Abstract
Background
The Candida parapsilosis complex species has emerged as an important cause of human disease. The molecular identification of C. parapsilosis isolates at the species level can be helpful for epidemiological studies and then for the establishment of appropriate therapies and prophylactic measures.
Methods
The present study was undertaken to analyze 13 short tandem repeat (STR) markers (7 minisatellites and 6 microsatellites) in a global set of 182 C. parapsilosis complex isolates from different origins including invasive and superficial clinical sites.
Results
Upon the analysis of 182 strains of C. parapsilosis complex species, 10–17 haplotypes were detected for each minisatellite marker. The combination of 7 minisatellite markers yielded 121 different genotypes with a 0.995 D value. Upon the analysis of 114 isolates (68 from invasive infections and 46 from superficial infections), 21–32 genotypes were detected for each microsatellite marker. The combination of all 13 markers yielded 96 different genotypes among 114 isolates with a high degree of discrimination (0.997 D value).
The same multilocus genotype was shared by isolates recovered from some patients and from the hand of theirs correspondent healthcare worker. For another patient, the same multilocus genotype of C. metapsilosis was detected in blood and skin confirming that candidemia usually arises as an endogenous infection following prior colonization.
Conclusions
These STR markers are a valuable tool for the differentiation of C. parapsilosis complex strains, to support epidemiological investigations especially studies of strain relatedness and pathways of transmission.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Clinically, Candida parapsilosis is an important pathogen among the non C. albicans emerging species, and its significance and prevalence have dramatically increased over the past two decades [1, 2].
Candida parapsilosis strains, though, are more heterogeneous than other Candida species. Tavanti et al. suggested a division of the C. parapsilosis group into three species named C. parapsilosis sensustricto, C. orthopsilosis, and C. metapsilosis [3]. Species of C. parapsilosis complex are important agents of nosocomial infections, because they can easily colonize hospital environments, essentially medical devices and hands of health care workers [1, 2].The rapid identification of the strains involved in the infection can contribute to the development of new strategies not only for treatment but also for the prevention of the infections [4]. Then, molecular identification of C. parapsilosis isolates at the species level is very important for optimal therapeutic options and studies of nosocomial cross-transmission [5, 6]. But, the vast majority of clinical isolates belonging to C. parapsilosis sensu stricto is characterized by low levels of nucleotide sequence variation, which constitutes a challenge for the development of genotyping techniques [5,6,7]. Instead, the trend in C. parapsilosis genotyping is toward reliance on length variations associated with microsatellites [4, 8, 9]. But, there are several studies from different investigation groups that have already described microsatellite markers for Candida parapsilosis genotyping with different degrees of specificity regarding C. orthopsilosis and C. metapsilosis [7, 8], which emphasize the need of a new set of markers for better genotyping of these two species.
Accordingly, the present study was undertaken to analyze 13 short tandem repeat (STR) markers (7 minisatellites and 6 microsatellites) in a global set of C. parapsilosis complex isolates from different origins including invasive and superficial clinical sites. We also aimed to analyze the genetic structure to distinguish epidemiologically related isolates, and to determine possible routes of acquisition of a strain in the hospital environment or identify possible reservoirs.
Materials and Methods
Fungal Strain
A total number of 182 C. parapsilosis complex species isolates analyzed in this study were recovered from clinical samples received by Department of Parasitology—Mycology-University Hospital—Sfax, Tunisia, during a 14-year period (from January 2002 to January 2016). They were 172 strains of C. parapsilosis sensu stricto, 6 strains of C. metapsilosis and 4 strains of C. orthopsilosis. The isolates of C. parapsilosis sensu stricto were obtained from various clinical specimens including blood (36.05%), urine (16.86%), auricular samples (22.68%), respiratory tract (2.33%), catheters (5.23%), and other sites such as skin (4.07%), nails (1.16%), oral cavity (1.74%), vagina (0.58%), peritoneal fluid (0.58%), and hand carriage (8.72%). C. metapsilosis was isolated from blood, urine, skin, and auricular samples. C. orthopsilosis were isolated from blood, urine, auricular sample, and hand carriage. Isolates were identified to the species level by standard methods [morphological characteristics and biochemical tests using ID 32C (BioMerieux, France)]. Molecular identification of the C. parapsilosis species complex was performed according to Tavanti et al. [3], and supplemented, as needed by internal transcribed spacer 1 (ITS1), 5.8S, and ITS2 region rRNA sequence analysis [10]. Eleven of these isolates were as reference strains, identified by ITS sequence analysis of the ribosomal DNA, and deposed in GenBank: C. parapsilosis sensu stricto (KT948326), C. metapsilosis (KU665248, KU665249, KU665250, KU665251, KU665252, and KX421284), and C. orthopsilosis (KU665253, KU665254, KU665255, and KU665256).
Four reference strains were included: C. parapsilosis (ATCC 22019), C. orthopsilosis (1219482, 1343124), and C. metapsilosis (1240011).
One Candida albicans (ATCC 3153), one Candida glabrata (CBS 138), one Candida tropicalis (ATCC 66029), and two Aspergillus flavus (CBS 12685.7 and JX 852615) strains were used in the specificity tests.
STR Design
To collect the published sequences of C. parapsilosis genome, the following websites were consulted: NCBI (http://ncbi.nlm.nih.gov), DDBJ (www.ddbj.nig.ac) and EMBL (http://www.embl.fr/) databases. The Tandem Repeats Finder software (http://tandem.bu.edu/trf/trf.html) was used to identify the short tandem repeats in C. parapsilosis genome. Selection criteria for the STR markers were: to have perfect repeat sequences with 100% identity between repeat units and to be highly repetitive sequences (> 20 repeats and ≥ 2 repeats if consensus size is more than 50 bp). Six microsatellites and seven minisatellites loci were then selected. The Primer (version 3) software (http://frodo.wi.mit.edu) was used to design primers. BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was consulted to verify the specificity of primers to C. parapsilosis.
DNA Extraction
DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN), as indicated by the manufacturer’s instructions.
PCR Amplification of STR Markers
Amplification reactions were performed in final volumes of 25 μl containing 1 ng of genomic DNA, 5 µl of 5X reaction buffer (pH 8.5), 3 mM MgCl2, 0.2 mM (each) dATP, dCTP, dGTP, and dTTP (Promega), 0.5 µM of each primer, and 2.5 U of GOTaq® DNA polymerase (Promega). PCR amplification was performed in a thermocycler (Bio-Rad). It consisted of initial denaturation for 5 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 56 °C, and 1 min at 72 °C, and a final extension for 10 min at 72 °C. Different markers were amplified using different fluorescent labels (FAM, HEX, TET). One microliter of the tenfold diluted amplification products (with formamide) was added to 15 µl of formamide and 0.5 µl of internal size marker LIZ 500 (Applied Biosystems Inc.). The PCR products were then denatured and injected with polymer POP-7 for run onto an ABI 310 genetic analyzer (Applied Biosystems Inc.).
For minisatellite analysis, the amplification products were then migrated on 3% agarose gel. The capture and analysis of the gels were performed by the Quantity One software package (Bio-Rad) coupled to the imaging system Gel Doc XR+™.
Data Analysis
Different DNA preparations of the same isolate with repeated analyses of the same DNA preparation were tested to evaluate the repeatability of the STR markers. The GENEPOP software version 4.2 was used to calculate genotype frequencies for each marker [11]. For each marker and for each possible marker combination, the Simpson index of diversity (D) [12] was calculated. A combination is considered as parsimonious if D value ≥ 0.95. The NTSYS-PC numerical taxonomy and multivariate analysis system (version 2.1; Exeter Software, Setauket, NY) was used to calculate the degree of similarity by applying the Dice coefficient test. The genetic distance between strains of C. parapsilosis complex species was determined by the dice index. An UPGMA dendrogram was generated. The ARLEQUIN software package calculated the fixation index (FST) on all loci [13]. The minimum spanning tree (MStree) method as implemented by Network V4.6 estimated the phylogenetic relationships [14].
Results
A total of 26041 STR were found among 13.0501 Mb of C. parapsilosis genomic sequences available in online databases (February 2015); 0.68.6% consisted of repeat units greater than 7 nucleotides (minisatellites) and 31.4% contained repeat units ranging from 1 to 6 nucleotides in length. For the microsatellites, they were: mononucleotide (12.2%), dinucleotide (14.4%), trinucleotide (37%), tetranucleotide (4.9%), pentanucleotide (4.6%), and hexanucleotide (26.9%). For the minisatellites, they ranged from 7 to 50 bp (94.1%), 51 to 100 bp (4%), 101 to 150 bp (1.2%), and ≥ 151 bp (0.7%). Thirteen markers were selected for further assays since they matched the selection criteria (Table 1): four dinucleotides, two trinucleotides, and seven minisatellites (with the following repeat unit sizes: 88, 12, 24, 81, 82, 134, and 138 bp).
Specificity of STR Markers
The amplification of the 13 STR markers, using the genomic DNA of 3 Candida species [1 C. albicans (ATCC 3153), 1 C. glabrata (CBS 138), 1 C. tropicalis (ATCC 66029)] and 2 Aspergillus flavus (CBS 12685.7 and JX 852615), revealed that the 13 new loci were specific for C. parapsilosis complex (no PCR product was generated).
All microsatellite–minisatellite markers reacted with DNA from C. orthopsilosis and C. metapsilosis isolates.
Genetic Diversity
Upon the analysis of 182 strains of Candida parapsilosis complex species isolated from 2002 to 2016, 5–8 distinct alleles and 10–17 genotypes were detected for each minisatellite marker (Table 2). The highest discriminatory power for a single minisatellite locus was obtained with the STR12 marker, which had 7 distinct alleles, 17 genotypes, and a D value of 0.858 (Table 2). The combination of 7 minisatellites markers yielded 121 different genotypes with a 0.995 D value (Table 3).
Upon the analysis of 114 isolates (68 from invasive infections and 46 from superficial infections were part of the 182 strains), 8–17 distinct alleles and 21–32 genotypes were detected for each microsatellite marker (Table 2). The highest discriminatory power for a single microsatellite locus was obtained with the STR-AC marker, which had 12 distinct alleles, 30 genotypes, and a D value of 0.960 (Table 2). The combination of 6 microsatellites markers yielded 87 different genotypes with a 0.995 D value (Table 3).
The combination of all 13 markers yielded 96 different genotypes among 114 isolates with a 0.997 D value. A 6-marker combination (STR-AC, STR-TG, STR-ACA, STR12, STR138, and STR88) yielded also 96 different genotypes with a 0.997 D value (Table 3). This six-marker combination was selected as the most parsimonious panel achieving D > 0.95.
The observed percent of heterozygosity was similar to estimated percent for four STR (STR-AC, STR-TC, STR-TTG, and STR138), superior for one STR (STR24) and inferior for all the others STR (Table 2).
Cluster Analysis
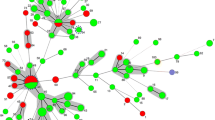
The dendrogram based on the Dice similarity coefficient upon analysis of 13 STR markers in 114 Candida parapsilosis species complex isolates revealed 8 clusters (Fig. 1). The majority of the strains belonging to cluster 1 (11 strains) were isolated from blood or catheter. Each of clusters 2 (20 strains) and 4 (27 strains) was subdivided in 2 subclusters: 1 for isolates from blood and the other for isolates from superficial sites. The cluster 3 (10 strains), 6 (23 strains), 7 (4 strains), and 8 (15 strains) contained strains from different clinical sites. Cluster 5 (four strains) contained strains isolated only from urine and auricular samples.
UPGMA dendogram based on the Dice similarity coefficient upon analysis of 13 new STR markers in 114 Candida parapsilosis species complex isolates. The isolates are identified by the culture collection number. Six strains of C. metapsilosis (dots) and four strains of C. orthopsilosis (times) were included. The bordered strains represent the cases of multiple strains for 11 patients with invasive infections (dotted rectangle P1, P4, P5, P7, P10, P13, P14, P15, P16, P17, and P18) and 3 patients with superficial infections (dashed rectangle P6, P11, and P12). For two patients (solid rectangle P8 and P9), they were strains isolated from blood culture and from hand carriage of hers health care workers
For C. orthopsilosis and C. metapsilosis, they were not grouped in homogeneous groups or independent clusters. But, two strains of C. orthopsilosis belonged to cluster 2 (subcluster of superficial sites) and five strains of C. metapsilosis and one C. orthopsilosis belonged to cluster 4 (subcluster of superficial sites). The genetic diversity per locus (FST) values were calculated for the three species of C. parapsilosis complex and were noted to range between 0.039 and 0.041, indicating a moderate differentiation between the three species (Table 4).
Similarity Dice Coefficient
Pairwise Dice coefficient of similarity values between strains ranged from 0.09 to 1 (Fig. 1). The dice value was 1 for cases with multiple strains.
They were eleven patients with invasive infections (P1, P4, P5, P7, P10, P13, P14, P15, P16, P17, and P18). For patient, 18 identical multilocus genotypes for both strains isolated from blood culture and catheter were obtained. For patient 10, the same genotype of C. metapsilosis was detected in blood and skin. It was noted also that patient 2 and patient 3 (hospitalized in two different departments) presented bloodstream infection with strains which had identical genotypes.
The same multilocus genotypes were obtained for each of the three patients (P6, P11, and P12) with multiple strains in superficial infections (urinary infection and otomycosis).
For each patient P8 and P9, the strains isolated from blood culture and from hand carriage of hers health care workers displayed the same multilocus genotypes. The strains causing these two infections were closely related in cluster analysis (Fig. 1).
Distance Interpopulation
A minimum spanning tree (MStree) was generated from the analysis of 182 Candida parapsilosis species complex isolates (74 strains from invasive infections and 108 from superficial infections) using Network V4.6 (Fig. 2). The findings revealed that cluster A (131 strains) was dominated by strains from the of the invasive infection population; cluster B (51 strains) was dominated by strains from the superficial infection population.
A factorial analysis correspondence graph was generated to illustrate if there is population structure associated with the period of isolation (Fig. 3). In this two-dimensional graphical representation of genetic similarity between individuals, the information for all loci is condensed into a smaller dimension (two factors, axis I and axis II) with a minimum loss of information. A graphical proximity between two individuals indicates a genetic similarity. This graph illustrated that the majority of isolates were grouped with the exception of some strains isolated in 2011 and 2015. The most distant strains were isolated in 2013 and 2016. In fact, the pairwise Fst values of 2016 compared to others years were the most important and ranged from 0.191 (with 2011) to 0.644 (with 2008).
Discussion
Opportunistic infections are an increasingly common problem in hospitals worldwide. The Candida parapsilosis complex species has emerged as an important cause of human disease with the ability to infect different body sites.
For a better understanding of the epidemiology of this complex, it is essential to discriminate exactly C. parapsilosis strains, and to identify rapidly the strains involved in the infections [2, 15]. So, a variety of strain typing methods have been used including electrophoretic karyotype analysis [16], DNA fingerprinting with probe Cp3-13 [17], RAPD analysis [18], RFLP analysis [19], multilocus sequence typing (MLST) [3], and pyrosequencing [20].
But, given the clonal nature of C. parapsilosis, most of the genotyping methods became not discriminant enough to distinguish closely related isolates and to establish routes of transmission [21, 22]. However, microsatellite analysis may be more able than others genotyping methods to distinguish between strains with low degrees of sequence variation, because microsatellite loci behave as codominant markers and evolve rapidly in a genome [8].
In fact, the STR markers proposed in the present study yielded promising results in terms of identifying intraspecies polymorphisms in C. parapsilosis complex strains. In a multilocus analysis, 96 genotypes were found among 114 isolates, resulting in a D of 0.997, similar than that published by Reiss et al. [7]. In 2006, Lasker et al. described a set of seven dinucleotide microsatellite markers, able to discriminate C. parapsilosis sensu stricto strains, with a discriminatory power of 0.97 [8]. In 2010, Sabino et al. reported a high degree of polymorphisms by microsatellite analysis in C. parapsilosis, with 192 different genotypes found among 233 isolates, based on 4 hyper variable loci (D of 0.99) [4]. In the present work, the combination of seven minisatellite markers yielded a 0.995 D value. This is very interesting because these markers showed a high discriminatory power using gel electrophoresis, which is less expensive and easy to implement in laboratories with basic molecular biology equipment.
The 13 STR markers presented in this work showed high levels of discrimination and specificity for the inter-strain differentiation of C. parapsilosis complex species. Although the strains studied are from the same geographical origin, a genetic heterogeneity was detected among the C. parapsilosis complex strains. In fact, microsatellite typing analysis was a very powerful technique to clarify epidemiologic associations, to detect micro-evolutionary variations, and to confirm nosocomial infection caused by C. parapsilosis [9, 15, 21].
Our study is consistent with other previous reports implicating the hands of healthcare workers as the source of fungemia [15, 22]. In fact, the same multilocus genotype was shared by isolates recovered from patients and from the hand of theirs correspondent healthcare workers, suggesting that these could be possible routes of transmission and that infections due to C. parapsilosis may be mainly related with exogenous horizontal transmission to the patient.
This study confirms that candidemia in intensive care unit patients is caused by strain colonizing hands of healthcare workers. So, it is necessary to implement surveillance program for better dissemination of information to health care workers which must be careful to the correct use of gloves and hand washing [23, 24].
For the same patient, the same multilocus genotype of C. metapsilosis was detected in blood and skin. This observation is in accordance with what is generally agreed stating that candidemia usually arises as an endogenous infection, following prior colonization of the gastrointestinal tract, skin, or vagina [22].
This property of C. parapsilosis complex species as a skin colonizer facilitates transmission from an exogenous source, the hands of healthcare workers, to patient during installation and maintenance of intravascular catheters [2, 7]. In fact, identical multilocus genotypes for both strains isolated from blood culture and catheter were obtained for another of our patients. Then, appropriate control measures must be implemented against both exogenous and endogenous infection reservoirs for prevention strategies aimed at C. parapsilosis [6].
In our work, all the 13 STR amplified DNA from C. orthopsilosis and C. metapsilosis. In a previous study realized by Lasker et al., DNA obtained from single representatives of these two species was not amplified by all seven primer pairs [8]. Reiss et al. tested also cross-hybridization of five STR markers with the species C. metapsilosis and C. orthopsilosis. Some markers were completely unreactive for some isolates [7].
In cluster analysis, we noted that C. orthopsilosis and C. metapsilosis were not grouped in homogeneous groups or independent clusters. This can be explained by the limited number of tested isolates which hinders a statistical analysis to prove/disprove the existence of cluster structure within these species. Moreover, the use of microsatellite markers for strain identification among C. parapsilosis complex species may be limited by the high degree of polymorphism observed with this molecular method which could be correlated with a high mutation rate [4]. Furthermore, it may be due to the homoplasy which is explained by constraints on the microsatellite size distribution leading to alleles with an identical size, but not necessarily with a common ancestor [8].
Distance interpopulation study revealed that cluster A was dominated by strains from that of the invasive infection population and cluster B was dominated by strains from the superficial infection population. So, it seems that not all C. parapsilosis isolates are equally as likely to cause human infections and some of them appear to be clinically most relevant and cause invasive infections significantly more often than others [25].
A factorial analysis correspondence graph illustrated that the majority of isolates were grouped with the exception of some strains isolated in 2011 and 2015. So it is clear that “some genotypes may have evolved over time by spontaneous changes”, and clonal complexes of closely related genotypes were than generated [6, 7, 24]. It was also suggested that strains have insufficient time to diversify completely, but show adaptation to the environment [26].
In our study, we found that the most distant strains were isolated in 2013 and 2016. A micro-evolutionary process might be involved between these strains, differentiating them from a common ancestry [24, 26]. But, microsatellites loci are inherently unstable, so they would not be useful for genetic relatedness studies on larger time scale or on population structure [4, 24].
Conclusion
STR analysis presents a high discriminatory power and may be very useful for studies of kinetics of the colonization-to-infection process and for epidemiological studies of C. parapsilosis complex species. Then, we emphasize the importance of appropriate control measures for prevention of these infections. The identification of genotypes and population structure of C. parapsilosis complex species could be also a significant marker for further investigations of virulence factors and drug resistance.
References
Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008;21(4):606–25.
van Asbeck EC, Clemons KV, Stevens DA. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit Rev Microbiol. 2009;35(4):283–309.
Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol. 2005;43(1):284–92.
Sabino R, Sampaio P, Rosado L, Stevens DA, Clemons KV, Pais C. New polymorphic microsatellite markers able to distinguish among Candida parapsilosis sensu stricto isolates. J Clin Microbiol. 2010;48(5):1677–82.
Garcia-Effron G, Canton E, Peman J, Dilger A, Roma E, Perlin DS. Assessment of two new molecular methods for identification of Candida parapsilosis sensu lato species. J Clin Microbiol. 2011;49(9):3257–61.
Diab-Elschahawi M, Forstner C, Hagen F, Meis JF, Lassnig AM, Presterl E, Klaassen CH. Microsatellite genotyping clarified conspicuous accumulation of Candida parapsilosis at a cardiothoracic surgery intensive care unit. J Clin Microbiol. 2012;50(11):3422–6.
Reiss E, Lasker BA, Lott TJ, Bendel CM, Kaufman DA, Hazen KC, Wade KC, McGowan KL, Lockhart SR. Genotyping of Candida parapsilosis from three neonatal intensive care units (NICUs) using a panel of five multilocus microsatellite markers: broad genetic diversity and a cluster of related strains in one NICU. Infect Genet Evol. 2012;12(8):1654–60.
Lasker BA, Butler G, Lott TJ. Molecular genotyping of Candida parapsilosis group I clinical isolates by analysis of polymorphic microsatellite markers. J Clin Microbiol. 2006;44(3):750–9.
Brillowska-Dabrowska A, Schon T, Pannanusorn S, Lonnbro N, Bernhoff L, Bonnedal J, Haggstrom J, Wistedt A, Fernandez V, Arendrup MC. A nosocomial outbreak of Candida parapsilosis in southern Sweden verified by genotyping. Scand J Infect Dis. 2009;41(2):135–42.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols Guide Methods Appl. 1990;18:315–22.
Rousset F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8(1):103–6.
Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–6.
Maiden M, Bygraves J, Feil E, Morelli G, Russell J, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D, Feavers I, Achtman M, Spratt B. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95(6):3140–5.
Polzin TDSV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20.
Vaz C, Sampaio P, Clemons KV, Huang YC, Stevens DA, Pais C. Microsatellite multilocus genotyping clarifies the relationship of Candida parapsilosis strains involved in a neonatal intensive care unit outbreak. Diagn Microbiol Infect Dis. 2011;71(2):159–62.
Shin JH, Shin DH, Song JW, Kee SJ, Suh SP, Ryang DW. Electrophoretic karyotype analysis of sequential Candida parapsilosis isolates from patients with persistent or pecurrent fungemia. J Clin Microbiol. 2001;39(4):1258–63.
Enger L, Joly S, Pujol C, Simonson P, Pfaller M, Soll DR. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J Clin Microbiol. 2001;39(2):658–69.
Zancope-Oliveira RM, James MJ, Derossi AP, Sampaio JL, Muniz MM, Li RK, Nascimento AS, Peralta JM, Reiss E. Strain characterization of Candida parapsilosis fungemia by molecular typing methods. Eur J Clin Microbiol Infect Dis. 2000;19(7):514–20.
Van Asbeck EC, Clemons KV, Markham AN, Stevens DA. Candida parapsilosis global epidemiology G. Molecular epidemiology of the global and temporal diversity of Candida parapsilosis. Scand J Infect Dis. 2008;40(10):827–34.
Borman AM, Linton CJ, Oliver D, Palmer MD, Szekely A, Odds FC, Johnson EM. Pyrosequencing analysis of 20 nucleotides of internal transcribed spacer 2 discriminates Candida parapsilosis, Candida metapsilosis, and Candida orthopsilosis. J Clin Microbiol. 2009;47(7):2307–10.
Romeo O, Delfino D, Cascio A, Lo Passo C, Amorini M, Romeo D, Pernice I. Microsatellite-based genotyping of Candida parapsilosis sensu stricto isolates reveals dominance and persistence of a particular epidemiological clone among neonatal intensive care unit patients. Infect Genet Evol. 2013;13:105–8.
Sabino R, Sampaio P, Rosado L, Videira Z, Grenouillet F, Pais C. Analysis of clinical and environmental Candida parapsilosis isolates by microsatellite genotyping—a tool for hospital infection surveillance. Clin Microbiol Infect. 2015;21(10):954-e1–958.
da Silva Ruiz L, Montelli AC, Sugizaki Mde F, Da Silva EG, De Batista GC, Moreira D, Paula CR. Outbreak of fungemia caused by Candida parapsilosis in a neonatal intensive care unit: molecular investigation through microsatellite analysis. Rev Iberoam Micol. 2013;30(2):112–5.
Delfino D, Scordino F, Pernice I, Lo Passo C, Galbo R, David A, Barberi I, Criseo G, Cascio A, Romeo O. Potential association of specific Candida parapsilosis genotypes, bloodstream infections and colonization of health workers’ hands. Clin Microbiol Infect. 2014;20(11):O946–51.
Chillemi V, Lo Passo C, van Diepeningen AD, Rharmitt S, Delfino D, Cascio A, Nnadi NE, Cilo BD, Sampaio P, Tietz HJ, Peman J, Criseo G, Romeo O, Scordino F. Multilocus microsatellite analysis of European and African Candida glabrata isolates. Eur J Clin Microbiol Infect Dis. 2016;35(6):885–92.
Pulcrano G, Roscetto E, Iula VD, Panellis D, Rossano F, Catania MR. MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur J Clin Microbiol Infect Dis. 2012;31(11):2919–28.
Acknowledgements
We thank Dr Stephane Ranque for Candida parapsilosis reference isolates.
Author information
Authors and Affiliations
Ethics declarations
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Handling Editor: Vishnu Chaturvedi.
Rights and permissions
About this article
Cite this article
Neji, S., Hadrich, I., Ilahi, A. et al. Molecular Genotyping of Candida parapsilosis Species Complex. Mycopathologia 183, 765–775 (2018). https://doi.org/10.1007/s11046-018-0278-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-018-0278-1