Abstract
Lung cancer has the highest incidence in the world. The standard tests for its diagnostics are medical imaging exams, sputum cytology, and lung biopsy. Computed Tomography (CT) of the chest plays an essential role in the early detection of nodules since it can allow for more treatment options and increases patient survival. However, the analysis of these exams is a tiring and error-prone process. Thus, computational methods can help the specialist in this analysis. This work addresses the classification of pulmonary nodules as benign or malignant on CT images. Our approach uses the pre-trained VGG16, VGG19, Inception, Resnet50, and Xception, to extract features from each 2D slice of the 3D nodules. Then, we use Principal Component Analysis to reduce the dimensionality of the feature vectors and make them all the same length. Then, we use Bag of Features (BoF) to combine the feature vectors of the different 2D slices and generate only one signature representing the 3D nodule. The classification step uses Random Forest. We evaluated the proposed method with 1,405 segmented nodules from the LIDC-IDRI database and obtained an accuracy of 95.34%, F1-Score of 91.73, kappa of 0.88, sensitivity of 90.53%, specificity of 97.26% and AUC of 0.99. The main conclusion was that the combination by BoF of features extracted from 2D slices using pre-trained architectures produced better results than training 2D and 3D CNNs in the nodules. In addition, the use of BoF also makes the creation of the nodule signature independent of the number of slices.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is one of the most preventable causes of death as long as it is detected early [23]. For both sexes, lung cancer is the most commonly diagnosed cancer in the world, with 11.6% of all cases, being also the leading cause of death, accounting for 18.4% of all cancer deaths [10]. The Computed Tomography (CT) exam is the preferred method by specialists to perform the non-invasive screening of patients with the disease [53], which has been available since 1975. This exam offers a sharper image with a more significant distinction between the body tissues [44].
The analysis of CT images is a challenging task, as the density of the nodules can be similar to other pulmonary structures, and specialists tend to analyze large numbers of exams, making the process repetitive and prone to errors. A pulmonary nodule is characterized as a rounded opacity, well or poorly defined, with a diameter equal to or less than 3cm [19]. Lesions greater than 3 cm are often classified as malignant [16].
Early diagnosis and treatment of lung cancer at an early stage increases the chance of patient survival [18]. In this scenario, automated computational tools such as Computer-Aided Diagnosis (CAD) are widely explored to increase the accuracy in diagnosing lesions, as they can provide the specialist with a second opinion [33].
In recent years, several techniques have been applied to detect abnormalities in chest CT images to help specialists search for an early diagnosis. Approaches such as texture descriptors, shape attributes, and Convolutional Neural Networks (CNN) explored different image properties. The most recent literature has focused on techniques based on 3D CNNs. However, they still have some limitations, mainly related to the need for large amounts of samples available for training. Thus, 2D CNNs are also commonly investigated, as they require fewer samples than 3D CNNs to obtain good performance.
Many studies to classify pulmonary nodules applied CNNs [1, 21, 22, 46, 52], which require a single input format for images, although pulmonary nodules have different sizes. In addition, some works used 3D CNNs [55], which require more images in the training stage than 2D CNNs, leading to increased computational cost. BoF is a technique widely used in different medical applications [6, 8, 35, 47]. However, to the best of our knowledge, this technique has not been used to classify pulmonary nodules on computed tomography images.
This paper proposes an automatic method to classify pulmonary nodules from CT images as benign or malignant. The proposed method is based on Pre-trained Networks, Principal Component Analysis (PCA) [25], and BoF [37]. To create this algorithm, we used different pre-trained 2D architectures to extract features from different slices of each 3D nodule. We applied PCA to reduce and standardize the dimensionality of the features, which were combined using BoF to create a single signature representing a nodule. Then, we evaluated the Random Forest (RF) classifier parameters to obtain a better combination of hyper-parameters that improved the prediction in the classification step.
The main contributions of this work are:
-
A BoF-based classification method combines features extracted from 3D nodes from different pre-trained 2D network architectures;
-
A combination of techniques capable of classifying three-dimensional images from two-dimensional architectures does not depend on a single depth size for all nodule samples;
-
The proposed method performed better than that presented by 2D and 3D CNNs trained using the nodules.
The rest of the paper is divided into four main sections: Section 2 comprises a synthesis of the related works. Section 3 details the BoF-based classification method proposed in this work. In Section 4, the results obtained in the experiments are presented and discussed. Finally, Section 5 presents the conclusions and limitations of this study and proposes future works.
2 Related works
In recent years, several computational techniques have been applied to detect abnormalities in CT images in the thoracic region to assist the work of specialists in the search for an early diagnosis. In this scenario, works that used texture descriptors, shape descriptors, 2D CNNs, 3D CNNs, and BoF will be presented.
In [14] the methodology presented the Mean Phylogenetic Distance and the Taxonomic Diversity Index for feature extraction. In the classification step, a genetic algorithm was used together with the Support Machine (SVM). In the work of [39], Multiple Instance Learning was used to classify nodules as malignant or benign. In [40], the authors proposed a method that uses Gray Level Co-occurrence Matriz for feature extraction and SVM to perform nodule classification. In [36], the method consists of four steps: 1 - extraction of the lung region; 2 - method of detecting candidate nodes based on geometric adjustment in parametric form; 3 - hybrid geometric texture highlighting; 4 - classification of nodes using a deep learning approach based on autoencoder and softmax.
In terms of shape descriptors, examples are the works of [42], and [16]. In [42] they present a method that combines appearance and shape descriptors, the 3D Ambiguity Local Binary Pattern, and the seventh order Markov Gibbs Random Field. The characteristics of the images extracted by the descriptors are classified separately using Denoising Autoencoder and SVM. In [16], functional Minkowski measures, distance measures, vector representation of point measures, triangulation measures, and Feret diameters were used to differentiate the patterns of malignant and benign forms. They used a genetic algorithm to select the best individuals to generate the model in the classification with SVM.
In [52], the authors proposed a semi-supervised adversarial classification model that can be trained using labeled and unlabeled data. In [21], was explored a new diagnostic method based on Deep Transfer Convolutional Neural Network and Extreme Learning Machine (ELM). In [1], transferable texture convolutional neural networks were proposed to improve classification performance. The study [46] proposed an efficient approach based on a deep neural network for automatic classification. Finally, in [55], an end-to-end classification of CT spots of raw 3D nodules was performed using CNNs.
In [17], four CNN architectures were proposed, including a basic 3D CNN, a new multi-output network, a DenseNet 3D, and an augmented DenseNet 3D with multiple outputs. In [56], a Regions with Convolutional Neural Net was used for nodule detection with dual-path 3D blocks and a U-net-like structure to learn the nodule characteristics effectively. In [22], the authors develop a self-supervised transfer learning based on the 3D CNN Domain Adaptation Framework to classify pulmonary nodules.
In addition, studies that used BoF showed promising results for other medical applications. In [6], the authors developed an ensemble-based BoF classification system for detecting COVID-19. In [35], an approach consisting of a BoF and a neural network was proposed to classify chest radiography images into non-COVID-19 and COVID-19. In [8], a model is presented for the classification of Dementia brain disease using magnetic resonance imaging. The BoF was used to extract features and SVM to distinguish exams into three categories such as demented, mild cognitive impairment, and normal controls. The work proposed in [47] presents a proficient content-based image retrieval framework based on Spark Map Reduce and BoF for different medical applications.
Table 1, presents the main related works divided by the techniques used. We also highlight the metrics, which are Accuracy (ACC), Sensitivity (SEN), Specificity (SPE), and Area under the ROC Curve (AUC).
By analyzing Table 1, it is possible to observe that texture and shape descriptors perform well in detecting pulmonary nodules. However, they need a good segmentation of the nodules region, which is challenging. In this way, CNNs have gained greater notoriety recently, mainly because of their promising results in many applications. However, some limitations are mainly related to the need for large amounts of samples available for training. Especially in deep architectures with many layers, 2D and 3D CNNs are sensitive to overfitting due to the reduced size of the available databases. Another disadvantage of 2D and 3D CNNs is that they require all nodules to have the same number of slices. Therefore, it is necessary to use techniques to replicate or remove nodule slices.
In this work, we investigated the combination by BoF of features extracted by different pre-trained 2D CNNs to represent a 3D nodule. Using pre-trained 2D CNNs was motivated because they required fewer samples for training [4, 50]. On the other hand, the motivation for using BoF was to make the method independent of the number of 2D slices of the nodules.
Some works apply resizing techniques to standardize the size of the images since CNNs require that all images have the same input size. This process can lead to distortion of essential features for the classification of nodules [21, 46]. Other works classify 3D images using 2D CNNs; this method considers the depth as the channels of the images. In this scenario, all 3D images need to have equal depth [55]. Other works perform the classification known as slice by slice without considering the 3D nodule [1]. Furthermore, some limitations in using 3D CNN architectures may be related to unbalanced sets that contain few samples [17]. With BoF, we guarantee the preservation of essential characteristics of the nodules, even in images with different depths. Another advantage is the fusion of features extracted by different architectures, which makes the method more robust and efficient. Furthermore, our method uses zero padding to avoid distortions, preserving the original size of the nodule in the image.
3 Proposed method
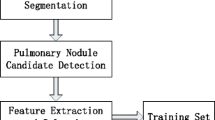
The proposed methodology is divided into five main steps: image acquisition, feature extraction, feature fusion, signature acquisition, and classification. A summary of the steps that are part of this methodology is presented in Fig. 1.
3.1 Image acquisition
The LIDC-IDRI image collection is used for diagnosing and screening lung cancer on CT scans with marked lesions. The data set contains 1018 exams, each including images from a clinical chest CT and an associated XML file, which records four experts’ results of a two-phase image annotation process. All images are in Digital Imaging and Communications in Medicine (DICOM) format and up to 16 bits per voxel. Its dimension is 512×512 with a variable number of cuts per exam. The CT images were acquired by different CT scanners, making detecting pulmonary nodules difficult [5]. In addition to the tags in the XML file, some characteristics are highlighted, such as sphericity, texture, and malignancy (all indicated by a value from 1 to 5). If the indicated value is closer to 1, the nodule is characterized as benign, and closer to 5, it is marked as malignant [16]. Classification as to malignancy or benignity is first obtained with calculations that summarize in a single value the nodular characteristics made by up to four experts, calculating the mode or median [24].
The method used to acquire the Volumes of Interest (VOI) was proposed by [16], 833 tomographic exams of the 1018 present in the LIDC-IDRI database were used in this step. A total of 185 exams were discarded, where some did not present nodules equal to or greater than 3 mm or had a divergence of information found in the notes file of an exam versus the information present in the DICOM header of the same exam. In the nodule segmentation step, the contour information provided in the XML file containing the nodule coordinates was used, together with expert analysis.
After segmenting the CT images, 1011 benign and 394 malignant nodules were obtained, which are available atFootnote 1. It was found that the VOIs extracted have different depths for each nodule. In addition, the cuts performed were not standardized in a fixed size, in this sense, the dimensions of the nodule slices are also different.
3.2 Extraction and merging of features
In this step we use the VGG16 [43], VGG19 [43], InceptionV3 [48], ResNet50 [20] and Xception [13] architectures, pre-trained in the base ImageNet [38] for extracting the characteristics of each of the nodule slices. The reason for the choice of these models is based on the good performance in ImageNet classification, in addition to promising results presented by these architectures in the medical literature included in several imaging works [12, 31, 41].
Although the networks are pre-trained on color images, they still produce good results for grayscale images. As they were trained on a general purpose database (Imagenet), their first layers formed by convolutional filters learn to extract generic features from images. This technique is commonly applied to medical imaging problems, as we can see in the literature [32, 50, 51]. Table 2 presents the pre-trained networks, layers used to extract the characteristics and quantity of characteristics of each network.
As noted in Table 2, the extracted features have different dimensions. Therefore, we use PCA to leave all feature vectors of the same size vectors to be combined, generating only one signature through BoF. PCA is widely used in large data sets, as it reduces the dimensionality of such sets, increases the interpretability of results, and at the same time minimizes the loss of information. It does this by creating new uncorrelated variables that successively maximize the variance [25] .
With PCA, we test different sizes of dimensionality, as few features may not represent the images; on the other hand, many features increase the computational cost and are more sensitive to overfitting. The tested dimensions were: 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, 1024 and 2048. After applying the PCA, the characteristics extracted by the different architectures were combined by Bof to generate a single signature per nodule. This signature contains information from all 2D slices extracted by different network architectures.
3.3 Bag of features
BoF is a method based on text classification known as Bag of Word (BoW). In its classic form, this method uses the SIFT [30] descriptor to represent key points of an image [26]. After detecting the points in the image, the detected points are grouped, generating a visual word vocabulary. Finally, the Bag of Visual Words is obtained, which is represented by a Visual-Word vector [54].
In the BOF, images are represented by the frequency of their pixel intensities that are obtained with the histogram. The image is described as a visual word frequency histogram based on a vocabulary that quantifies the spatial characteristics of all image samples. Such a method facilitates the visualization of patterns dependent on the image collection in the content of each sample. Furthermore, the method is adaptable and allows, for example, the use of different feature extractors and clustering algorithms [9].
Our method replaces the point detection step that uses SIFT descriptor. Instead, we use pre-trained networks to extract features of each slice of a nodule. Compared with the classic BoF model, in our method, each 2D slice of a 3D nodule represents a detected point, while the description step uses pre-trained networks. With BoF, it is possible to represent the 3D nodule, generating signatures from the features extracted from the 2D slices of the nodule. The BoF technique is composed of two stages: the construction of a visual dictionary of words and the generation of signatures. The database is divided into training and testing sets. Visual dictionary words are created with training images. After generating the dictionary, each image is represented by a vector, which is the image signature [45].
Once the feature vector of the images is extracted and standardized with the same size as the features, the most representative patches need to be identified, which will constitute the visual words of the system [2]. To create the dictionary, it is necessary to define the size k, which is the number of representative words. The dictionary must be large enough to distinguish relevant differences between images, but it cannot include irrelevant variations [49]. To this end, we use k-means [29], which is a clustering algorithm that places data into separate groups based on their similarity, where the value of k represents the number of clusters.
The tests performed with the individual networks were necessary to evaluate the influence of the number of clusters in the proposed method. The number of clusters is the size of the signature, that is, the feature vector. The k parameter values tested of K-means were: 2, 4, 8, 16, 32, 64, 128, 256, 512 and 1024. We emphasize that the PCA was not applied in the tests carried out with the individual networks, whose objective was to find the best k value. After defining a default value for the cluster number obtained in the individual tests, we applied the PCA to standardize the dimensionality of the CNNs and combine the features. Thus, we estimated the dimensionality of the features with the PCA, we used a default value for k, we applied the BoF, and we carried out the classification.
3.4 Classification
In this step, we use the RF classifier [11]. RF is a combination of tree predictors, so each tree depends on the values of an independently sampled random vector with the same distribution for all trees in the forest. A point to be highlighted is that all experiments were performed with fixed RF parameter values. Thus, after defining all the steps of our method for the description of the nodules, we estimated the parameters: number of trees in the forest and the maximum depth of the RF tree.
3.5 Metrics evaluation
To evaluate the results, we used the following metrics: Accuracy (1), Sensitivity (2), Specificity (3), Kappa coefficient (4), F1-Score (7) and Area Under ROC Curve (AUC) [3, 4]. These metrics are based on the four values: True Positive (TP), correct classification of the malignant class; True Negative (TN), correct classification of the benign class; False Positive (FP), prediction errors of the benign class; False Negative (FN), prediction errors of the malignant class. Being, the positive class malignant and the negative class benign.
The ACC (1), considered by several researchers, one of the simplest metrics to assess the quality of a classification, was used in this work to measure the number of correct predictions without taking into account positives and negatives [34].
The SEN (2), or rate of true positives, is the metric computed by the ratio between true positives and all positive cases [34].
The SPE (3), or false positive rate, corresponds to the proportion of false positives in relation to all other negative data [34].
The KAP (4), is a challenging metric when working with multi-class problems or when problem classes are unbalanced. Although there is no standard for interpreting its data, [27] presents a way to achieve understanding, which makes it possible to verify the degree of agreement between evaluators, where: a value less than 0 indicates non-agreement, between 0 and 0 .20 mild agreement, between 0.21 and 0.40 as fair agreement, between 0.41 and 0.60 as moderate agreement, between 0.61 and 0.80 as substantial agreement and between 0.81 and 1 as almost agreement perfect.
where,
and
The F1-Score (7) metric measures a binary classifier’s success, and it is calculated by the harmonic mean of precision and recall [28].
where,
and
Finally, the AUC is used when the classification in problem of binary solution, as is the case of this work. In practice, it corresponds to the probability that a positive example chosen at random is higher than that of a negative example being chosen also at random, and demonstrates the contrast in the prediction [7, 34].
4 Results
To evaluate the results, we divided the set of 1,011 benign and 394 malignant nodules into training (70%), validation (10%), and testing (20%). The nodule slices were resized to the fixed size of 80×80, which was the value of the largest slice of the nodules. The resizing was done using the zero-padding technique; that is, we added borders with a value of 0 until all the slices had the same dimension. The training process was carried out with the RF classifier. The values used in the first experiments were: 100 for the number of trees in the forest, the Gini function to measure the quality of a division, and unlimited depth of the tree.
The tests were carried out in three stages, which are:
-
1.
Individually pre-trained networks: In this stage, experiments were carried out by varying the number of clusters, keeping the same values for each tested architecture;
-
2.
Combining the characteristics: in this step, we use different dimensions of dimensionality through the PCA technique, to find the best dimensionality value. In addition, we used the value of k found in experiment 1.
-
3.
Search for best RF parameters: Here, we performed a search to find the best RF parameters. In this step, the search was carried out to find the ideal depth of the tree and the number of trees in the forest.
We considered kappa as the main metric for the analysis of the results, as it is more suitable for unbalanced sets. Thus, the choice of parameters was made based on this metric. We performed each experiment five times, and for each result, the mean and standard deviation are presented. Figure 2 presents the graphs of the kappa metric, containing the results obtained from the individual networks and the general average of the networks, for each value of k. Tables 3, 4, 5, 6 and 7 show the values for all metrics. The results were obtained by varying the number of clusters, and each table represents an architecture.
By analyzing the results of the Tables and Fig. 2, it is possible to observe that the k variation had no great influence on the results. The kappa results had a lower performance for k values less than 16 and greater than 512. For values less than 16, it is probably not possible to represent all the characteristics of the nodules with this smaller number of clusters. However, for k values greater than 512, the classifiers are more sensitive to overfitting due to the greater number of features, mainly due to the unbalance of the classes. Thus, values between 32 and 256 produced more consistent results, and the value 128 was the one with the highest average of results and the smallest deviation, so we chose this value to be used in next steps. The architectures present similar results and, according to Fig. 2, the one that stood out the most individually was Inception, with Acc of 87.07, F1-Score of 77.60 Kap of 0.69, Sen of 78.42, Spe of 90.74, and AUC of 0.89. The Dashed line confirms that the highest average of the architectures was obtained with 128, so this value was selected for the next step.
In Table 8 and Fig. 3 the results are presented after combining features of the VGG16, VGG19, Inception, Resnet50 and Xception architectures. In this step, we used the PCA to reduce the dimensionality of the feature vectors, the value of k = 128, and the BoF to generate signatures for each nodule. In this experiment, different sizes of dimensionality were tested, and the best results were obtained with the dimensionality of 1,024. Another highlight is that the best result was maintained in almost all metrics (Acc, Kap, Spe, and AUC), performing less only in Sen metric for dimensionality 16. However, if we analyze the Kap metric referring to the PCA equal to 1,024, it was greater than the PCA equal to 16. The combined features of different architectures provide a better representation of each nodule, especially for unbalanced sets. Also, due to the use of BoF, the signature calculation is independent of the number of slices of the nodule.
Step 3 was performed to estimate the parameters of the RF classifier. We adopted as default k = 128 and PCA= 1024 for this experiment. We used the validation set to perform this experiment to use it in the test set later. We evaluated values from 10 to 200 for maximum tree depth and 100 to 500 for the number of trees in the forest. The jump in the variation of parameters was 10 out of 10. At the end of the search, we obtained 250 for the number of trees in the forest and 100 for the maximum depth of the tree.
The result obtained in the test set is presented in Table 9. Comparing the Tables 8 (Step 2) and 9 (Step 3), we observed that the method with the new parameters found for the RF improved the values of Acc, F1-Score, Sen, Spe and AUC. As for the Kap metrics, the values were maintained about the best experiment in Step 2. The correct prediction of benign nodules caused the increase in values for the Acc, F1-Score, Sen, Spe and AUC metrics.
Based on the analysis of the results obtained in each phase of the work, we can conclude:
-
1.
The use of individually pre-trained networks did not perform well compared with works presented in the literature. However, the combination of features extracted by different architectures presented promising results.
-
2.
The cluster number variation had no great influence on the results, but it is clear that for the dataset used, the ideal value was between 32 and 512, and that 128 performed better.
-
3.
The PCA used for dimensionality reduction and the BoF to the combination of features present promising results in the representation of 3D nodules.
4.1 Discussions
The comparison of our results with other works in the literature is presented in Table 10.
Analyzing the results presented in Table 10, it is possible to observe that the works [36] and [1] have a slightly higher accuracy than our method. However, our method performed better than other works presented in Table 10. It is important to mention that the results presented in this table for each method were obtained from the original articles. Therefore, they were evaluated with different evaluation criteria and for a different number of images, using public and/or private databases.
5 Conclusion
This paper presented an approach for classifying pulmonary nodules as benign or malignant. The most recent literature focuses on using CNNs (2D and 3D) techniques in image classification problems. However, in this work we investigated the combination by BoF of features extracted by different pre-trained 2D CNNs to represent a 3D nodule. The use of pre-trained 2D CNNs was motivated by the fact that they required fewer samples for training. On the other hand, the motivation for using BoF was to make the method independent of the number of 2D slices of the nodules.
The proposed methodology presents promising results when compared to other related works. Another advantage of our method is that it works for nodules with different numbers of slices. On the other hand, methods based on training 2D and 3D CNNs require that all nodes have the same number of slices. Therefore, they use techniques to replicate or remove slices, which can decrease their performance due to the addition of noise or removal of important features.
In our work, we do not investigate the influence of pre-processing and data augmentation techniques. Thus, in future works, we will investigate the influence of these techniques, which may increase the results obtained by the proposed method.
Data Availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Ali I, Muzammil M, Haq IU, Khaliq AA, Abdullah S (2020) Efficient lung nodule classification using transferable texture convolutional neural network. IEEE Access 8:175859–175870
Anthimopoulos MM, Gianola L, Scarnato L, Diem P, Mougiakakou SG (2014) A food recognition system for diabetic patients based on an optimized bag-of-features model. IEEE J Biomed Health Inform 18(4):1261–1271
Araujo FH, Santana AM, de A Santos Neto P (2016) Using machine learning to support healthcare professionals in making preauthorisation decisions. Int J Med Inform 94:1–7
Araujo FH, Silva RR, Medeiros FN, Parkinson DD, Hexemer A, Carneiro CM, Ushizima DM (2018) Reverse image search for scientific data within and beyond the visible spectrum. Expert Syst Appl 109:35–48
Armato-III SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA et al (2011) The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Med Phys 38 (2):915–931
Ashour AS, Eissa MM, Wahba MA, Elsawy RA, Elgnainy HF, Tolba MS, Mohamed WS (2021) Ensemble-based bag of features for automated classification of normal and covid-19 cxrimages. Biomed Signal Process Control 68:102656. https://doi.org/10.1016/j.bspc.2021.102656. https://www.sciencedirect.com/science/article/pii/S1746809421002536
Avelar A (2019) O que é auc e roc nos modelos de machine learning. Disponível em: https://medium.com/@eam.avelar/o-que-%C3%A9-auc-e-roc-nos-modelos-de-machine-learning-2e2c4112033d. Accessed 2020 Feb 15
Bansal D, Khanna K, Chhikara R, Dua RK, Malhotra R (2020) Classification of magnetic resonance images using bag of features for detecting dementia. Procedia Comput Sci 167:131–137. https://doi.org/10.1016/j.procs.2020.03.190. https://www.sciencedirect.com/science/article/pii/S1877050920306554. International Conference on Computational Intelligence and Data Science
Bhatt SD, Soni HB (2021) Image retrieval using bag-of-features for lung cancer classification. In: 2021 6th International conference on inventive computation technologies (ICICT), pp 531–536. https://doi.org/10.1109/ICICT50816.2021.9358499
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 68(6):394–424
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Carvalho ED, Antonio Filho O, Silva RR, Araujo FH, Diniz JO, Silva AC, Paiva AC, Gattass M (2020) Breast cancer diagnosis from histopathological images using textural features and cbir. Artif Intell Med 105:101845
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
Costa RWD, Silva G, Filho A, Silva A, Paiva A, Gattass M (2018) Classification of malignant and benign lung nodules using taxonomic diversity index and phylogenetic distance. Med Biol Eng Comput 56. https://doi.org/10.1007/s11517-018-1841-0
da Nóbrega RVM, Peixoto SA, da Silva SPP, Rebouças Filho PP (2018) Lung nodule classification via deep transfer learning in ct lung images. In: 2018 IEEE 31st International symposium on computer-based medical systems (CBMS). IEEE, pp 244–249
de Carvalho Filho AO et al (2017) Computer-aided diagnosis system for lung nodules based on computed tomography using shape analysis, a genetic algorithm, and svm. Med Biol Eng Comput 55(8):1129–1146
Dey R, Lu Z, Hong Y (2018) Diagnostic classification of lung nodules using 3d neural networks. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp 774–778
Flehinger BJ, Kimmel M, Melamed MR (1992) The effect of surgical treatment on survival from early lung cancer: implications for screening. Chest 101 (4):1013–1018
Hansell DM, Bankier AA, MacMahon H, McLoud TC, Muller NL, Remy J (2008) Fleischner society: glossary of terms for thoracic imaging. Radiology 246(3):697–722
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Huang X, Lei Q, Xie T, Zhang Y, Hu Z, Zhou Q (2020) Deep transfer convolutional neural network and extreme learning machine for lung nodule diagnosis on ct images. arXiv:2001.01279
Huang H, Wu R, Li Y, Chao P (2022) Self-supervised transfer learning based on domain adaptation for benign-malignant lung nodule classification on thoracic ct. IEEE J Biomed Health Inform:1–1. https://doi.org/10.1109/JBHI.2022.3171851
Inca (2019) Instituto Nacional do Cancer̂ - ministério da saúde, câncer de pulmão. https://www.inca.gov.br/tipos-de-cancer/cancer-de-pulmao. Accessed 08 Feb 2019
Jabon SA, Raicu DS, Furst JD (2009) Content-based versus semantic-based retrieval: an lidc case study. In: Medical imaging 2009: image perception, observer performance, and technology assessment, vol 7263. International Society for Optics and Photonics, p 72631L
Jolliffe IT, Cadima J (2016) Principal component analysis: a review and recent developments. Philos Trans Royal Soc A: Math Phys Eng Sci 374 (2065):20150202
Ke Y, Sukthankar R (2004) Pca-sift: a more distinctive representation for local image descriptors. In: Proceedings of the 2004 IEEE computer society conference on computer vision and pattern recognition, 2004. CVPR 2004., vol 2. IEEE, pp II–II
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics:159–174
Lipton ZC, Elkan C, Narayanaswamy B (2014) Thresholding classifiers to maximize f1 score. arXiv:1402.1892
Lloyd S (1982) Least squares quantization in pcm. IEEE Trans Inf Theory 28(2):129–137
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. Int J Comput Vision 60(2):91–110
Luz DS, Costa RJ, Ricardo de Andrade LR, Rodrigues JJ, Araujo FH (2021) Automatic identification of metastasis in histopathological images using deep learning. In: 2020 IEEE International conference on e-health networking, application & services (HEALTHCOM). IEEE, pp 1–6
Luz DS, Lima TJ, Silva RR, Magalhães DM, Araujo FH (2022) Automatic detection metastasis in breast histopathological images based on ensemble learning and color adjustment. Biomed Signal Process Control 75:103564
Masood A et al (2018) Computer-assisted decision support system in pulmonary cancer detection and stage classification on CT images. J Biomed Inform 79:117–128
Mishra A (2018) Metrics to evaluate your machine learning algorithm. Disponível em: https://towardsdatascience.com/metrics-to-evaluate-your-machine-learning-algorithm-f10ba6e38234 Accessed 15 Feb 2020
Nabizadeh-Shahre-Babak Z, Karimi N, Khadivi P, Roshandel R, Emami A, Samavi S (2021) Detection of covid-19 in x-ray images by classification of bag of visual words using neural networks. Biomed Signal Process Control 68:102750. https://doi.org/10.1016/j.bspc.2021.102750. https://www.sciencedirect.com/science/article/pii/S1746809421003475
Naqi SM, Sharif M, Jaffar A (2020) Lung nodule detection and classification based on geometric fit in parametric form and deep learning. Neural Comput Applic 32(9):4629–4647
O’Hara S, Draper BA (2011) Introduction to the bag of features paradigm for image classification and retrieval. arXiv:1101.3354
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M et al (2015) Imagenet large scale visual recognition challenge. Int J Comput Vision 115(3):211–252
Safta W, Frigui H (2018) Multiple instance learning for benign vs. malignant classification of lung nodules in ct scans. In: 2018 IEEE International symposium on signal processing and information technology (ISSPIT). IEEE, pp 490–494
Safta W, Farhangi MM, Veasey B, Amini A, Frigui H (2019) Multiple instance learning for malignant vs. benign classification of lung nodules in thoracic screening ct data. In: 2019 IEEE 16th International symposium on biomedical imaging (ISBI 2019). IEEE, pp 1220–1224
Santos JD, de MS Veras R, Silva RR, Aldeman NL, Araújo F. H., Duarte AA, Tavares JMR (2021) A hybrid of deep and textural features to differentiate glomerulosclerosis and minimal change disease from glomerulus biopsy images. Biomed Signal Process Control 70:103020
Shaffie A, Soliman A, Khalifeh HA, Taher F, Ghazal M, Dunlap N, Elmaghraby A, Keynton R, El-Baz A (2019) A novel ct-based descriptors for precise diagnosis of pulmonary nodules. In: 2019 IEEE International conference on image processing (ICIP), pp 1400–1404. https://doi.org/10.1109/ICIP.2019.8803036
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Sluimer I, Schilham A, Prokop M, Van Ginneken B (2006) Computer analysis of computed tomography scans of the lung: a survey. IEEE Trans Med Imaging 25(4):385–405
Sousa LP, Veras RMS, Vogado LHS, Britto Neto LS, Silva RRV, Araujo FHD, Medeiros FNS (2020) Banknote identification methodology for visually impaired people. In: 2020 International conference on systems, signals and image processing (IWSSIP), pp 261–266
Sundar AJA (2020) Automatic 2d lung nodule patch classification using deep neural networks. In: Proceedings of the 2020 4th international conference on inventive systems and control (ICISC), pp 500–504. https://doi.org/10.1109/ICISC47916.2020.9171183
Sunitha T, Sivarani T (2022) Novel content based medical image retrieval based on bovw classification method. Biomed Signal Process Control 77:103678. https://doi.org/10.1016/j.bspc.2022.103678. https://www.sciencedirect.com/science/article/pii/S1746809422002002
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Veras R, Silva R, Araujo F, Medeiros F (2015) Surf descriptor and pattern recognition techniques in automatic identification of pathological retinas. In: 2015 Brazilian conference on intelligent systems (BRACIS). IEEE, pp 316–321
Vieira P, Sousa O, Magalhes D, Rablo R, Silva R (2021) Detecting pulmonary diseases using deep features in x-ray images. Pattern Recognit, p 108081
Vogado L, Veras R, Aires K, Araújo F, Silva R, Ponti M, Tavares JMRS (2021) Diagnosis of leukaemia in blood slides based on a fine-tuned and highly generalisable deep learning model. Sensors 21(9). https://doi.org/10.3390/s21092989. https://www.mdpi.com/1424-8220/21/9/2989
Xie Y, Zhang J, Xia Y (2019) Semi-supervised adversarial model for benign–malignant lung nodule classification on chest ct. Med Image Anal 57:237–248
Yan X et al (2017) Classification of lung nodule malignancy risk on computed tomography images using convolutional neural network: a comparison between 2D and 3D strategies. In: Lecture notes in computer science, pp 91–101
Yang J, Jiang YG, Hauptmann AG, Ngo CW (2007) Evaluating bag-of-visual-words representations in scene classification. In: Proceedings of the international workshop on workshop on multimedia information retrieval, pp 197–206
Zhang Q, Wang H, Yoon SW, Won D, Srihari K (2019) Lung nodule diagnosis on 3d computed tomography images using deep convolutional neural networks. Procedia Manuf 39:363–370
Zhu W, Liu C, Fan W, Xie X (2018) Deeplung: deep 3d dual path nets for automated pulmonary nodule detection and classification. In: 2018 IEEE Winter conference on applications of computer vision (WACV). IEEE, pp 673–681
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lima, T., Luz, D., Oseas, A. et al. Automatic classification of pulmonary nodules in computed tomography images using pre-trained networks and bag of features. Multimed Tools Appl 82, 42977–42993 (2023). https://doi.org/10.1007/s11042-023-14900-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14900-5