Abstract
Multi-class brain tumor classification is an important area of research in the field of medical imaging because of the different tumor characteristics. One such challenging problem is the multiclass classification of brain tumors using MR images. Since accuracy is critical in classification, computer vision researchers are introducing a number of techniques; However, achieving high accuracy remains challenging when classifying brain images. Early diagnosis of brain tumor types can activate timely treatment, thereby improving the patient’s chances of survival. In recent years, deep learning models have achieved promising results, especially in classifying brain tumors to help neurologists. This work proposes a deep transfer learning model that accelerates brain tumor detection using MR imaging. In this paper, five popular deep learning architectures are utilized to develop a system for diagnosing brain tumors. The architectures used is this paper are Xception, DenseNet201, DenseNet121, ResNet152V2, and InceptionResNetV2. The final layer of these architectures has been modified with our deep dense block and softmax layer as the output layer to improve the classification accuracy. This article presents two main experiments to assess the effectiveness of the proposed model. First, three-class results using images from patients with glioma, meningioma, and pituitary are discussed. Second, the results of four classes are discussed using images of glioma, meningioma, pituitary and healthy patients. The results show that the proposed model based on Xception architecture is the most suitable deep learning model for detecting brain tumors. It achieves a classification accuracy of 99.67% on the 3-class dataset and 95.87% on the 4-class dataset, which is better than the state-of-the-art methods. In conclusion, the proposed model can provide radiologists with an automated medical diagnostic system to make fast and accurate decisions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain tumors are one of the most serious health problems in the world that can affect anyone. Cancer is the second leading cause of death because one in six deaths is caused by cancer. Early classification of cancer can be life-saving, but this is not always possible. Brain tumors are one of the deadliest cancer types due to their aggressiveness and low survival rate. Since 2000, June 8 has been considered World Brain Tumor Day, the purpose of which is to raise awareness and inform people about brain tumors. The brain is a very complex and sensitive organ. It contains about 100 billion nerve cells that control the human nervous system [41]. It may be affected by the tumor. Tumors can change brain behavior. Therefore, any abnormality in the brain is dangerous to human health. Brain tumors are the uncontrolled spread of abnormal cell populations in or around the brain. Brain tumors can generally be classified as malignant and benign. A benign tumor can be removed by surgery because it will not spread to other parts of the brain, Malignant tumors are larger than benign tumors and can spread to other parts of the body. Therefore, early detection of brain tumors is essential to improve the survival rate of patients. According to the American Brain Tumor Society, about 700,000 patients in the United States suffer from brain tumor disease. Patients reportedly have a survival rate of only 36%. In the last year of 2020, approximately 87,000 patients have been diagnosed with brain tumors. In 2021, 84,170 patients from all over the world were diagnosed with brain tumors [62]. There are over 120 types of brain tumors. However, the most common types of brain tumors are glioma, pituitary gland and meningioma. Among all brain tumors, the incidence rate of glioma is 45%, pituitary tumors is 15% and meningioma is 15% [67]. Meningioma is the most common benign tumor, and it develops in the membrane that surrounds the brain and central nervous system. Pituitary tumors primarily affect the pituitary gland of the brain. On the other hand, glioma originates from the brain tissue within the substance of the brain. The main difference is that gliomas are malignant, while meningiomas and pituitary tumors are usually benign. According to the type of tumor, doctors can diagnose and predict the survival rate of patients. Therefore, tumor grading is an important part of the treatment of patients with brain tumors.
Medical imaging techniques are used to detect tumors. Medical imaging is the most economical and accurate method for diagnosing and detecting dangerous human diseases such as brain tumors detection [54], classification of skin cancer [30], stomach cancer [31] and lung cancer [32]. There are different ways to treat a brain tumor, depending on the size and type of tumor. Computed tomography (CT), magnetic resonance imaging (MRI), and other diagnostic imaging methods are used to look inside the human body. MRI is considered the first choice for brain tumors because it is the only painless medical imaging method used to provide excellent images of brain tumors. However, due to the large number of patients, viewing these images manually is time-consuming and can cause errors. MRI makes it easy to calculate the size, shape, and location of detective tissue. According to tissue characteristics, different MRI protocols are used, such as T1W1, CE-T1W1, and T2W2.
For early detection and classification of brain tumors, computer-aided diagnosis (CAD) systems may be helpful and can be used as a tool to help radiologists and doctors [3]. Automatic detection of brain tumors is necessary not only for accurate assessment and timely diagnosis, but also for saving radiologist time. Some efforts have been made to develop powerful solutions for the automatic classification of brain tumors. Over the past few years, many machine learning (ML) and deep learning (DL) methods based on feature selection and learning techniques have been proposed to classify brain tumors. There are many ways to classify brain tumors, such as machine learning methods [39, 74], fusion vectors [56], deep networks [46], and transfer learning (TL) [68]. Deep learning is much better at dealing with more complex classification problems than traditional machine learning techniques [51]. With the recent development of deep networks, there are several studies that have adopted Convolution Neural Network (CNN) for the diagnosis of brain tumors [17, 71]. The essence of this work is to find the best deep learning framework for the classification of brain cancer. In this article, an enhanced deep learning model is proposed to examine brain MRI and provide early diagnosis. Most of the research in previous work has focused on classifying binary classes. However, the binary classification is simple because the shape of the tumor can be easily interpreted. Multiclassification is difficult due to the high similarity between tumor types. We used publicly available three-class and four-class brain MRI dataset for performance analysis of our proposed model. The main findings of this study are as follows.
-
1.
We have proposed a novel and robust deep learning-based system for multiclass brain tumor classification on two benchmark datasets exploiting five state-of-the-art architectures, Xception, DenseNet201, DenseNet121, ResNet152V2, and InceptionResNetV2.
-
2.
The performance of the proposed model using deep dense block based on the Xception architecture is compared with the state-of-the-art methods. The proposed model uses various preprocessing techniques, data augmentation and deep dense block to improve classification performance. Various techniques are used to avoid overfitting, such as dropout, batch normalization, global average pooling, early stopping method, and L2 regularization.
-
3.
We also implemented 3-class and 4-class versions of the proposed model and compared the results with other studies in the literature.
2 Related work
In recent years, there have been many attempts to create an accurate and effective classification system for brain tumors. Many methods have been proposed to automatically classify MRI of the brain based on traditional machine learning and deep learning methods such as convolutional neural networks (CNNs) and transfer learning. Therefore, we conducted a detailed study of the previously proposed methods for classifying brain tumors from various sources such as Springer, IEEE Explore, and Elsevier. In the literature, most methods focus on binary classification. However, the binary classification is simple because the shape of the tumor can be easily interpreted. Due to the high degree of similarity between tumors, the multi-class classification of brain tumors is difficult.
Several authors used traditional ML methods to obtain the final output through sequential stages. Different feature extraction schemes are used, such as DWT [18, 43, 50], GLCM [37, 45], and genetic algorithm [7]. Several authors use support vector machines because it is the most popular technique for classification problems [10, 18]. Other authors used various classification methods such as Random Forest [38], Extreme Learning Machines [63], and Sequential Minimal Optimization [16]. Ullah et al. [72] extracted the approximation, used color moments (CM) to reduce the coefficients, and finally used a feedforward artificial neural network to classify the brain tumors. Zang et al. [73] used the ML paradigm to conduct brain tumor classification research, where binary classification is the main focus. In addition, it is difficult to distinguish between glioblastoma multiforme (GBM) and brain metastases (MET) using MRI. Therefore, this is another challenge faced by researchers in the field of brain tumors. Yang et al. [70] used morphological features to study MET and GBM tumor classification. Rajan and Sundar [52] proposes hybrid energy saving method consisting of 7 long stages for automatic tumor detection and reports 98% accuracy. Hence, it is noticeable that there is manual feature extraction in the traditional machine learning method, which is time consuming and error prone. Traditional ML methods relies on hand-crafted functions that require reliable upfront information, such as the location of the tumor, and the potential for human error is high. Therefore, it is necessary to develop a robust and effective method that does not use manual features. The DL method has recently been widely used in the fields of medical imaging and brain tumor classification [35]. The DL method does not require hand-crafted features; however, sometimes it is necessary to perform preprocessing operations and use the correct architecture to achieve improved classification performance. CNNs are a type of deep neural network that is widely used for classification and detection. Recently, various researchers have proposed CNN to classify brain tumors using MRI [23, 61]. Several authors used a brain tumor dataset called Figshare [12] generated by Cheng to obtain an efficient method for the classification of brain tumors. We also used the same data set for experiments in this work. Cheng et al. [11] tried to use this data set to solve three types of problems in detecting brain tumors. They used GLCM and BoW model for feature extraction and SVM to improve the classification accuracy of brain tumors to 91.28%. In 2018, the Figshare dataset was used to classify brain tumors in [26, 61]. Anaraki et al. [4] proposed CNN based on genetic algorithm to classify brain tumor types. They achieved 94% classification accuracy in brain tumor datasets by using traditional neural networks. CNN is also used by Ahmad et al. [59] for the purpose of classifying brain tumors. The proposed method uses DWT and CNN model. The overall accuracy of the experimental results reaches 99.3%. Deepak and Ameer [17] used GoogleNet model and applied transfer learning technique for the purpose of extracting MRI features. They used Figshare dataset to train and test the proposed method, and the SVM classifier is used for classification and achieves an accuracy of 97%. Saxena et al. [60] applies the transfer learning method on three deep learning models, namely Inception V3, ResNet-50 and VGG-16 models, and classifies brain tumor data in their research. The Resnet-50 model achieves the highest accuracy rate of 95%. Francisco et al. [19] proposed a multi-path CNN architecture for automatic segmentation of brain tumors. They tested their proposed model using a publicly available MRI dataset and achieved an accuracy of 97.3%. Sajjad et al. [57] uses a deep convolutional neural network (CNN) and uses data augmentation to classify brain tumors. The overall accuracy of the proposed CNN model reached 94.5%. Maharjan et al. [42] has published a multi-class brain tumor classification study to avoid overfitting. The proposed CNN claims a 2% improvement in accuracy using a modified softmax loss function. Citak et al. [14] stated that they used SVM, multilayer perceptrons and logistic regression in the study of brain tumors. As a result, they achieved 93% accuracy and 96.4% sensitivity. Khwaldeh et al. [33] proposed a CNN model to classify brain tumors by modifying the architecture of the alexnet model, with an accuracy rate of 91%. Badža and Barjaktarović [6] proposed a 4-layer CNN for extracting features from brain tumor images and performed classification. They classified brain tumors with 97.39% accuracy. Zar et al. [68] has proposed a block-by-block fine-tuning strategy based on the TL paradigm using CNN. This method was more common because it achieved an average accuracy of 94.82% without the use of handcrafted features. Sultan et al. [65] proposed a deep learning model that relies on CNNs to classify brain tumors. The proposed model achieves 96.13% and 98.7% accuracy separately. In another study, eight CNN models [28] were designed and trained on a brain MRI dataset to classify a brain tumor. The proposed CNN models have achieved an accuracy of 90% to 99%. In the research conducted by Ruba et al. [55], the semantic segmentation network was first used to segment brain images, and then the GoogleNet transfer learning model was used to classify the images. They produced almost 99% classification performance for each category. Jaeyong Kang et al. [29] used 13 different pretrained deep convolutional neural networks and 9 different ML classifiers. They experimented with three different brain tumor datasets and achieved the highest classification accuracy of 98.50%. Naseer et al. [47] proposed a CNN model for the early diagnosis of brain tumors using MR images of the brain. They used different enhancement techniques and six different datasets to train and validate the proposed model. They achieved a classification accuracy of 98.8% for the detection of brain tumors. Ercan Avşar et al. [58] proposed a deep learning model based on Faster Region-based Convolutional Neural Networks (faster R-CNN). The authors trained and tested the model using 3064 MR images of the brain, achieving 91.66%. Tanzila Saba et al. [56] used VGG-19 model for brain tumor detection by applying transfer learning techniques. The proposed method was evaluated on different datasets such as BRATS 2015–17. they achieved 98.78%, 99.63% and 99.67% accuracy on BRATS 2015, BRATS 2016 and BRATS 2017. Aderghal et al. [1] proposed a CNN model using transfer learning techniques to classify brain scans focusing only on a small ROI. they used a shallow CNN architecture with fewer layers. They used two different transfer learning techniques, namely cross-domain and cross-modal, and achieved good results even on small datasets. Various classification schemes for brain tumors are presented in the literature. Achieving better classification accuracy is a difficult task in classifying brain tumor images. It can be seen from the above research that compared with the traditional ML techniques, the accuracy of brain tumor classification using DL is much higher. In addition, it was noticed that none of the models proposed above were validated. Thus, we identified a clear gap in studies of the multiclass classification of brain tumors using the TL method. we have proposed a new DL method using the TL technique to classify brain tumors. We use brain MRI to study five unique DL models, such as Xception, DenseNet201, DenseNet121, ResNet152V2, and InceptionResNetV2, and apply TL method on two publicly available benchmark datasets. Finally, we use various important parameters for brain tumor classification to investigate and compare the above models.
3 Materials and methods
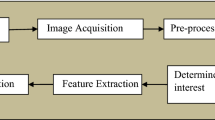
This section describes the methods and materials used in this study. Figure 1 shows the proposed approach for classifying brain tumor disease based on deep transfer learning technique. Section 3.1 details the brain tumor imaging datasets used to train the proposed method. MR images are preprocessed and cropped in Section 3.2. Section 3.3 introduces data augmentation procedures to solve the problem of limited datasets and improve classification performance. The deep transfer learning model for feature extraction and classification is introduced in Section 3.4. Finally, Section 3.5 presents various performance indicators to analyze the effectiveness of the proposed method.
3.1 Datasets for this study
Most of the latest models use the Figshare benchmark brain tumor dataset [11] to assess performance. Therefore, we also considered the same dataset to evaluate the effectiveness and robustness of the proposed method. Two different MRI datasets that are publicly available have been used to perform a set of experiments. The first data set used in this article contains 3064 T1-weighted contrast-enhanced MRI images obtained from the Nanfang Hospital and General Hospital of Tianjin Medical University, China between 2005 and 2010. It was developed by Cheng [11] in 2017 to create a classification model for brain tumors. The dataset includes 3064 brain MRI slices of anonymous 233 cancer patients. It contains three types of brain tumors: glioma (1426 images), meningioma (708 images), and pituitary tumor (930 images). The second dataset, called the Brain Tumor Classification, was downloaded from the Kaggle open source data source repository [9]. The data set contains 3264 brain MRI slices, divided into four classes: normal (500 images), glioma (926 images), meningioma (937 images), and pituitary (901 images). Figure 2 shows an example of brain MR images in 3 and 4 class datasets. In the images, the tumor is marked with a red outline.
3.2 MRI data preprocessing and cropping
Before processing the image into the proposed structure, both datasets were preprocessed at various stages to ensure maximum accuracy. Almost all images in our brain MRI dataset contain unwanted space and noise, which can reduce performance. Our goal is to crop the image to remove unwanted areas, make sure that all images are of the same type, and the focus is only on the central part of the brain. Extreme point calculation and finding contour are used to perform the above preprocessing. Figure 3 shows the process of cropping the MR image at each step. First, we load the original MR image from the dataset. After that, the MR image is converted to a binary image by applying a threshold. Then erosion and dilation operations are performed to remove any small noisy parts of the MR image, then the largest contour is selected and the four extreme points of the image (extreme right, extreme bottom, extreme top and extreme left) are calculated. Finally, we cropped the images after combining contour points and extreme points to ensure that the brain parts were in focus in each image.
The MR images from the dataset are of different sizes, and it is recommended to adjust them to the same height and width for best results. Different models have different input requirements. For example, the DenseNet201, DenseNet121, and Resnet152v2 architectures expect an image size of 224 × 224, while the Xception and InceptionResNetv2 architectures expect an input size of 299 × 299. The resize function is used to resize all brain tumor images to the shape (224 × 224) so that all architectures used in this study can accept a common size. Data partitioning also plays an important role in image classification. To start the training phase of a deep learning model, the image data is divided into three parts; training, testing and validation. According to the Pareto principle [20, 66], 80% of the images are reserved for training and validation purposes, and 20% are reserved for testing purposes. This dataset split ratio (80–20) is one of the most common split ratios in deep learning and has been used in similar studies on medical images [2, 29]. Table 1 shows the details of the images in the dataset and the distribution of the data used to train and test the model.
3.3 Data augmentation
We use image augmentation to ensure that each model receives enough input images to avoid overfitting problems due to limited images in the data set. By augmenting existing data instead of collecting new data, the classification performance of the DL model can be significantly improved. However, this study used three augmentation strategies to generate a new training set: 1) The images was rotated by an angle of 90 degrees, 2) All images are horizontally flipped, and 3) Random contrast is used to randomly adjust the contrast during training by a factor of 0.2.s.
3.4 Proposed deep transfer learning models for feature extraction and classification
Designing a CNN from scratch is a challenging task. This process requires multiple iterations, a lot of experience to ensure correct convergence, and involves careful setting of many hyperparameters (such as architecture depth). Therefore, leveraging existing recognized pre-trained models (Xception, DenseNet201, DenseNet121, ResNet152V2, InceptionResNetV2, etc.) for the classification of brain tumors is an alternative solution. In the field of medical imaging, there is a lack of labelled data and this is a major challenge for a reliable and accurate detection system. Therefore, using pre-trained models with TL technique to quickly learn new jobs and solve these challenges. In this research, data augmentation technique and deep transfer learning are carried out to overcome the problem of insufficient training data and reduce the problem of overfitting. In TL, a CNN trained for a specific task can be reused for another related task. Moreover, the TL approach was found to be much faster and simpler than a network trained from scratch. Here, we examine five pre-trained models Xception, DenseNet201, DenseNet121, ResNet152V2, InceptionResNetV2 using MR images and apply TL technique on the given dataset. Figure 4 shows the details of the layers and their order in the deep dense block (DDB). First, we remove the fully connected layer from these architectures, leaving only the convolutional layer and the pooling layer. These two types of layers are responsible for extracting features. In Table 2, we also provide architectural details of the model, such as the required input image size and the number of spatial feature maps extracted from the convolution base. All parameters are initialized with weights obtained from the ImageNet dataset. We have introduced a deep dense block to improve the accuracy of the brain tumor classification. In the deep dense block, first we added a global average pooling layer as a better alternative to the flattening layer. It transforms the (H × W × N) feature map into a (1 × N) feature map, where (H × W) represents the size of the image and N represents the number of filters. Instead of adding a fully connected layer, it is more meaningful and interpretable because it enforces the correspondence between feature maps and categories. Other benefits of the global average pooling layer are that it better solves the overfitting problem and enables a direct mapping between output channels and feature categories, reducing the number of parameters and eliminating the need for parameter optimization [40]. Then, three layers of batch normalization, dropout, and three dense layers are added to the network, where the first and second dense layers are composed of 512 and 256 neurons with ReLU activation functions. The parameters used in the dense layer are learned at each epoch and incorporate features of brain tumors that help improve classification accuracy. ReLU is a commonly used activation function in dense layers because it can improve training and testing performance. Overfitting is a big problem in deep networks, which occurs when a model is over trained on the training data and negatively affects the test data. The dropout layer prevents the model from being overfitted. We drop 20% of the neurons after the first dropout layer. In addition, such an operation greatly helps to speed up the training process of the models [64]. L2 regularization [15] was used in the first dense layer with a value of 0.0001. The batch normalization layer is used after each dense layer to normalize the extracted features to the mean and standard deviation, which plays an important role in our classification model. Batch normalization performs very well when used immediately after dense layers [25]. It is used to train models faster and balance activation values, and to achieve better generalization performance. In the deep dense block, the last dense layer contains 3 and 4 neurons for the classification of 3 and 4 classes of brain tumor. The softmax activation function is used to classify the image into its corresponding class. The softmax function converts the resultant values between 0 and 1 so that they can be interpreted as probabilities. It is defined as the following equation:
Figures 5, 6, 7, 8 and 9 highlights the basic architecture and customization of deep transfer learning models, which have finally been deployed to obtain the classification results of brain MR images. Below we briefly describe the architecture of these models.
3.4.1 Xception
Xception takes Inception’s method to the extreme, developed by Francois Chollet [13]. It is proposed as an improved version of Inception V3. This architecture completely relies on the depth of the separable convolutional layer, and strongly assumes that the spatial and cross-channel correlations can be separated. A depthwise separable convolution consists of a deep convolution, which runs independently on all input channels., followed by a pointwise convolution to map the correlation between the channels. The network consists of 14 modules. There are linear residual connections around all other modules except the first and last modules. When training on the ImageNet dataset, the top-1 accuracy reported by the Xception framework is 79.0%, and the top-5 accuracy is 94.5%. The classification performance of Xception networks on ImageNet datasets is slightly better than InceptionV3. Due to its outstanding performance in different image classification tasks, we use the Xception model to classify brain tumors.
3.4.2 DenseNet
DenseNet, an abbreviation for Dense Convolutional Network, requires fewer parameters than traditional CNNs because it does not learn redundant feature maps. Huang et al. [24] introduced the DenseNet network, which connects each layer of the network to each other layer in a feed-forward manner. DenseNets has the same advantages as ResNets and has some attractive properties. For example, the problem of vanishing gradients, achieving high performance and a significant reduction in the overall training parameters of the network. Deep DenseNet is built with multiple dense blocks, where each layer is a sequence of convolution operations, batch normalization, and ReLU activation. DenseNet introduces a bottleneck layer to prevent the number of feature maps from growing exponentially. In order to resolve the difference in the size of feature maps, DenseNet applies a transition layer between dense blocks. DenseNet has four different variants: DenseNet264, DenseNet201, DenseNet169, and DenseNet121. In our research, we experimented with two DenseNet variants: 121-layer and 201-layer architectures. We use the DenseNet121 CNN model, which requires fewer parameters and is computationally efficient. It can improve the training time by finding the gradient values directly from the loss function. DenseNet169 has over 14 million parameters and a model size of 57 MB, while the DenseNet121 network has about 8 million parameters and a model size of 33 MB, which significantly reduces the computational cost and makes it a superior choice.
3.4.3 ResNet152V2
ResNet152V2 is a deep residual network developed by He et al. [22] as an updated version of ResNet152. ResNet contains a large number of layers and has powerful performance. We choose ResNet152V2 because it has the highest accuracy in the ResNet family [22]. Although the depth is greatly increased, ResNet with 152 layers is still less complex than many other architectures such as VGG16 and VGG19. Deeper models will lead to better feature extraction performance. But due to back propagation, very deep models are difficult to train due to vanishing gradients. ResNet solves this problem by adding residual connections to reduce the impact of vanishing gradient. The significant difference between ResNet-V1 and ResNet-V2 is that ResNet-V2 uses batch normalization and ReLU activation before each weight layer. When using this architecture to train on the ImageNet dataset, it reported a top-1 error rate of 21.1% and a top-5 error rate of 5.5%.
3.4.4 InceptionResNetV2
InceptionResNetV2 [69] is a modified version of the Inception model that includes the idea of residual learning to improve model performance. Residual connections also shorten training time. This network is built by integrating a combination of Inception and ResNet architectures. The batch normalization is only used on the top of the traditional layer. InceptionResNetV2 replaces the filter concatenation stage with residual connections to take advantage of the two approaches (i.e. get deeper and wider) while maintaining the same computational efficiency. A 1 × 1 convolutional layer follows each inception block, and no activation is performed to match the dimensionality of various feature maps. The model contains three different types of blocks, namely. The InceptionResNet block, the Reduction block and the Stem block. These blocks contain convolutional layers, pooling layers and activation functions. The stem block accepts the input and computes three 3 × 3 convolutions on the input data. This is followed by three inception blocks, where the first and third blocks consist of two paths: 3 × 3 convolution operations and max pooling. The second block includes two paths with 1 × 1 and 3 × 3 convolution operations, while the other path has 3 × 3, 1 × 7 and 7 × 1 convolution operation. InceptionResNetV2 has three types of inception modules, i.e. InceptionResNet-A uses 35 × 35 grid modules, Inception-ResNet-B uses 17 × 17 grid modules, and Inception-ResNet-C uses 8 × 8 grid blocks. Finally, there are two reduction modules in InceptionResNetV2 that use convolution and maximum pooling to reduce the number of features. The Reduction-A block has two paths for convolution and one path for maximum pooling. Reduction-B block has 3 paths for convolution operations and 1 max pooling.
3.5 Performance evaluation metrics
The performance of the models for classifying brain tumors was assessed based on several indicators: accuracy, sensitivity, precision, specificity, and F1-score. Correspondingly, a confusion matrix is introduced to visualize diagnostic instances of MR images from the proposed model. In the equations below, the overall performance of the trained model using the proposed method was calculated using test data.
In the equations above, TP means true positives, FP means false positives, TN means true negatives, and FN means false negatives.
4 Results and experiments
This section presents the experimental setup and results of the five architectures used in the study. We analyzed the effectiveness of the deep transfer learning models proposed in this study along with the competitive model. Two publicly available brain tumor datasets are considered for evaluation of the proposed method. These datasets are more visible and most useful for this area. The main goal of this task is to improve the accuracy of multiclass brain tumor classification.
4.1 Experimental settings
We trained the proposed deep transfer learning models using the Python programming language and the Keras framework. All experimental studies are performed on the Google Colaboratory notebook using the GPU runtime type. This software is provided by Google for research activities and is free to use. The model was trained using the NVIDIA Tesla K80 with 12 GB of memory and 16 GB of RAM.
4.2 Hyperparameter and optimization techniques
The main goal of this task is to design the optimal model for multiclass brain tumor classification. This can be achieved by finding the best hyperparameter configuration so that the model can have increased recognition capability. The set of parameters that can affect model training are called hyperparameters. Parameters including the number of layers, learning rate, number of epochs and activation functions play a vital role in the performance of the model. In this study, we trained five deep transfer learning models and adjusted hyperparameters for optimal configuration. In the training process, we first trained only the DDB that were added on top of the pre-trained models. The convolutional base of the pre-trained models was completely frozen, so the weights of these layers did not change during training. It is necessary to freeze the convolutional base by setting the model trainable parameters to false in order to avoid destroying the pre-learned filters. We use the Adam optimizer with a learning rate of 1e-2 to train our new DDB on 50 epochs using our data augmentation method. The initial training of the model runs fast because we keep the convolution base frozen and only train the DDB. After training the DDB, we unfreeze some layers of the convolutional base of the models and jointly train these unfrozen layers and the novel DDB. This time the models was trained once more on the same dataset for 50 epochs using the Adam optimizer with a low learning rate of 1e-3. Fine-tuning the entire network is not recommended because the risk of overfitting is high due to the large number of parameters and the small dataset. Table 3 shows the complete details of the hyperparameters used to train the models.
Most of the above selected hyperparameters that we used were motivated by related work on the classification of brain tumors [5, 21, 29, 36, 44, 48, 49, 53]. In order to avoid overfitting during training, a regularization function (L2) is used. We use L2 regularization with a fixed value of 0.001 in the first dense layer. This means using function solvers appropriately to prevent the network from overfitting. We have used a variety of techniques to prevent overfitting of the models. As discussed earlier, TL is an effective method when there is a risk of overfitting. The image data is also augmented to avoid overfitting due to the limited data size. After that, batch normalization and global average pooling are also applied to prevent overfitting of the models. We chose Adam as the optimizer function because it combines the advantages of the AdaGrad and RMSProp algorithms, and it has been found to work quite well in practice. AdaGrad is suitable for computer vision problems and works well for sparse gradients, and RMSProp works well for non-stationary settings. The Adam optimizer is quite computationally efficient and is specifically designed for training deep models [34]. Most similar studies have also used the Adam optimizer and have achieved better results than other optimizers. Also, Adam is currently recommended as the default algorithm as it generally performs better than other algorithms. Therefore, we use the Adam optimizer with an initial learning rate of 1e-3 to train the proposed model. In this work, a categorical cross-entropy loss function is utilized since our work is based on 3 and 4 class classification of brain MRI datasets. Overfitting can also be reduced by introducing dropout during training. We dropped 20% of the neurons in the dropout layer and found this to be the best. All models are trained for 50 epochs. we use the early stopping method to stop training if the accuracy of the validation dataset does not change within a predefined number of epochs to avoid overfitting the system. It improves generalization and helps the model to avoid overfitting by doing useless epochs, which would take a long time and reduce accuracy [8]. We chose this number of epochs because experimental observations showed that the proposed model converged well and achieved the desired accuracy within 50 epochs. The code used in this work is available to facilitate future research (https://github.com/sohaibasif1592/M-R-I).
4.3 Experimental results: Three class classification
In our study, a total of five deep transfer learning models were developed, and the performance of each model was assessed in terms of the indicators described in Section 3.5. We report a comparative analysis of each individual architecture. The main purpose of this study is to test the success of the deep transfer learning model proposed in this study for the multiclass classification of brain tumors and compare it with the performance of the most advanced CNN model in the literature. As described in Section 3.4, TL was used to train all deep learning models. All experimental studies were performed on both the original and cropped datasets. In this context, the average accuracy, sensitivity, specificity, precision, and F1-score obtained by all models on the test dataset for the original and cropped datasets are given in Table 4, respectively. The values marked in bold in Table 4 represent the best model for the relevant performance criteria. In the original dataset, the average accuracy values for all models are very close to each other, as shown in Table 4. The original dataset shows that the Xception + DDB and ResNet152V2 + DDB outperform other models in almost every performance metric. As shown in Table 4, in the cropped dataset, the Xception + DDB model achieves the best overall performance with an accuracy of 99.67% Moreover, the model achieves the highest sensitivity, specificity, precision and F1-score of 99.54%, 99.83%, 99.69% and 99.62%. The Xception + DDB model also showed good sensitivity, which is important because we want to limit the rate of misdiagnosis of brain tumors as much as possible. The results show that Xception + DDB can more accurately distinguish between brain tumor types. The possible reason is that in Xception, the depth-wise separable convolution is replaced by the general convolution, thus making the model computationally efficient. Depth-wise separable convolutions are more productive and have a stronger expressive ability than classical convolutions. The depth-wise separable convolution in the Xception model makes the model highly efficient in learning several distinct and high-level features that some simpler models may ignore. DenseNet201 + DDB performed quite well, with an accuracy rate of 97.06%, while the sensitivity, specificity, and F1-score reached 96.28%, 98.24%, and 97.10%, respectively. While all the models used in the study increased the accuracy with varying differences on the cropped dataset, the Xception + DDB model, which offered the best performance on the cropped dataset, improved in accuracy and sensitivity with 4.06% and 4.13%, respectively. This showed us that cropping consistently outperformed the original dataset. Therefore, we only continue to experiments with the cropping strategy.
Figure 10 shows the accuracy obtained in the test set of the original dataset and the cropped dataset of all models. The test accuracy shown in this figure was calculated as the ratio of the number of correctly classified patients to the number of all patients. It can be clearly seen from the figure that the Xception + DDB model is superior to the other four proposed models in terms of accuracy. It can be seen from the test accuracy curve that the success rate of all models on the cropped dataset is higher than that of the original dataset. Our results show that cropping images is the best strategy to provide superior classification over the original dataset.
The class-wise performance of the models is presented in Table 5 with best result highlighted in bold. A total of 2451 images were used for training and 613 images were used for testing. Three different classes are analyzed such as glioma, meningioma and pituitary. From the results table, we can observe that the Xception + DDB model performed well in all classes. The model achieves an average precision of 1.0, an average sensitivity of 1.0, and an average F1-score of 1.0. It can be seen that this model achieves a precision of 1.0 for the glioma and meningioma classes and 0.99 for the pituitary class. An ideal sensitivity of 1.0 is achieved for the pituitary and glioma classes, and a sensitivity of 0.99 is achieved for the meningioma class. When considering the macro-average scores of all evaluation indicators, we observe that the Xception + DDB model provides better performance than other models. The confusion matrix in Fig. 11 summarizes the details class-wise results of the Xception + DDB model. By observing the confusion matrix, we get an idea of the results for specific classes in terms of the number of correctly classified and misclassified images. From the confusion matrix, we can conclude that the Xception + DDB model only made 2 misclassifications on the test dataset. These misclassifications occurred in the class of meningioma. We see that out of 613 tests, and our model is correct on 611 tests. Therefore, from the evaluation metrics achieved by the proposed model, we can conclude that Xception + DDB performs better than other models in all aspects. The Xception + DDB model shows an average increase of 2–3% in all evaluation parameters such as accuracy, precision, sensitivity, and F1-score.
4.3.1 Performance comparison with baseline models
To highlight the advantages of the proposed model, we compared our proposed method by benchmarking its performance against five base models: Xception, DenseNet121, DenseNet201, InceptionResNetV2, and ResNet152V2. We use the same dataset to build the base model for comparison, but without DDB to train the network. Table 6 reports the classification performance between the baseline and the proposed model in terms of classification accuracy, sensitivity, precision and F1-score with best result highlighted in bold. It was found that the accuracy of the proposed Xception + DDB model is 7.15% higher than the baseline model, the accuracy of DenseNet201 + DDB is 7.79% higher than the baseline, and the accuracy of DenseNet121 + DDB is 5.03% higher than the baseline, which proves that the multiple dense layers used in DDB lead to enhanced learning ability of the model, thus improving the accuracy of the model. It can be seen that DDB significantly improves the detection rate and has better stability than the baseline for the classification of brain tumors. The baseline model performs very poorly on the test set, providing accuracies between 86% and 93%. The sensitivity and F1-scores are also below the acceptable range. There are several reasons why base models perform worse: (a) different classes in the dataset, (b) The biggest reason behind their poor performance is overfitting, and (b) difficulty in extracting features from MRI images due to the high degree of similarity between tumors. The results in Table 6 show that after adding the DDB to the models, the classification performance has improved significantly compared to the baseline. It is clear that the approach proposed in this work is much better than that of the baseline models.
4.4 Experimental results: Four class classification
This section presents the classification results of the four classes. There are a total of 2611 images for training and 653 images for testing. Four different classes are analyzed, such as glioma, meningioma, pituitary and normal. Table 7 summarizes the average evaluation metrics of the competitive deep transfer learning models on the test dataset with best result highlighted in bold. All values are shown as percentages and best results are shown in bold. The Xception + DDB model outperforms other models in almost every performance metric, including accuracy, precision, sensitivity, F1-score and specificity. In this case, we see that the proposed model based on Xception architecture achieves a very impressive average classification accuracy of about 95.87% on the test dataset for classifying glioma, meningioma, pituitary, and normal patients. The model also achieved an average sensitivity rate of 95.60% and an average specificity rate of 98.56%, which are two very important performance indicators in medical applications. ResNet152V2 + DDB performed poorly on the test dataset and gave the lowest accuracy values of 93.11%. Compared to other models, the proposed model delivers satisfactory performance with an average improvement of about 1% on all metrics.
Table 8 shows the class-wise performance of the models with best result highlighted in bold. The different classes used in the study are glioma, meningioma, pituitary and normal. It can be seen from the result table that the Xception + DDB model performs well in all classes. As shown in Table 8, the model achieves an average precision of 0.96, an average sensitivity of 0.96, and an average F1-score of 0.96. It can be seen that this model achieves 0.99 precision in both the pituitary and normal classes and 0.94 and 0.93 precision in the glioma and meningioma classes. However, the model receives a sensitivity rate of 0.98 for the pituitary class. The model also has a high sensitivity to the others classes. When considering the macro-average scores of all evaluation indicators, we observe that the proposed model provides better performance than other models. The DenseNet121 + DDB network was found to be the second-best predictor of brain tumors, with an average precision of 0.95, an average sensitivity of 0.95, and an F1-score of 0.95. The confusion matrix is the main tool for evaluating errors in classification problems. We have constructed a confusion matrix for the proposed model based on Xception as shown in Fig. 12. The figure shows that the proposed model can successfully classify four patient classes (glioma, meningioma, pituitary, and normal) with the highest ratio to pituitary images (0.9824), then meningioma (0.9551), then glioma (0.9476) and finally normal (0.9375). By observing the confusion matrix, the results obtained from the test dataset are good. We see that out of 653 test images, the proposed model correctly classified 626 cases and misclassified 27 cases. It produced acceptable results with an overall accuracy of 95.87%. This result ensures that the classification is performed correctly for the four classes. Real-time detection of the presence of a tumor in the human brain can be performed using this model.
4.4.1 Performance comparison with baseline models
To show the effectiveness of our proposed method in classifying brain tumors into 4 classes, we compared the performance of the proposed model with the base models. Table 9 shows the detailed comparison results of the proposed model with the baseline models. The performance analysis shows that the proposed model shows a performance improvement over the baseline models in terms of accuracy, sensitivity, precision, and F1-score. It was found that the accuracy of the proposed Xception + DDB model was 6.44% higher than the Xception baseline model, and the accuracy of DenseNet121 + DDB was 6.74% higher than the DenseNet121 baseline, which proved that DDB can significantly improve the accuracy of the model. It can be seen that DDB significantly improves the detection rate and has better stability than the baseline for classifying brain tumors.
4.5 Proposed model comparison with different optimizers
In this experimental setting, various optimizers, namely Adam, SGD and RMSProp, are explored to obtain the superior classification accuracy of the proposed Xception + DDB model. Initially, the training phase of the proposed model is carried out by empirically selecting the Adam optimizer. To evaluate the effectiveness of the proposed model using the Adam optimizer, its results are compared with two popular optimization methods, namely RMSProp and SGD. Table 10 shows the effect of different optimizers on the proposed model using two different brain MRI datasets, i.e., 3-class and 4-class datasets. The proposed model with Adam optimizer achieves 99.67% classification accuracy on the 3-class brain MRI dataset. Then, using RMSProp and SGD, the classification accuracy of brain MRI was 95.61% and 96.75%, respectively. Adam has better classification accuracy compared to RMSProp and SGD. However, the proposed model achieved a similar classification accuracy of 95.87% on a 4-class brain MRI dataset using the Adam and RMSProp optimizers. The classification accuracy of brain MRI using SGD was 93.11%. We can see that the proposed model adapted all three optimizers very well for 3-class and 4-class classification of MRI brain images.
4.6 Computational cost
In addition to the classification performance of the models proposed for brain tumor classification, comparisons were also made in terms of computational costs. Table 11 compares the performance of the proposed models with the base models in terms of total training time, time per epoch, test time, and number of parameters. The total training time of the Xception + DDB model proposed for 3-class and 4-class classification took 3950.63 seconds and 1772.87 seconds. On the other hand, although the base models finished training before our method, they did not get the best classification results. As can be seen from the table, our proposed method showed the best performance among the base models, except for the training time. However, our aim here is to improve the accuracy of the system. The results show that after adding DDB to the model, the performance is significantly improved compared to the baseline, while the number of parameters is only slightly increased, not exceeding 1 M. Small increase in model parameters is acceptable compared to large increase in the diagnostic outcome.
4.7 Comparison with state-of-the-art methods
The performance of the proposed model based on Xception architecture is compared with the most recent competitive models. Several papers use the same dataset to classify brain tumors. To compare our results with those of previous studies, we selected only those articles that used the Figshare brain tumor dataset. It is noted that accuracy is the main metric used to compare classification results. Table 12 contains a comparison of the proposed model with benchmark studies in the literature using the same dataset. This table shows that the proposed model is superior to the existing models with 99.67% accuracy on 3-class dataset and 95.87% on 4-class dataset. In addition, most of the existing work focuses on three or four class classifications. As far as the author knows, there is no similar study that addresses both three- and four-class classification of brain tumors. The proposed model can effectively solve the three and four classes problem.
4.8 Strengths, limitations and future work
Until now, most brain tumor research has focused on binary or three-class problems. As previously described, experimental studies of the system were conducted using two publicly available datasets that classified brain tumors into three or four tumor classes. According to the Pareto principle, 80% of the images are reserved for training and validation, and 20% are reserved for testing. We use image augmentation to ensure that each model receives enough input images to avoid overfitting problems due to the limited number of images in the dataset. For best performance, these datasets were trained using transfer learning based on five deep neural networks: Xception, DenseNet201, DenseNet121, ResNet152V2, and InceptionResNetV2 for predicting brain tumors in MR images. The last layer of these architectures has been modified with our deep dense block and the softmax layer as the output layer. The deep dense block contains three dense layers with output layer to improve the classification accuracy of the deep transfer learning model proposed for multi-class classification. The dense layer used in the deep dense block adapts the features of brain tumors that significantly improves classification performance. The proposed model does not require separate feature extraction because the model uses a deep neural network. We use the global averaging pooling in our deep dense block as a flattening layer to convert the multidimensional feature map into a one-dimensional feature vector. It helps to reduce overfitting by reducing the total number of parameters and does not require parameter optimization. We use batch normalization immediately after each dense layer to increase the stability of the model. The motivation for using the early stop method is to end training if there is no improvement in order to avoid the system being overfitted and to prevent poor generalization performance. Dropout layers and L2 regularization are used to minimize overfitting and allow it to produce meaningful predictions with reasonable accuracy. The proposed model uses ADAM, which is one of the most popular gradient descent optimization algorithms, as it combines the advantages of AdaGrad and RMSProp. It is computationally efficient for deep neural networks. The classification results of the proposed models are calculated using various evaluation metrics such as accuracy, sensitivity, precision, and F1-score. This study compares five DL models, and it turns out that Xception + DDB has advantages over the other models. On the one hand, the deep separable convolution in Xception is more productive than the general convolution. It makes the model highly efficient in learning high-level features that some simpler models may ignore. On the other hand, point wise convolution, i.e. 1 × 1 convolution, is performed on each channel before using the depth-wise convolution to make the model computationally efficient. Another reason is the use of depth-wise separable convolutional layers and residual connections, which enables the model to learn richer representations from brain MR images. In addition, the absence of any non-linearity makes the model highly efficient in all performance measures. As shown in Tables 4 and 7, the proposed model Xception + DDB provides good accuracy, reaching 99.67% for 3-class classification of brain tumors and 95.87% for 4-class classification. Furthermore, the obtained results are compared with some existing methods. As shown in Table 12, it is clear that our proposed model outperforms the benchmark studies in terms of classification accuracy. Based on these encouraging results, we believe our “Xception + DDB” model will benefit doctors in diagnosing and detecting brain tumors. We believe that the model is effective in classifying MRI brain tumors with low misdiagnosis rates and helps doctors make accurate decisions. The method proposed above for classifying brain tumors is a major strength of this article.
In the absence of brain tumor data, this study is limited to single-institutional data. MR images need to be increased for better model training. In addition, the study used single protocol T1W MRI data. The system can be made more robust by merging multiple MRI protocols.
5 Conclusion
The article focuses on the development of an automated deep learning system for the multiclass classification of brain tumors. Due to the high similarity between tumor types, the multi-class classification of brain tumors is a complex task. In earlier works, the four-class paradigm was absent for the classification of brain tumors. In this work, we conducted experiments on the classification of 3 and 4 types of brain tumor patients; and all images were enhanced using image processing techniques in the pre-processing stage. Five popular deep learning architectures utilizing deep transfer learning technique are adopted for brain tumor detection by analyzing MR images. The last layer of these architectures has been modified with our deep dense block along with softmax layer to improve classification performance. We propose a deep learning model based on the Xception architecture to detect brain tumor cases using MR images. The proposed model uses depthwise-separable convolution, which makes the model highly efficient in learning several distinct and high-level features, while the deep dense block significantly improves the performance. The proposed model demonstrates fast learning through the use of Adam optimizer, while batch normalization, data augmentation, global average pooling and dropouts avoid the model overfitting issues. The proposed model achieves 99.67% and 95.87% overall classification accuracy on 3-class and 4-class dataset. From the results, it is clear that the proposed method gives the best performance on the selected dataset among all other models used in this study. Our proposed model is superior to the existing models in terms of classification accuracy. Therefore, the proposed model can be used as a tool that can accurately identify multiple types of brain tumors. In future work, we will extend this work to experiment with more brain MRI data without compromising performance.
References
Aderghal K, Afdel K, Benois-Pineau J, Catheline G, Alzheimer's Disease Neuroimaging Initiative (2020) Improving Alzheimer's stage categorization with Convolutional Neural Network using transfer learning and different magnetic resonance imaging modalities. Heliyon 6(12):e05652
Alanazi MF, Ali MU, Hussain SJ, Zafar A, Mohatram M, Irfan M, AlRuwaili R, Alruwaili M, Ali NH, Albarrak AM (2022) Brain tumor/mass classification framework using magnetic-resonance-imaging-based isolated and developed transfer deep-learning model. Sensors 22(1):372. https://doi.org/10.3390/s22010372
Amin J, Sharif M, Yasmin M, Fernandes SL (2020) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn Lett 139:118–127. https://doi.org/10.1016/j.patrec.2017.10.036
Anaraki AK, Ayati M, Kazemi F (2019) Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybernetics Biomed Eng 39(1):63–74. https://doi.org/10.1016/j.bbe.2018.10.004
Ansari M, Mehrotra R, Agrawal R (2020) Detection and classification of brain tumor in MRI images using wavelet transform and support vector machine. J Interdiscipl Math 23(5):955–966. https://doi.org/10.1080/09720502.2020.1723921
Badža MM, Barjaktarović MČ (2020) Classification of brain tumors from MRI images using a convolutional neural network. Appl Sci 10(6):1999. https://doi.org/10.3390/app10061999
Bangare SL, Pradeepini G, Patil ST (2017) Brain tumor classification using mixed method approach. In: 2017 International Conference on Information Communication and Embedded Systems (ICICES), Chennai, pp 1–4. https://doi.org/10.1109/ICICES.2017.8070748
Bengio Y (2012) Practical recommendations for gradient-based training of deep architectures, In Neural networks: Tricks of the trade. Springer, Berlin, Heidelberg, pp 437–478
Bhuvaji S, Kadam A, Bhumkar P, Dedge S, Kanchan S (n.d.) Brain tumor classification (MRI) dataset. Available: https://www.kaggle.com/sartajbhuvaji/brain-tumor-classification-mri. Accessed 5 Aug 2021
Chauhan S, More A, Uikey R, Malviya P, Moghe A (2017) Brain tumor detection and classification in MRI images using image and data mining. In: 2017 International Conference on Recent Innovations in Signal processing and Embedded Systems (RISE), Bhopal, pp 223–231. https://doi.org/10.1109/RISE.2017.8378158
Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, Wang Z, Feng Q (2015) Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS One 10(10):e0140381. https://doi.org/10.1371/journal.pone.0140381
Cheng J, Yang W, Huang M (2017) Brain Tumor Dataset. https://doi.org/10.6084/m9.figshare. 2017;1512427:v5
Chollet F (2017) Xception: Deep learning with depthwise separable convolutions. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, pp 1800–1807. https://doi.org/10.1109/CVPR.2017.195
Citak-Er F, Firat Z, Kovanlikaya I, Ture U, Ozturk-Isik E (2018) Machine-learning in grading of gliomas based on multi-parametric magnetic resonance imaging at 3T. Comput Biol Med 99:154–160. https://doi.org/10.1016/j.compbiomed.2018.06.009
Cortes, C, Mohri, M, Rostamizadeh, A (2012) L2 regularization for learning kernels. arXiv preprint arXiv:1205.2653
Deepa SN, Aruna Devi B (2011) Neural networks and SMO based classification for brain tumor. In: 2011 World Congress on Information and Communication Technologies, Mumbai, pp 1032–1037. https://doi.org/10.1109/WICT.2011.6141390
Deepak S, Ameer P (2019) Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med 111:103345. https://doi.org/10.1016/j.compbiomed.2019.103345
Devi T Menaka, GR, Arockiaraj SX (2018) MR brain tumor classification and segmentation via wavelets. In: 2018 International Conference on Wireless Communications, Signal Processing and Networking (WiSPNET), Chennai, pp 1–4. https://doi.org/10.1109/WiSPNET.2018.8538643
Díaz-Pernas FJ, Martínez-Zarzuela M, Antón-Rodríguez M, González-Ortega D (2021) A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare 9(2):153. https://doi.org/10.3390/healthcare9020153
Dunford R, Su Q, Tamang E (2014) The Pareto Principle. Plymouth Student Sci 7(1):140–148
Gumaei A, Hassan MM, Hassan MR, Alelaiwi A, Fortino G (2019) A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 7:36266–36273. https://doi.org/10.1109/access.2019.2904145
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Hemanth DJ, Anitha J, Naaji A, Geman O, Popescu DE (2018) A modified deep convolutional neural network for abnormal brain image classification. IEEE Access 7:4275–4283. https://doi.org/10.1109/access.2018.2885639
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, pp 2261–2269. https://doi.org/10.1109/CVPR.2017.243
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. Int Conf Mach Learn PMLR 37:448–456
Ismael MR, Abdel-Qader I (2018) Brain tumor classification via statistical features and back-propagation neural network. In: 2018 IEEE International Conference on Electro/Information Technology (EIT), Rochester, pp 0252–0257. https://doi.org/10.1109/EIT.2018.8500308
Ismael SAA, Mohammed A, Hefny H (2020) An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif Intell Med 102:101779
Kalaiselvi T, Padmapriya ST (2021) Brain tumor diagnostic system—a deep learning application. Machine Vision Inspection Systems, Volume 2. Mach Learn Based Approaches 27:69–90
Kang J, Ullah Z, Gwak J (2021) MRI-based brain tumor classification using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 21(6):2222. https://doi.org/10.3390/s21062222
Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS (2020) Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recogn Lett 129:293–303. https://doi.org/10.1016/j.patrec.2019.11.034
Khan MA, Sarfraz MS, Alhaisoni M, Albesher AA, Wang S, Ashraf I (2020) StomachNet: optimal deep learning features fusion for stomach abnormalities classification. IEEE Access 8:197969–197981. https://doi.org/10.1109/access.2020.3034217
Khan MA, Rubab S, Kashif A, Sharif MI, Muhammad N, Shah JH, Zhang Y-D, Satapathy SC (2020) Lungs cancer classification from CT images: An integrated design of contrast based classical features fusion and selection. Pattern Recogn Lett 129:77–85. https://doi.org/10.1016/j.patrec.2019.11.014
Khawaldeh S, Pervaiz U, Rafiq A, Alkhawaldeh RS (2018) Noninvasive grading of glioma tumor using magnetic resonance imaging with convolutional neural networks. Appl Sci 8(1):27. https://doi.org/10.3390/app8010027
Kingma, DP, Ba, J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980
Kleesiek J, Urban G, Hubert A, Schwarz D, Maier-Hein K, Bendszus M, Biller A (2016) Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. Neuroimage 129:460–469. https://doi.org/10.1016/j.neuroimage.2016.01.024
Kokkalla S, Kakarla J, Venkateswarlu IB, Singh M (2021) Three-class brain tumor classification using deep dense inception residual network. Soft Comput 25(13):8721–8729. https://doi.org/10.1007/s00500-021-05748-8
Kumar PMS, Chatteijee S (2016) Computer aided diagnostic for cancer detection using MRI images of brain (Brain tumor detection and classification system). In: 2016 IEEE Annual India Conference (INDICON), Bangalore, pp 1–6. https://doi.org/10.1109/INDICON.2016.7838875
Latif G, Butt, MM, Khan, AH, Butt, O, Iskandar, DA (2017) Multiclass brain Glioma tumor classification using block-based 3D Wavelet features of MR images. In: 2017 4th International Conference on Electrical and Electronic Engineering (ICEEE), Ankara, pp 333–337. https://doi.org/10.1109/ICEEE2.2017.7935845
Lavanyadevi R, Machakowsalya M, Nivethitha J, Niranjil Kumar A (2017) Brain tumor classification and segmentation in MRI images using PNN. In: 2017 IEEE International Conference on Electrical, Instrumentation and Communication Engineering (ICEICE), Karur, pp 1–6. https://doi.org/10.1109/ICEICE.2017.8191888
Lin, M, Chen, Q, Yan, S (2013) Network in network. arXiv preprint arXiv:1312.4400
Louis DN, Perry A, Reifenberger G, Von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. https://doi.org/10.1007/s00401-016-1545-1
Maharjan S, Alsadoon A, Prasad P, Al-Dalain T, Alsadoon OH (2020) A novel enhanced softmax loss function for brain tumour detection using deep learning. J Neurosci Methods 330:108520. https://doi.org/10.1016/j.jneumeth.2019.108520
Mathew AR, Anto PB (2017) Tumor detection and classification of MRI brain image using wavelet transform and SVM. In: 2017 International Conference on Signal Processing and Communication (ICSPC), Coimbatore, pp 75–78. https://doi.org/10.1109/CSPC.2017.8305810
Mehrotra R, Ansari M, Agrawal R, Anand R (2020) A transfer learning approach for AI-based classification of brain tumors. Mach Learn Appl 2:100003. https://doi.org/10.1016/j.mlwa.2020.100003
Minz A, Mahobiya C (2017) MR image classification using adaboost for brain tumor type. In: 2017 IEEE 7th International Advance Computing Conference (IACC), Hyderabad, pp 701–705. https://doi.org/10.1109/IACC.2017.0146
Muhammad K, Khan S, Del Ser J, de Albuquerque VHC (2020) Deep learning for multigrade brain tumor classification in smart healthcare systems: a prospective survey. IEEE Trans Neural Netw Learn 32(2):507–522. https://doi.org/10.1109/tnnls.2020.2995800
Naseer A, Yasir T, Azhar A, Shakeel T, Zafar K (2021) Computer-aided brain tumor diagnosis: performance evaluation of deep learner CNN using augmented brain MRI. Int J Biomed Imaging 5513500. https://doi.org/10.1155/2021/5513500
Pashaei A, Ghatee M, Sajedi H (2020) Convolution neural network joint with mixture of extreme learning machines for feature extraction and classification of accident images. J Real-Time Image Proc 17(4):1051–1066. https://doi.org/10.1007/s11554-019-00852-3
Polat, Ö, Güngen, C (2021) Classification of brain tumors from MR images using deep transfer learning. J Supercomput, 1–17. https://doi.org/10.1007/s11227-020-03572-9
Polly FP, Shil SK, Hossain MA, Ayman A, Jang YM (2018) Detection and classification of HGG and LGG brain tumor using machine learning. In: 2018 International Conference on Information Networking (ICOIN), Chiang Mai, pp 813–817. https://doi.org/10.1109/ICOIN.2018.8343231
Qiu Y, Yan S, Gundreddy RR, Wang Y, Cheng S, Liu H, Zheng B (2017) A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. J X-ray Scie Technol 25(5):751–763. https://doi.org/10.3233/xst-16226
Rajan P, Sundar C (2019) Brain tumor detection and segmentation by intensity adjustment. J Med Syst 43(8):1–13. https://doi.org/10.1007/s10916-019-1368-4
Rehman A, Naz S, Razzak MI, Akram F, Imran M (2020) A deep learning-based framework for automatic brain tumors classification using transfer learning. Circ Syst Signal Process 39(2):757–775. https://doi.org/10.1007/s00034-019-01246-3
Rehman A, Khan MA, Saba T, Mehmood Z, Tariq U, Ayesha N (2021) Microscopic brain tumor detection and classification using 3D CNN and feature selection architecture. Microsc Res Tech 84(1):133–149. https://doi.org/10.1002/jemt.23597
Ruba T, Tamilselvi R, ParisaBeham M, Aparna N (2020) Accurate classification and detection of brain cancer cells in MRI and CT images using nano contrast agents. Biomed Pharm J 13(3):1227–1237. https://doi.org/10.13005/bpj/1991
Saba T, Mohamed AS, El-Affendi M, Amin J, Sharif M (2020) Brain tumor detection using fusion of hand crafted and deep learning features. Cogn Syst Res 59:221–230. https://doi.org/10.1016/j.cogsys.2019.09.007
Sajjad M, Khan S, Muhammad K, Wu W, Ullah A, Baik SW (2019) Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J Comput Sci 30:174–182. https://doi.org/10.1016/j.jocs.2018.12.003
Salçin K (2019) Detection and classification of brain tumours from MRI images using faster R-CNN. Tehnički glasnik 13(4):337–342. https://doi.org/10.31803/tg-20190712095507
Sarhan AM (2020) Brain tumor classification in magnetic resonance images using deep learning and wavelet transform. J Biomed Sci Eng 13(06):102–112. https://doi.org/10.4236/jbise.2020.136010
Saxena P, Maheshwari A, Maheshwari S (2021) Predictive modeling of brain tumor: A Deep learning approach. In: Innovations in Computational Intelligence and Computer Vision, pp. 275–285. Springer, Singapore
Seetha J, Raja SS (2018) Brain tumor classification using convolutional neural networks. Biomed Pharma J 11(3):1457. https://doi.org/10.13005/bpj/1511
Sharif MI, Li JP, Khan MA, Saleem MA (2020) Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recogn Lett 129:181–189. https://doi.org/10.1016/j.patrec.2019.11.019
Sornam M, Kavitha MS, Shalini R (2016) Segmentation and classification of brain tumor using wavelet and Zernike based features on MRI. In: 2016 IEEE International Conference on Advances in Computer Applications (ICACA), Coimbatore, pp 166–169. https://doi.org/10.1109/ICACA.2016.7887944
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
Sultan HH, Salem NM, Al-Atabany W (2019) Multi-classification of brain tumor images using deep neural network. IEEE Access 7:69215–69225. https://doi.org/10.1109/access.2019.2919122
Suthaharan S (2016) Machine learning models and algorithms for big data classification. Integr Ser Inf Syst 36:1–12
Swati ZNK, Zhao Q, Kabir M, Ali F, Ali Z, Ahmed S, Lu J (2019) Content-based brain tumor retrieval for MR images using transfer learning. IEEE Access 7:17809–17822. https://doi.org/10.1109/access.2019.2892455
Swati ZNK, Zhao Q, Kabir M, Ali F, Ali Z, Ahmed S, Lu J (2019) Brain tumor classification for MR images using transfer learning and fine-tuning. Comput Med Imaging Graph 75:34–46. https://doi.org/10.1016/j.compmedimag.2019.05.001
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI Conference on Artificial Intelligence 31(1). https://doi.org/10.1609/aaai.v31i1.11231
Taheri S, Gasparovic C, Shah NJ, Rosenberg GA (2011) Quantitative measurement of blood-brain barrier permeability in human using dynamic contrast-enhanced MRI with fast T1 mapping. Magn Reson Med 65(4):1036–1042. https://doi.org/10.1002/mrm.23165
Toğaçar M, Ergen B, Cömert Z (2020) BrainMRNet: brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med Hypotheses 134:109531. https://doi.org/10.1016/j.mehy.2019.109531
Ullah Z, Farooq MU, Lee S-H, An D (2020) A hybrid image enhancement based brain MRI images classification technique. Med Hypotheses 143:109922. https://doi.org/10.1016/j.mehy.2020.109922
Zhan T, Feng P, Hong X, Lu Z, Xiao L, Zhang Y (2017) An automatic glioma grading method based on multi-feature extraction and fusion. Technol Health Care 25(S1):377–385. https://doi.org/10.3233/thc-171341
Zhou M, Scott J, Chaudhury B, Hall L, Goldgof D, Yeom KW, Iv M, Ou Y, Kalpathy-Cramer J, Napel S (2018) Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. AJNR Am J Neuroradiol 39(2):208–216. https://doi.org/10.3174/ajnr.a5391
Funding
This work was supported by the Natural Science Foundation of Hunan Province, China (Grant No. 2020JJ4757).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Asif, S., Zhao, M., Tang, F. et al. An enhanced deep learning method for multi-class brain tumor classification using deep transfer learning. Multimed Tools Appl 82, 31709–31736 (2023). https://doi.org/10.1007/s11042-023-14828-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14828-w