 ng of the genomic DNA, 2.0 μM of each primer (forward primer Para I and reverse primer Para II), and 12.5 µl of PCR Master Mix 2X (Promega, Madison, USA), with the remaining volume made up of sterile distilled water. The PCR protocol was carried out with an initial 5-min denaturation step at 94 °C coupled to a repeating cycle at 94 °C (30 s), 55 °C (30 s), and 72 °C (30 s) for 35 cycles, followed by a 5-min completion step at 72 °C.
The products of the first PCR were used as templates in a second round of PCR with the inner primer set comprising the forward primer Para III and the reverse primer Para IV (Table 1). The reaction mixture (25 μl) consisted of 1 µl of the amplification products of the first reaction, 2.0 μM of each primer, and 12.5 µl of PCR Master Mix 2X (Promega, Madison, USA), with the remaining volume made up of sterile distilled water. The nested PCR conditions were 94 °C for 5 min, followed by 40 cycles at 94 °C (30 s) and 72 °C (1 min), and one final cycle of 72 °C (5 min). PCR amplifications were performed in a thermal cycler GeneAmp (PCR System 9700). In both reactions, DNA from Histoplasma capsulatum (closely related to Paracoccidioides spp.) and human (no PCM carrier) were used as negative controls. PCR products were visualized with ethidium bromide using 1.5% agarose gel electrophoresis.
PCR products of the second PCR were purified from the agarose gel using the Pure Link® Quick Gel Extraction Kit (Invitrogen®, Carlsbad, USA). DNA sequencing was performed using the DYEnamic™ ET Dye Terminator Kit (GE Healthcare, Piscataway, USA), with Thermo Sequenase® II DNA polymerase (ThermoFisher Scientific, USA), in the MegaBACE 1000 capillary sequencer. Sequences on both strands were analyzed using the software Bioedit [41], and the sequencing quality was accessed by the Chromas program (http://technelysium.com.au/wp/chromas/).
Haplotype Network, Phylogenetic Inferences, and Protein Structure Analyses
All sequences obtained in the present study were aligned to other PbGP43 sequences available in the GenBank database (Supplementary Material 1) with MAFFT v.7 [42], using default settings. Regarding the sequences coming from the database, we considered for analysis only the gene region amplified in this study. We used DnaSP v.6 [43] to create a genotype network file (Roehl data file). The haplotype network was generated by the median-joining method [44] with Network v.10.2 (http://www.fluxus-engineering.com/), using the criterion connection cost, epsilon 0, and the MJ square option active.
We inferred the phylogenetic relationships among the sequences from the same alignment used to generate the haplotype network. With the software MEGA-X [45], we estimated the best-fitting nucleotide model of substitution and used this information to construct a phylogenetic tree by the Maximum Likelihood (ML) method. The robustness of branches was assessed by bootstrap analysis of 1.000 replicates [46].
We also investigated, using the TreeSAAP v.3.2 software package [47], whether amino acid replacements found in the haplotypes can alter physicochemical properties in the respective proteins. For that, we randomly selected one sequence from each haplotype (Supplementary Material 2) and considered the significant positive z-scores in categories six to eight (p < 0.05), which are the most extreme categories of structural or functional changes.
Results
Using nested PCR, we were able to successfully amplify a 196-bp fragment of PbGP43 exon 2 from 23 FFPE samples out of 31 obtained from 20 PCM patients (74.2% of the total) (Table 2). All samples were also used as template for amplification of the human β-globulin housekeeping gene (data not shown), yielding the expected amplicon of 268 bp in 25 of the samples, including all 23 yielding PbGP43 amplification. Two samples were amplified only for the β-globulin gene, and six samples did not yield any product. We were successful in amplifying fungal DNA from different FFPE tissue samples: skin (n = 8), liver (n = 1), oral mucosa (n = 5), tonsil (n = 1), salivary gland (n = 1), subcutaneous cyst (n = 1), skullcap (n = 1), lymph node (n = 1), oropharynx (n = 1), lung (n = 1), and floor of the mouth (n = 1). Another point worth mentioning is the gender ratio: among the 23 samples amplified for the PbGP43 gene, 21 samples were from male patients and two from female patients, corresponding to a male:female ratio of 10.5:1.
We sequenced all 196-bp fragment from the PbGP43 exon 2 resulting from the nested PCR. To maintain the translation frame of the fragment, we excluded the first nucleotide from the sequences before the analysis. The 23 FFPE sequences were aligned to other 61 PbGP43 sequences (Supplementary Material 1), including representatives of the four species belonging to the P. brasiliensis complex [19]. The excluded nucleotide consisted of a guanine in all sequences analyzed, including sequences from the database, and therefore, its exclusion did not interfere with the results obtained. The nucleotide alignment showed 163 invariable sites, 27 singleton variable sites (containing two variants), and five variable parsimony-informative sites (83.59%, 13.85%, and 2.56% of the sites analyzed, respectively). Nucleotide diversity was estimated as 0.01287. The variable parsimony-informative sites were located at positions 1, 124, 136, 172 and 194, of the nucleotide alignment (Table 3).
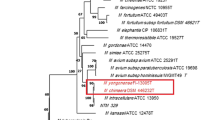
We identified seven haplotypes in our dataset (Supplementary Material 3), six of them belonging to the P. brasiliensis complex (Fig. 1A). Haplotype 1 (H1), Haplotype 2 (H2), Haplotype 3 (H3), and Haplotype 6 (H6) include representatives of the P. brasiliensis sensu stricto (S1), P. restrepiensis (PS3), P. venezuelensis (PS4), and P. americana (PS2), respectively. Haplotype 4 (H4) comprises only FFPE samples, and Haplotype 5 (H5) comprises three database sequences annotated as P. brasiliensis, but that did not group with any species of the complex. Haplotype 7 (H7) comprises the outgroup P. lutzii. Besides, our results indicated another haplotype belonging to the P. brasiliensis complex represented by the median vector (mv) in the haplotype network. Haplotype diversity was estimated as 0.7008.
Genetic diversity of the gene coding for the 43-kilodalton glycoprotein from Paracoccidioides brasiliensis (PbGP43) among lineages of the P. brasiliensis complex. A Median-joining haplotype network. The size of circumferences is proportional to the haplotype frequency in the dataset; mutational steps are represented by black lines; mv = median vector. B Maximum-likelihood phylogenetic tree. Numbers above the branches indicate the support of each branch. Scale bar represents substitutions per site
The ML phylogenetic tree (Fig. 1B) was built from the same nucleotide alignment used to infer the haplotype network and considering the Kimura 2-parameter model as the best-fitting model. The tree was rooted with P. lutzii. Our results indicated P. americana as the most basal species of the complex, followed by the H5 sequences and a polytomy involving P. brasiliensis sensu stricto, P. restrepiensis, P. venezuelensis, and H4. Phylogenetic inferences results were consistent with results of the haplotype network.
According to our results, 14 of the 23 FFPE samples studied were clustered in H4 (named FFPE clade). The other nine samples were clustered in H6 (P. americana clade). From the 14 samples belonging to the FFPE clade, 13 were collected in the São Paulo State and one in Minas Gerais State (Table 2). On the other hand, the P. americana clade was composed of samples collected not predominantly in the São Paulo State (five samples) but also collected in the states of Rio de Janeiro (one sample), and Rio Grande do Sul (three samples).
We identified a coinfection in Patient 3, the patient for which more than one sample could be amplified for the PbGP43 gene (Table 2). Among the four sequences obtained from this patient, three (GR5, GR6, and GR7 from skin, liver, and salivary gland, respectively) were clustered in the FFPE clade. The other sample (GR56, also collected from the skin but in a different area) was clustered in the P. americana clade.
The five variable parsimony-informative sites identified among the haplotypes consisted of non-synonymous changes (Table 3). One of them, the site at the amino acid position 1, is part of the P10 T-cell epitope. This was the only amino acid change detected in the P10 peptide sequence among the P. brasiliensis lineages, being the inner core of the P10 peptide identical among them [31]. We analyzed whether these non-synonymous changes promote structural and functional changes in the proteins. We observed that three sites showed radical non-synonymous changes. Substitution at the amino acid position 42, identified in the haplotypes H1, H2, H3, and H4, is associated with a change in isoelectric point (z-score = 8; p < 0.001). Substitution at the amino acid position 58, found in H1, is associated with the property alpha-helical tendencies (z-score = 6; p < 0.05). Substitution at the amino acid position 65, found in H3, is associated to three properties: alpha-helical tendencies (z-score = 6; p < 0.05), power to be at the N-terminal (z-score = 8; p < 0.05), and turn tendencies (z-score = 7; p < 0.05).
Discussion
In the present study, we identified phylogenetic lineages of the P. brasiliensis complex from the sequencing of the nested PCR products of FFPE samples. The successful use of this type of material for PCM diagnosis has been demonstrated in several clinical investigations to detect the fungus DNA as a marker of infection, highlighting the value of these samples for molecular diagnosis and genotyping [17, 33,34,35,36,37]. These 23 sequenced samples were collected in four Brazilian states located in the Southeast and South regions, which are recognized as the mainly endemic areas of PCM in the country [3]. The majority of the samples (91.3%) were obtained from male patients. It is in agreement with the previously described in a study that analyzed 584 PCM patients (575 of them from the Southeast region of Brazil), in which 84.2% of the samples were from male patients [48]. A predominance of male patients is characteristic of PCM, irrespective of phylogenetic species [49, 50], and the reasons include the fact that circulating estrogens in women can inhibit the transformation of the inhaled conidia into yeast cells, and the greater proportion of men involved in agricultural activities in endemic areas [3, 51].
The alignment of our sequences with other PbGP43 sequences available in the GenBank database showed the presence of five parsimony-informative sites in the gene region analyzed. We analyzed a fragment of the second exon of PbGP43 because it is the most polymorphic region among P. brasiliensis genes already studied [20, 28, 52]. A highly polymorphic gene region (more informative) is critical in FFPE samples, since the extracted DNA is more prone to be fragmented, challenging the amplification of long PCR products [53]. The limitations in getting DNA with good quality from FFPE tissue biopsies are formalin fixation and paraffin embedding procedures, resulting in DNA damage. The difficulty of amplifying DNA from FFPE samples is well discussed and explored in the literature [32, 54, 55], but despite this difficulty, only six samples did not show any amplification products, which indicates that for most of the samples, we obtained a good quality DNA from the FFPE blocks.
Sequences were assigned to seven haplotypes, six of them belonging to the P. brasiliensis complex. The other haplotype comprises P. lutzii, which was first considered as P. brasiliensis, but later proposed as a new species in the genus Paracoccidioides [8]. Our results also indicated the existence of an unsampled or extinct haplotype in the P. brasiliensis complex, as represented by the median vector in the haplotype network. We found a relatively high value of haplotype diversity and a low value of nucleotide diversity, a pattern commonly observed in recently diverged species. Indeed, that is the case for the P. brasiliensis complex, since it has been demonstrated that the divergence times within P. brasiliensis lineages range between 23,000 and 575,000 years ago [21].
Each recognized species of the P. brasiliensis complex was assigned to a distinct haplotype (H1, H2, H3, and H6), but sequences of two haplotypes (H4 and H5) could not be assigned to any species of the complex. The ML phylogenetic tree supported the haplotype network results and indicated that P. americana was the most basal species of the complex, in agreement with previous studies [19, 21, 56]. The expected phylogeny for the other taxa in the complex is P. restrepiensis and P. venezuelensis as sister species, with P. brasiliensis sensu stricto as the sister group of these dyad [19]. However, we found H5 located between the P. americana clade and a polytomy involving the other haplotypes. The isolates that comprise H5 were obtained from human patients in Costa Rica, and as far as we know, only Takayama et al. [57] reported a phylogeny considering these isolates. They analyzed a different region of the PbGP43 gene and also obtained an exclusive clade for these isolates, which prevented the assignment of a phylogenetic species. We, therefore, considered these isolates as an indeterminate species of the P. brasiliensis complex.
Due to the limitations in getting DNA with good quality from FFPE tissue biopsies, we could not obtain long PCR fragments. Even considering the most polymorphic PbGP43 gene region (the exon 2), only five nucleotide substitutions were observed among the P. brasiliensis lineages in the gene region analyzed. Consequently, the sequence data do not provide sufficient information to resolve the relationships among some lineages in the phylogenetic tree, which caused the observed polytomy and relatively low bootstrap values. In these cases, haplotype networks have the potential to be graphically more informative, although we also have observed a loop involving H2, H4 and H5. Despite that, the substitutions found in the sequenced gene region were sufficient to identify the P. brasiliensis lineages previously described, and even enabled us to identify a new lineage.
FFPE samples were collected in an area of sympatry between P. brasiliensis sensu stricto and P. americana. The former is usually reported as the most frequent species, associated with most PCM cases, while the latter is found at a considerably lower frequency [10, 20, 56, 58, 59]. Our results showed that nine out of 23 FFPE samples (39.1%) were assigned to P. americana, a higher proportion than expected. In comparison, a recent study analyzing samples collected in the southwest region of Brazil reported six out of 39 (15.4%) samples assigned to P. americana [59]. The other 14 FFPE sequences were grouped in H4 and could not be assigned to any species of the complex, as observed for H5. Unexpectedly, no FFPE samples were assigned to the species P. brasiliensis sensu stricto. Since H4 comprises only FFPE samples, we named the clade that contains these sequences as FFPE clade.
Our findings indicated genotypes not covered by the four species currently recognized, suggesting a higher genetic diversity in the P. brasiliensis complex than that described so far. Similar findings had already been described for the genus Paracoccidioides in a study that reported isolates obtained from environmental samples clustering apart from the clinical referenced strains of P. lutzii and P. brasiliensis [60]. By analyzing the rRNA universal fungal region ITS1-5.8S-ITS2, the authors found two new genotypes in soil samples and one in aerosol samples that indicated a higher genetic variation than the previously reported for the ITS in Paracoccidioides. Our study expands the knowledge regarding the genetic diversity in Paracoccidioides by finding new genotypes inside the P. brasiliensis complex and considering FFPE human tissue samples. Moreover, the higher genetic diversity found in FFPE and environmental samples, compared with isolates maintained in culture collections, may suggest that some lineages of Paracoccidioides are not successful in culture adaptation. Further studies are needed to evaluate this issue.
We obtained four PbGP43 sequences from the same patient (named Patient 3), an initially healthy male, 22 years-old, ex-rural worker living in Botucatu (São Paulo State, Brazil). He was hospitalized with a diagnosis of pneumonia, whose symptoms lasted for over one month and aggravated in the last days before hospitalization. He showed multiple cutaneous lesions, fever, pneumonia, and was later diagnosed with the acute juvenile form of PCM, established by both the microscopic findings (histopathology) and skin test. Serology was negative for HIV and he died a week after being admitted to the hospital. It is noteworthy that we detected a coinfection in Patient 3, for which three samples were clustered in H4, and one sample was clustered with P. americana. Coinfection by Paracoccidioides has already been reported in armadillos [61], and a recent study reported a case of coinfection in humans, showing P. brasiliensis sensu stricto and P. lutzii in the same patient [37]. Here, for the first time, we report a coinfection involving two different lineages of the P. brasiliensis complex. Pinheiro et al. [37] hypothesize that the use of non-selective media most likely favors the isolation of a faster-growing or a major representative genotype in Paracoccidioides, while FFPE samples have a higher potential to detect coinfections. Likewise, our results reinforce the high potential of FFPE samples in Paracoccidioides coinfection detection.
The PbGP43 gene region analyzed comprises part of the P10 T-cell epitope, including its essential inner core formed by the conserved amino acid sequence HTLAIR. We found one amino acid substitution in the P10 peptide among the haplotypes H1 to H6, which corresponded to a variable parsimony-informative site. However, this substitution occurred between two amino acids with similar characteristics, and did not represent a radical non-synonymous change, according to our results. On the other hand, the essential inner core was identical among the six haplotypes comprising the P. brasiliensis complex, including H4 and H5 that could not be assigned to any recognized species. Our results corroborate the high conservation level of the P10 inner core among species in the complex and reinforce its potential to be used in the PCM treatment and possible vaccines. The gene region analyzed also includes the N-glycosylation site of this glycoprotein [28], which showed no nucleotide substitutions among the species of the complex, but showed one non-synonymous substitution in P. lutzii. This substitution prevents glycosylation, as already proven experimentally [62].
Interestingly, the five variable parsimony-informative sites identified among the haplotypes consisted of non-synonymous changes in one or more species of the P. brasiliensis complex, in relation to P. lutzii. Genes encoding pathogenesis-related proteins, as PbGP43, are likely to evolve in response to selective pressure from the host’s immune system, which explains why non-synonymous substitutions are common and have been maintained by positive selection in genes that code for virulence factors [29]. Three out of five sites showed radical non-synonymous changes, i.e., amino acid changes that can promote structural and/or functional changes in the protein.
We highlight the change from lysine (basic amino acid) to glutamic acid (acidic amino acid) observed in H1, H2, H3, and H4, associated with changes in the isoelectric point (pI). Changes related to this property are essential because the electrical charge of the protein affects its solubility. That is especially relevant in the gp43 isoforms since they are primarily secreted to the extracellular environment and bound to major histocompatibility complex (MHC) molecules [63]. Indeed, a variety of pIs have been reported in gp43 [26, 28, 52, 64, 65], which have the potential to influence immunogenicity, i.e., the ability of an antigen to induce an adaptive immune response. We also found substitutions associated with alpha-helical tendencies (in H1 and H3), power to be at the N-terminal, and turn tendencies (both in H3), affecting the protein tertiary structure.
Our data shed additional light to the genetic diversity existing in the P. brasiliensis complex and contribute to the knowledge of the geographic distribution of lineages. In addition, we confirmed PbGP43 as a remarkable informative gene for further species identification in the complex, even considering a short gene fragment. It is also important to point out the correlation between molecular and phylogenetic analyzes of Paracoccidioides found in FFPE human biopsies, which is rare in the literature and shows a high potential to detect cases of coinfection, an interesting aspect that is worth exploring in the future.
References
Shikanai-Yasuda MA, Mendes RP, Colombo AL, et al. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Epidemiol Serv Saúde 2017;20.
Restrepo A, Tobón A. Paracoccidioides brasiliensis. In: Mandell GL, Bennett JE, Dollin R, editors. Principles and practice of infectious diseases. 2005. pp 3062–3068.
Martinez R. New trends in paracoccidioidomycosis epidemiology. J Fungi (Basel). 2017;3(1).
Wanke B, Londero A. Epidemiology and paracoccidioidomycosis infection. In: Paracoccidioidomycosis. Epidemiology and paracoccidioidomycosis infection; 1994. pp 109–120.
McEwen JG, Garcia AM, Ortiz BL, Botero S, Restrepo A. In search of the natural habitat of Paracoccidioides brasiliensis. Arch Med Res. 1995;26(3):305–6.
Coutinho ZF, da Silva D, Lazéra M, et al. Paracoccidioidomycosis mortality in Brazil (1980–1995). Cad Saúde Pública. 2002;18:1441–54.
Dantas KC, Mauad T, de André CDS, Bierrenbach AL, Saldiva PHN. A single-centre, retrospective study of the incidence of invasive fungal infections during 85 years of autopsy service in Brazil. Sci Rep. 2021;11(1):3943.
Teixeira MM, Theodoro RC, de Carvalho MJA, et al. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol. 2009;52(2):273–83.
Salgado-Salazar C, Jones LR, Restrepo Á, McEwen JG. The human fungal pathogen Paracoccidioides brasiliensis (Onygenales: Ajellomycetaceae) is a complex of two species: phylogenetic evidence from five mitochondrial markers. Cladistics. 2010;26(6):613–24.
Teixeira MM, Theodoro RC, Nino-Vega G, Bagagli E, Felipe MSS. Paracoccidioides species complex: ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 2014;10(10).
Gonzalez A, Hernandez O. New insights into a complex fungal pathogen: the case of Paracoccidioides spp. Yeast. 2016;33(4):113–28.
Montenegro M, Franco M. Pathology. In: Franco M, Lacaz CS, Restrepo-Moreno A, Del Negro G, editors. Paracoccidioidomycosis; 1994. pp. 131–150.
Martinez R. Epidemiology of paracoccidioidomycosis. Rev Inst Med Trop São Paulo. 2015;57:11–20.
Franco M, Bagagli E, Scapolio S, da Silva LC. A critical analysis of isolation of Paracoccidioides brasiliensis from soil. Med Mycol. 2000;38(3):185–91.
Bagagli E, Franco M, Bosco SDMG, Hebeler-Barbosa F, Trinca LA, Montenegro MR. High frequency of Paracoccidioides brasiliensis infection in armadillos (Dasypus novemcinctus): an ecological study. Med Mycol. 2003;41(3):217–23.
Ono MA, Bracarense AP, Morais HS, Trapp SM, Belitardo DR, Camargo ZP. Canine paracoccidioidomycosis: a seroepidemiologic study. Med Mycol. 2001;39(3):277–82.
Ricci G, Mota FT, Wakamatsu A, Serafim RC, Borra RC, Franco M. Canine paracoccidioidomycosis. Med Mycol. 2004;42(4):379–83.
de Oliveira HC, Assato PA, Marcos CM, et al. Paracoccidioides-host interaction: an overview on recent advances in the Paracoccidioidomycosis. Front Microbiol. 2015;6.
Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet Biol. 2017;106:9–25.
Matute DR, McEwen JG, Puccia R, et al. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol. 2006;23(1):65–73.
Muñoz JF, Farrer RA, Desjardins CA, et al. Genome diversity, recombination, and virulence across the major lineages of Paracoccidioides. mSphere. 2016;1(5).
Mattos K, Cocio TA, Chaves EGA, et al. An update on the occurrence of Paracoccidioides species in the Midwest region, Brazil: Molecular epidemiology, clinical aspects and serological profile of patients from Mato Grosso do Sul State. PLoS Negl Trop Dis. 2021;15(4):e0009317.
Teixeira MM, Cattana ME, Matute DR, et al. Genomic diversity of the human pathogen Paracoccidioides across the South American continent. Fungal Genet Biol. 2020;140:103395.
Cocio TA, Nascimento E, Kress MRVZ, Bagagli E, Martinez R. Characterization of a Paracoccidioides spp strain from southeastern Brazil genotyped as Paracoccidioides restrepiensis PS3 and review of this phylogenetic species. Genet Mol Biol. 2020;43(2):201.
Roberto TN, De Carvalho JA, Beale MA, et al. Exploring genetic diversity, population structure, and phylogeography in Paracoccidioides species using AFLP markers. Stud Mycol. 2021;100:100131.
Puccia R, Schenkman S, Gorin PA, Travassos LR. Exocellular components of Paracoccidioides brasiliensis: identification of a specific antigen. Infect Immun. 1986;53(1):199–206.
Cisalpino PS, Puccia R, Yamauchi LM, Cano MIN, da Silveira JF, Travassos LR. Cloning, characterization, and epitope expression of the major diagnostic antigen of Paracoccidioides brasiliensis. J Biol Chem. 1996;271(8):4553–60.
Morais FV, Barros TF, Fukada MK, Cisalpino PS, Puccia R. Polymorphism in the gene coding for the immunodominant antigen gp43 from the pathogenic fungus Paracoccidioides brasiliensis. J Clin Microbiol. 2000;38(11):3960–6.
Matute DR, Quesada-Ocampo LM, Rauscher JT, McEwen JG. Evidence for positive selection in putative virulence factors within the Paracoccidioides brasiliensis species complex. PLoS Negl Trop Dis. 2008;2(9):e296.
Oliveira AF, Coelho PSR. Yeast expressing gp43 protein as a vaccine against Paracoccidioides brasiliensis infection. Methods Mol Biol. 2017;1625:213–24.
Taborda CP, Juliano MA, Puccia R, Franco M, Travassos LR. Mapping of the T-cell epitope in the major 43-kilodalton glycoprotein of Paracoccidioides brasiliensis which Induces a Th-1 response protective against fungal infection in BALB/c mice. Infect Immun. 1998;66(2):786–93.
Guarner J, Brandt ME. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev. 2011;24(2):247–80.
Endo S, Komori T, Ricci G, et al. Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol Lett. 2004;234(1):93–7.
Sano A, Yokoyama K, Tamura M, et al. Detection of gp43 and ITS1-58S-ITS2 ribosomal RNA genes of Paracoccidioides brasiliensis in paraffin-embedded tissue. Nippon Ishinkin Gakkai Zasshi. 2001;42(1):23–7.
Ricci G, da Silva IDCG, Sano A, Borra RC, Franco M. Detection of Paracoccidioides brasiliensis by PCR in biopsies from patients with paracoccidioidomycosis: correlation with the histopathological pattern. Pathologica. 2007;99(2):41–5.
Ricci G, Zelck U, Mota F, Lass-Flörl C, Franco MF, Bialek R. Genotyping of Paracoccidioides brasiliensis directly from paraffin embedded tissue. Med Mycol. 2008;46(1):31–4.
Pinheiro BG, Hahn RC, de Camargo ZP, Rodrigues AM. Molecular tools for detection and identification of Paracoccidioides species: current status and future perspectives. J Fungi. 2020;6(4):293.
Greer CE, Wheeler CM, Manos MM. PCR amplification from paraffin-embedded tissues: sample preparation and the effects of fixation. In: Dieffenbach CW, Dveksler GS, editor. PCR primer: a laboratory manual; 1995. pp. 99–112
Gattás GJ, Soares-Vieira JA. Cytochrome P450–2E1 and glutathione S-transferase mu polymorphisms among Caucasians and mulattoes from Brazil. Occup Med (Lond). 2000;50(7):508–11.
Bialek R, Ibricevic A, Aepinus C, et al. Detection of Paracoccidioides brasiliensis in tissue samples by a nested PCR assay. J Clin Microbiol. 2000;38(8):2940–2.
Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80.
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–302.
Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–91.
Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19(5):671–2.
Blotta MH, Mamoni RL, Oliveira SJ, et al. Endemic regions of paracoccidioidomycosis in Brazil: a clinical and epidemiologic study of 584 cases in the southeast region. Am J Trop Med Hyg. 1999;61(3):390–4.
Nery AF, de Camargo ZP, Rodrigues AM, et al. Puzzling paracoccidioidomycosis: factors associated with the severity of Paracoccidioides lutzii infections. Int J Infect Dis. 2021;107:284–90.
Hahn RC, Rodrigues AM, Terra PPD, et al. Clinical and epidemiological features of paracoccidioidomycosis due to Paracoccidioides lutzii. PLOS Negl Trop Dis. 2019;13(6):e0007437.
Shankar J, Restrepo A, Clemons KV, Stevens DA. Hormones and the resistance of women to paracoccidioidomycosis. Clin Microbiol Rev. 2011;24(2):296–313.
Puccia R, McEwen JG, Cisalpino PS. Diversity in Paracoccidioides brasiliensis. The PbGP43 gene as a genetic marker. Mycopathologia. 2008;165(4–5):275–87.
Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61(1):64–71.
Bialek R, Feucht A, Aepinus C, et al. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J Clin Microbiol. 2002;40(5):1644–7.
Bonnet E, Moutet M-L, Baulard C, et al. Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples. PLoS ONE. 2018;13(4):e0195471.
Cocio TA, Nascimento E, von Zeska KMR, Bagagli E, Martinez R. Phylogenetic species of Paracoccidioides spp. isolated from clinical and environmental samples in a hyperendemic area of paracoccidioidomycosis in Southeastern Brazil. J Fungi. 2020;6(3):132.
Takayama A, Itano EN, Sano A, Ono MA, Kamei K. An atypical Paracoccidioides brasiliensis clinical isolate based on multiple gene analysis. Med Mycol. 2010;48(1):64–72.
Hrycyk MF, Garcia GH, Bosco SMG, de Oliveira SL, Marques SA, Bagagli E. Ecology of Paracoccidioides brasiliensis, P. lutzii and related species: infection in armadillos, soil occurrence and mycological aspects. Med Mycol. 2018;56(8):950–62.
de Macedo PM, Teixeira MM, Barker BM, Zancopé-Oliveira RM, Almeida-Paes R, do Valle ACF. Clinical features and genetic background of the sympatric species Paracoccidioides brasiliensis and Paracoccidioides americana. PLOS Negl Trop Dis. 2019;13(4):e0007309.
Arantes TD, Theodoro RC, Teixeira MM, Bosco SMG, Bagagli E. Environmental mapping of Paracoccidioides spp. in Brazil reveals new clues into genetic diversity, biogeography and wild host association. PLOS Negl Trop Dis. 2016;10(4):4606.
Sano A. Pathogenicities and GP43kDa gene of three Paracoccidioides brasiliensis isolates originated from a nine-banded armadillo Dasypus novemcinctus. Mycopathologia. 1999;144(2):61–6.
Leitão NP Jr, Vallejo MC, Conceição PM, Camargo ZP, Hahn R, Puccia R. Paracoccidioides lutzii Plp43 is an active glucanase with partial antigenic identity with P. brasiliensis gp43. PLOS Negl Trop Dis. 2014;8(8):e3111.
Straus AH, Freymüller E, Travassos LR, Takahashi HK. Immunochemical and subcellular localization of the 43 kDa glycoprotein antigen of Paracoccidioides brasiliensis with monoclonal antibodies. J Med Vet Mycol. 1996;34(3):181–6.
Moura Campos MCR, Gesztesi J-L, Vincentini AP, Lopes JD, Camargo ZP. Expression and isoforms of gp43 in different strains of Paracoccidioides brasiliensis. J Med Vet Mycol. 1995;33(4):223–7.
Souza MC, Gesztesi J-L, Souza AR, Moraes JZ, Lopes JD, Camargo ZP. Differences in reactivity of paracoccidioidomycosis sera with gp43 isoforms. J Med Vet Mycol. 1997;35(1):13–8.
Acknowledgements
We thank Prof. Marcello F. Franco, MD, who was mentor to GR in her Master and PhD program and that through his passion for fungus, mainly Paracoccidioides brasiliensis showed her the importance of the mycology in the world. Prof. Arnaldo L. Colombo, MD (Laboratório Especial de Micologia – LEMI – Disciplina de Infectologia, Departamento de Medicina, UNIFESP) for making part of the molecular analyses performed in this study available and Prof. Zoilo P. Camargo (Disciplina de Biologia Celular, Departamento de Microbiologia, Imunologia e Parasitologia, UNIFESP) for providing Histoplasma capsulatum DNA as a negative control. The authors are also grateful to Prof. Sigrid de Sousa Santos, MD (Departamento de Medicina, Universidade Federal de São Carlos, SP, Brasil) and the following pathologists: Clovis Klock, MD; Eduardo Haruo Suguiama, MD; Fernando Tito Motta, MD; Flavio Lima, MD; Gisela Nascimento Silva, MD; Margarida M. F. Moraes, MD; Paulo Faria,MD; Rogério Mendes Grande, MD for sending paraffin-embedded tissue blocks from PCM patients.
Funding
This work was supported by the grant from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [finance code 001 to G.R. and E.B.C.]; the Conselho Nacional de Desenvolvimento Científico e Tecnológico [Grant No. 313375/2017-8 to R.P.]; and the Fundação de Amparo à Pesquisa do Estado de São Paulo [Grant No. 06-05095-6 to R.P.].
Author information
Authors and Affiliations
Contributions
GR: conceptualization, funding acquisition, methodology, formal analysis, writing-original draft, writing-review and editing, supervision; EBC: conceptualization, methodology, formal analysis, writing-original draft, writing-review and editing, supervision; ASN: methodology, writing-original draft, writing-review, and editing; RP: resources, writing-original draft, writing-review and editing; MM: resources, writing-original draft, writing-review, and editing; RB: conceptualization, methodology, writing-original draft, writing-review, and editing; AMR: methodology, formal analysis, writing-original draft, writing-review, and editing; MF: conceptualization; WLB: conceptualization, methodology, formal analysis, writing-original draft, writing-review and editing, supervision. All the authors approved the final version of the manuscript, including the authorship list.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest. The authors alone are responsible for the content and the writing of the paper.
Ethical approval
This study was approved by the Ethics in Research Committee of the Universidade Federal de São Paulo under protocol number 0782/07.
Ethical responsibilities of authors
The authors confirm that this material is original and has not been published in whole or in part elsewhere and that the manuscript is not currently being considered for publication in another journal and that all authors have been personally and actively involved in substantive work leading to the manuscript and will hold themselves jointly and individually responsible for its content.
Additional information
Handling Editor: Ferry Hagen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ricci, G., Campanini, E.B., Nishikaku, A.S. et al. PbGP43 Genotyping Using Paraffin-Embedded Biopsies of Human Paracoccidioidomycosis Reveals a Genetically Distinct Lineage in the Paracoccidioides brasiliensis Complex. Mycopathologia 187, 157–168 (2022). https://doi.org/10.1007/s11046-021-00608-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-021-00608-3