Abstract
Wireless capsule endoscopy is a noninvasive wireless imaging method that has grown in popularity over the last several years. One of the efficient and effective ways for examining the gastrointestinal system is using WCE. It sends a huge number of images in a single examination cycle, making abnormality analysis and diagnosis extremely difficult and time-consuming. As a result, in this research, we provide the Expectation maximum (EM) algorithm, a revolutionary deep-learning-based segmentation approach for GI tract recognition in WCE images. DeepLap v3+ can extract a variety of features including colour, shape, and geometry, as well as SURF (speed-up robust features). Thus the Lenet 5 based classification can be made in the extracted images. The effectiveness of the performances is carried out on a publicly available Kvasir-V2 dataset, on which our proposed approach achieves 99.12% accuracy 98.79% of precision, 99.05% of recall and 98.49% of F1- score when compared to existing approaches. Effectiveness benefits are demonstrated over multiple current state-of-the-art competing techniques on all performance variables we evaluated, especially mean of Intersection Over Union (IoU), IoU for background, and IoU for the entire class.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
A non-invasive method for finding irregularities in the human gastrointestinal (GI) tract is called wireless capsule endoscopy (WCE) [17]. It is a capsule-like structure and measures 26 mm in length by 11 mm in diameter. Its components include an imaging sensor, an optical dome, a battery, an illuminator and an RF transmitter [15]. During the examination, the patient ingests the WCE, and it passes slowly through the small intestine and takes images of the entire GI tract while it is shifting. Finally, these images are transferred wirelessly to a data-recording device so that doctors can subsequently study the images for diagnosis [5].
Infections in the gastrointestinal system, like bleeding, Crohn’s disease, cancer, and ulcer polyps have become more common in recent years, while ulcers and bleeding are widespread disorders [2]. Cancer in the lungs (1.1 million fatalities), stomach cancer (765,000 fatalities), rectum cancer (525,000 fatality), liver cancer (505,000 alive), and breast cancer (385,000 deaths) are the top causes of death, according to a WHO survey [26]. According to a study conducted in the United States, a huge number of gastrointestinal tract infection disorders have been reported in the United States since 2017, with roughly 200,000 new cases reported each year since 2011. If found and diagnosed early enough, this gastrointestinal tract infection can be treated. WCE is the main technology utilized by specialist physicians to detect GI tract infections [7, 23, 27].
For an accurate diagnosis, the work is time-consuming and difficult due to the volume of data. Additionally, it can frequently be very challenging to see some little bleeding areas with the naked eye [1]. These are some of the factors driving the numerous research projects being done to speed up reading by automatically identifying anomalies or other regions of interest in images (ROIs) [3, 22].
One of the WCE’s producers, Given Imaging, offers a technology called suspected bleeding indicator (SBI) for automatically identifying frames with potentially bleeding regions [16, 20, 25]. However, research has revealed that SBI is insufficient to test for all GI tract illnesses. This opened the way for researchers to create methods for the more accurate automatic detection of various kinds of anomalies in WCE images [4, 19].
Even though these studies have greatly improved defective area segmentation, the accuracy is still insufficient because of the limitations of conventional techniques or network capabilities [8, 12,13,14]. They lack a tailored architecture for WCE photos, and they haven’t completely exploited the enormous potential of using semantic segmentation techniques on WCE images [18, 21]. This research offers a unique deep-learning based system for automatic detection and classification that extends the state-of-the-art in order to address the aforementioned issues. Our main contributions include developing an effective deep-learning architecture for precise detection, as well as comprehensively evaluating it and comparing it to the state-of-the-art, showing consistent improvements in a range of performance criteria.
The key contributions of this paper are as follows,
-
For WCE image defect area segmentation, we propose a new EM method that employs a multi-stage design with attention blocks at every step.
-
We use a multi-stage architecture, which allows every level to be treated as a separate network. As a result, every layer in every stage is trained to extract meaningful features for our target using DeepLab V3+.
-
LeNet 5 is employed to classify the GI tract in WCE images, and to find the presence stage of images.
-
The Kvasir-V2 dataset has undergone various ablation experiments. Our proposed network outperforms the state efficiency concerning all other approaches, according to the experimental data.
The remainder of the paper is laid out as follows: The state of the art on how the problem of energy limitation has been addressed is presented in part 2. Our method is presented in Section 3. In Part 4, the findings are presented and discussed. At last, Section 5 contains the conclusion.
2 Literature review
Here is a review of some previous studies on the automatic detection of anomalies in WCE images.
Semi-supervised Outlier Detection Model (SODM), a mix of semi-supervised deep-structured CNN-LSTM and outlier assessment mode (OAM), was introduced by authors in [6] to handle the issue of small intestinal disease outlier detection in WCE images. By evaluating the spatial-scale trends of successive image sections, irregular graphical patterns were discovered in the image. SODM also includes an outlier assessment model, which reduces the number of false alarms while making outlier declarations. When compared to the prior paper, the suggested scheme comes out ahead.
The authors of [19] introduced a method called BIR that classifies WCE bleedy images by combining the MobileNet with a custom-built CNN approach. Due to its low computation capacity, BIR uses the MobileNet approach to do initial phase computations. The results are then forwarded to CNN for further processing. For categorization, Ensemble is more accurate than either the MobileNet or CNN framework. In terms of effectiveness, the proposed BIR model surpasses current strategies.
In [3], CNN, GoogLeNet and AlexNet are used to classify ulcers and non-ulcers in object classification. The GoogLeNet and AlexNet methods were pre-trained on a section of the ImageNet database to find the optimum network parameter combination that allows these two CNNs to detect ulcers with good accuracy. When compared to other techniques, it had the highest detection rate.
The authors of [8] suggested using a deep CNN to detect hookworms in WCE images. Pooling layers are presented to blend the tube sections obtained from the extracted network with the feature maps created by the hookworm categorization in terms of improving the feature maps relating to tubular regions. The suggested methodologies outperformed other existing methods.
Based on the fusing and choosing of DL characteristics, the authors of [12] described a fully automated system for detecting stomach illnesses. The array-based technique is used to collect features from two consecutive fully linked layers and merge them into a re-trained model. The greatest entities were chosen using the PSO technique, which determines the best candidates based on a mean value-based fitness function. When compared to other feature selection techniques, it was discovered that the suggested method produced the greatest results.
In [9], the differential box-counting approach was utilized to extract the fractal dimension (FD) of Wireless Capsule Endoscopy images, and the RF-based hybrid classifier was employed to identify anomalous frames. The purpose of this analysis is on block-level local characteristics. To address the issue of class imbalance, they adopted the SMOTE method. The suggested method outperformed some of the existing state-of-the-art approaches.
By creating masks around polyps detected from still frames, [24] created a modified R-CNN. Feature extraction is performed using the Resnet-50 and Resnet-101 models. Polyps in images were recognized and segmented accurately using the provided approach. When compared to other methods, the suggested technique efficiently segments the polyps. Table 1 lists all of the existing relevant works.
3 Proposed methodology
In our strategy, the work is processed based on four phases such as preprocessing, segmentation, feature extraction, and classification. The raw images were first passed through the preprocessing stage. Data resizing, gray scale conversion and image enhancements were done in preprocessing. By using the power law transformation the image can be enhanced. Once the WCE images are preprocessed we fed them into the.
segmentation phase to segment the defected area after completing this process, the next phase is feature extraction. The DeepLap V3+ descriptor is used to extract feature representations from every given image during the feature extraction stage. The obtained features are sent into the deep learning classifier Lenet 5 during the training phase. Finally, test photos are utilized to evaluate the suggested method’s efficiency. Figure 1 represents the architecture diagram of the proposed methodology.
3.1 Preprocessing
In medical science, image processing is critical for enhancing images and adding more information. Image quality is an important aspect of achieving an optimum solution. To achieve a better result, an image that has distortion or loudness or is of poor quality must be upgraded. In the proposed model the input size of the image is 720*576. The two values indicate the height and width of the image. The images were first decreased in size from 720*576 to 256 × 256 pixels. The image quality was then enhanced by using power law transformation.
3.1.1 Image enhancement based on power law transformation
Image enhancement can be done in the Fourier or spatial domains, with contrast enhancement being one of the most significant parameters to consider. The methods utilized in the spatial domain can be further classed as grey-level transformations, histogram processing, and so on. As previously stated, histogram equalization has the disadvantage of reducing the contrast in some cases.
Histogram matching or specification can be tailored to the images, but it needs a significant amount of input data. Power law transformations functions, on the other hand, necessitate a huge number of input data. The coefficient occurring in the transfer function must be chosen in the first case, while the gradients and varies of the parallel lines that make up the scaling factor must be chosen in the second case.
Typically, the basic equation is described as follows:
The individual grey intensities in the output and input images are s and r, respectively and constancy is given as c. Because RGB color information is generally noisier than HSV color information, the input image is first transformed into its HSV color space in this paper.
For the multiple exposure images, a power law transformation is done to the V-channel of the input image. Gamma is supposed to have an ideal value of one. However, if the gamma value varies beyond a certain point, the image is entirely degraded. As a result, the value of gamma should be kept within a specific range to prevent degeneration. The gamma value range is chosen such that the entropy of multiple exposure images does not fall below a certain level.
Linear stretching is applied to the generated images after creating different exposure images to make the most of the complete dynamic range.
3.2 Data augmentation
Due to the unavailability of a significant number of annotated images, image augmentation plays a key role in medical image analysis. Data augmentation improves data quality and reliability. Zoom, rotation, flip, shear, and shifting are some of the transformations that are triggered. Furthermore, by shortening the distance between the testing and training datasets, augmented datasets help to increase data points. In the training dataset, the overfitting problem can be prevented.
3.3 Segmentation based on EM algorithm
The maximum likelihood estimations of unknown parameters are computed using the expectation-maximization (EM) algorithm, which uses probabilistic models. An algorithm is an iterative approach for solving the problem of maximization. By selecting random values, the maximum-likelihood approach is employed to discover the “best fit” for a data set. An iterative approach using the EM method in incomplete/missing data points is used to calculate the best fit in an unsupervised way. For a set of complete data θ = ξ, the best fit is determined through maximum likelihood estimation. The complicated EM technique is efficient enough to catch model parameters in the absence of a data set. The EM method chooses the lost data points at random and uses them to predict the previous batch of data.
The gained new values are then utilized to enhance the initial set’s prediction, and the method eventually converges to a fixed point for the best fit. With an understanding wi, the value of θcan is enhanced. The procedure begins with a forecast forθ, then calculates z, then updatesθ using this new value for z, and so on until convergence is achieved.
The EM algorithm can be thought of as a training set{a(1), a(2), ……, a(m)}, with m independent samples for an estimation issue. These training parameters must be used to fit a model P(m, z), with the corresponding likelihood defined as
The maximum likelihood estimation εis challenging, but it is possible if the latent variable z(1) can be observed. The EM algorithm can then be broken down into two steps, the first of which is the E step, which involves constructing a lower constraint on j. We also optimize the lower bound, sometimes known as the M step.
The EM technique has been successfully utilized to find a likelihood function for missing values in a data collection. The sample density is calculated as follows:
The likelihood function is defined asL(ε ∣ m), where x represents the sample data of size N. The procedure’s goal is to find an estimate of j that maximizes the value of L. Assume z = (m, h) for a complete data set, and a joint density function represents the relationship between the lacking and detectable values.
We can establish a new likelihood function using this new cumulative distribution function.
Initially, the technique uses log-likelihood logp(m, h, ξ) to calculate the estimated value for the complete data set, taking into account the unidentified data, m be detected data, and the currently estimated parameter. The current parameter’s estimation is represented as:
Where ξ(i − 1)are the estimated total parameters utilized to compute the expectation value, and ξ denotes the most recent parameters optimized to raise the Q value. We need to find a way to enhance the expectations.
The image was segmented periodically using the EM algorithm to achieve a satisfactory outcome in this research. The pixels from normal and afflicted tissues are sorted into separate categories based on their likelihood of being comparable in intensity. The cluster’s center point is reconstructed through the iteration process until it reaches a fixed cluster’s center point. The following equations give a brief description of the EM process.
To compute, set the mean, covariance, and mixing co-efficient tomi,σi andci, respectively.
The log-likelihood function that corresponds to it I(ξ) is defined as follows: We can also estimate the probability as follows:
Perform maximization as
The improved co-variance is calculated as follows:
The probability density function is represented by k, and the characteristic vector is x. The combination model has the following structure:
3.4 Feature extraction
The extraction of features is an important phase in the model construction process. A system can be built using a variety of features, including color, shape, and geometry, as well as SURF (speeded-up robust features).
To extract spatial domain information and integrate them with spectral domain features, we use DeepLab v3+ as the neural network structure. DeepLab v3+ is one of Google’s fourth-generation DeepLab semantic segmentation networks, and it offers the best overall performance to date. The ASPP module has been altered from the own approach of independent branches in the DeepLabv3+ network topology. The dense connection method allows for denser pixel sampling, which improves the algorithm’s capacity to extract features.
More pixels can be used in the calculation with the densely coupled ASPP module. Atrous convolution’s pixel sampling is sparser than regular convolution’s. This issue is extended to two dimensions, the single-layer atrous convolution only involves 9 pixels, whereas the cascaded atrous convolution involves 49 pixels. As an outcome, the least ASPP output is densely coupled to the high ASPP rate layers, enhancing the potential of network feature extraction.
The densely linked ASPP module provides a bigger receptive field as well as denser pixel sampling. Without increasing the feature map’s resolution, atrous convolution can produce a broader receptive field. The volume of the receptive field R is the following for ASPP convolution with ASPP rate r and k denotes the size of the kernel:
By stacking two atrous convolutions, a bigger receptive field can be generated, and the volume of the stacked particular area can be calculated using the equation below:
The sizes of the receptive fields of the two atrous convolutions are R1 and R2, respectively. The dense connection method’s receptive field size is calculated by stacking atrous convolutions. The basic ASPP module runs in parallel, with every branch sharing no information, whereas the upgraded module allows information to be shared by layer. The distinct ASPPs are dependent on one another, increasing the receptive field’s range. Utilizing \( {R}_r^k \) to represent the particular area the maximum receptive field Rmaxof the atrous method is:
The maximal particular area of the modified ASPP module, as shown in Eq. (17), is
Whereas the intensively linked ASPP module allows for a larger area in a particular manner, it increases the effectiveness of parameters, decreasing the network’s efficiency. To tackle this issue, 1 × 1 CNN is applied before every ASPP CNN following dense connection, reducing the number of channels in the feature map and improving the network’s expression ability. s represents the number of channels the equation is shown below:
Whenever the improved Xception network is used as the backbone network, the ASPP component’s inputs are the network’s dimensional vector with 2048 output size, so each ASPP module has 256 channels, and the parameter quantity S of the enhanced ASPP module can be computed as follows:
While L denotes the volume of ASPP CNN. DeepLabV3+ employs a false class cross-entropy equation, as regards:
The outcome is a softmax categorization function for the pixel level, x is the position of the two-dimensional pixel point, and \( {a}_{k_{(x)}} \) denotes the location of pixels point x in the size k. The output is the confidence of each pixel x in the k class. The certainty of every image pixel x within k class is provided. Equation (21) shows that DeepLab’s total loss is based on cross-entropy loss and \( {p}_{l_{(x)}} \)defines the tag’s efficiency likelihood.
To avoid overfitting and improve model robustness, regularization terms are frequently added after the loss function. The L2 regularization term is used to punish the loss of function. The following is the definition of L2 regularization, often known as ridge regularization:
Where regularization coefficient is η and weight is θ. The loss of the standard phrase is minimized in backpropagation optimization as the degradation of the error function is reduced.
3.5 Classification
The purpose of classification is to predict the label. We employed the multi-class classification technique in the proposed approach to categorize the input image based on the selected features. LeNet5 is a comprehensive deep categorization network that categorizes extracted features and saves processing time through techniques including convolution, parameter sharing, and pooling. The standard LeNet-5 CNN has an input layer, two convolutional layers, two max pooling, fully connected layers, and a softmax layer. The input size for the architecture is 224 pixels by 224 pixels.
A 32 × 32 grayscale image serves as the input for LeNet-5 and is processed by the first convolutional layer comprising six feature maps or filters with a stride of 1. The LeNet-5 then adds an average pooling layer or sub-sampling layer with a filter size of 2 × 2 and a stride of 2. On the result of the Conv1 layer, the Pool1 layer applies a 2× 2 max-pooling procedure to build six 14× 14 pixel image features. To construct 16 feature maps of 10 × 10 pixels, the Conv2 layer employs 16 convolution operations of size 5× 5. Pool2 generates 16 image features of 5× 5 pixels by performing a 2× 2 max-pooling function on the Conv2 layer’s outcome. The FC1 layer is a 120-neuron comprehensive layer that is fully coupled to the Pool2 layer and generates 120 1-pixel image features. The FC2 layer is an 84-neuron full-connection layer that computes the dot-product of the input vector and support vectors, applies the bias value, and outputs the findings via the sigmoid function. The FC3 layer, also known as the softmax layer, has ten neurons and separates all input pictures into ten sections equivalent to integers 0–9. Architecture details of LeNet 5 is represented in Table 2.
It is decided to use the dynamic adaptive pooling method. The Relu activation function is efficiently utilized to solve the gradient vanishing issue. In softmax, the final classifier was trained. The training time and detection rate have both improved as a result of the optimization of these three strategies.
The input data is normalized before being transferred to the next layer, which is unaffected by the altered data distribution. Because the normalization layer is a trainable and customizable interface, whitening is the optimum data preprocessing technique. With pre-treatment, the number of computations is very big. To make computations easier, the pre-treatment equation for approximation whitening is
The SD of each set of input data is shown in the numerator of the preceding formula (23) and E(m(s))is the sum amount of each sample of input data. The variance and mean of every batch should be recorded during the training process so that the sum and difference of the full database can be calculated after the training is completed.
The resulting high-level feature map can minimize the original feature map’s dimension and resolution while also avoiding overfitting and other concerns. Mean and maximum pooling are two instances of pooled procedures.
As a result, a mathematical method based on the max pool technique is built to replicate the function using the interpolation approach. If the pooling factor is denoted as μ, then after upgrading the pooling models, the equation is represented as
The pool μ is employed to improve the max pool procedure, the improved characteristics can represent features correctly. The balanced is identical to the maximum pooling model’s parameter values.
The tanh function is used to update the m > 0 part of the ReLU, and a new activation function is created, which is described in formula (28)
Formally, a set of N images {Xi,yi} Ni = 1 are taken, where Xi is the original image data and yi is a class category of the image (i.e., 0 and 1). The difference between the predicted label \( {\hat{y}}_i \) and the real label isyicalculated using the categorical cross-entropy function, defined as follows:
The biases and weights of the conventional LeNet-5, respectively are represented by and b. K is the number of class categories, and ylkis the softmax value of the kth class category, which is calculated as:
Where zi denotes the outcome of the last fully connected output’s associated ith class category. The convolution layers downsample at a rate of 2 strides per layer. The loss equation concerning parameters throughout the entire network, the convolution organization’s parameters, and fully linked layers were all trained using BP. The batch size, momentum, learning rate, and weight decay are 1, 0.9, 0.03, and 0.001, respectively. The initial learning rate is 0.01. In the ReLu layer the learning rate is saturated. Due to the possibility of the network being either under- or over-fitted, the number of epochs is also a crucial training parameter. For this dataset, we trained the network for 50 epochs. The proposed model’s training and testing accuracy varies, it ranges from 0.98 to 0.99 and loss values ranges from 0.001 to 0.004. Figure 2 represents the proposed method flowchart.
4 Result and discussion
This portion explores the details of the analytical outcomes employing the proposed approach, which is based on assessment criteria like precision, accuracy, recall, IOU, and F1 score. Dataset description, preprocessing, segmentation, feature extraction, classification method, and results in the analysis are the six processes in this research. In the selected data, images with sizes varying from 720*576 to 1920*1072 pixels were translated into pixels. The Kvasir version 2 database was utilized to classify GIT disorders in this study. The entire dataset is split into two parts: a training set with 80% of the data and a validation set with 20% of the data.
For our classification challenge, we will utilize an implementation of LeNet-5 Convolutional Neural Network. There are three Convolutional Layers each with 5 × 5 filters and followed by average pooling with 2 × 2 patches. For activation, we employ the ReLU function because it allows us to learn faster. Then we add the Dropout Layer with a factor of 0.2 to deal with overfitting. This means that 20% of the input will be neutralized at random to avoid significant dependencies across layers. Flattening and two Dense Layers are the final products. In the final Dense Layer, we will use the Softmax activation function to generate probabilities between 0.0 and 1.0 by assigning the same amount of neurons to each class. There are 70,415 parameters in this network as a consequence. The Adam learning rate optimization algorithm is a variation of the Stochastic Gradient optimization process. In terms of improving speed, Descent is a good option. The Categorical Cross-Entropy loss function is appropriate for this issue since it provides us with a multiclass probability distribution for our classification task. To assess the suggested method’s effectiveness, we utilized the following accuracy measures.
4.1 Experimental setting
Using 4 GB RAM and an Intel i5 2.60 GHz processor, it runs Windows 10. The studies are carried out in the Anaconda3 environment using Python and KERAS with the Tensor flow as a backdrop.
4.2 Dataset description
GT images collected in VV health trust were used in these investigations as the dataset. The training data comes from one of the trust’s main gastrointestinal departments. Medical specialists analyzed the dataset extensively and entitled it Kvasir-V2. As a component of the Mediaeval Medical Multimedia Challenge, a benchmarking initiative that assigns tasks to the research team, this dataset was made accessible in the fall of 2017. The dataset is divided into eight groups, each with 1000 photos, including anatomical markers, pathological findings, and polyp removal. The resolution of the images in the collection ranges from 720*576 to 1920*1072. Sample images from the dataset are shown in Fig. 3.
The Kvasir-V2 dataset is utilized for validation in this paper to estimate the effectiveness of our proposed approach. The data samples are split into two sections, one of which is utilized to create a classifier and is referred to as the training dataset. The testing dataset is used in the second step to evaluate the classifier.
4.3 Performance metrics
Several performance criteria can be used to evaluate gastrointestinal tract detection and categorization. The detection rate, or the ratio between the total number of pixels and infected pixels, has a significant impact on disease classifier performance. The likelihood of detection is another term for the detection rate. Several researchers analyzed the GI tract classification outcomes using keywords like recall, accuracy, True Negative (TN), precision, False Negative (FN), False Positive (FP), and True Positive (TP).
Accuracy
The predicted values accuracy score reflects the model’s accuracy. The accuracy score is calculated by dividing the number of accurate predictions by the overall number of recommendations. It can be summarized as follows:
Here,
-
The value is TP when the classifier is trained that the photo will be Normal and the image’s reality description is also Normal.
-
TN is the number when the classifier is trained that an image is affected and the image’s genuine label is also degraded.
-
FP is the value whenever the classifier is trained that the photo will be regular but the image’s real caption is contaminated.
-
The quantity FN is used when the classifier is trained that a photo is contaminated but the true caption is Normal.
Precision
Precision is defined as the ratio of precisely anticipated positive occurrences to all anticipated positive observations. Precision is the capacity to do the following things:
Recall
The True Positive Rate (TPR) and Sensitivity are both terms for recall. The recall score reflects the classifier’s ability to locate all positive samples. It’s the total of TP and FN divided by TP. It can be described in the following terms:
Intersection over union (IoU)
Intersection over Union is a statistic for comparing the total of the predicted and observed pixels in the image in the same category to accurately described pixels. Output of dyed-lifted-polyps, dyed-resection-margins, esophagitis, polyps and ulcerative-colitis is shown in Figs. 4, 5, 6, 7 and 8.
4.4 Segmentation evaluation
In this portion, we explain the outcome of segmentation using the Expectation Maximization Algorithm. Six assessment metrics are estimated using a segmented image: IoU_A, accuracy, mIoU recall, IoU_B, and precision. All evaluation parameters are.
based on four factors: TP, TN, FP, and FN. The comparison can be made with the existing segmentation approaches such as Superpixel-based segmentation, Gaussian mixture model, and VGG-19. We also compared the outcomes based on precision, accuracy, and recall [11]. The results of the Segmentation using the EM algorithm are displayed in Table 3.
We observed that our proposed method outperforms existing approaches, showing 99.59% accuracy, followed by Gaussian mixture, Super pixel-based, and VGG-19 with 99.29%, 98.76%, and 97.7%, respectively. When compared with other metrics like precision and recall our proposed approach yield a greater solution. And the second approach that yields a better performance is super pixel-based segmentation. Among the four approaches VGG-19 gains lower performance [10]. Figure 9 represents the differentiation of proposed vs existing segmentation approaches. Table 4 shows the segmentation approaches comparison.
Other metrics such as mIoU, IoU A, and IoU B can then be used to compare. IoU stands for intersection over union; IoU B stands for IoU for background; IoU A stands for IoU for all classes; mIoU stands for mean IoU. Our technique outperforms more advanced solutions. The suggested framework improves image segmentation, as demonstrated by the positive results of all metrics for both training and testing images.
Segmentation is based on an EM algorithm. It noted that the proposed approach presented a superior performance over all metrics rather than the existing approaches. Moreover, the proposed model has a good vision for differentiating between defected fields and backgrounds. IOU-based segmentation methods comparison is represented in Fig. 10.
4.5 Evaluation of training results
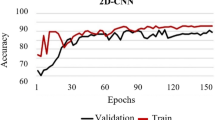
Train Accuracy and Validation Accuracy curves converge in the end, and after 50 epochs we received an accuracy of 99.9%, which is quite good. Figure 11 shows the training and testing accuracy. The validation Loss curve jumps up and down a bit. It implies that more validation data might be beneficial. Validation Loss exceeds Train Loss after around 25 epochs, indicating that there is some overfitting. However, because the curve does not rise over epochs and the variance between Validation and Train Loss is small, this could be acceptable. Figure 12 shows the training and testing loss.
4.6 Classification evaluation
The existing approaches like Alex Net, ResNet, and LSTM-CNN are compared with our proposed methods.
Differentiation of proposed classification approaches with existing techniques is represented in Table 5 in terms of f1-score, recall, accuracy, and precision on the dataset. It illustrates that when compared to the competitive method, the proposed model performs much better.
The graphical representation of classification performance differentiation is shown in Fig. 13. When comparing the proposed approach with the other methods, our presented approach yields the highest outcome. In terms of accuracy, our proposed approach gains the best result. Similarly, recall, precision, and f1-score also achieve the highest result. In terms of all metrics, the overall performance of the proposed method achieves a greater result (99.12% of accuracy, 98.79% of recall, 99.05% of precision, and 98.49% of F1-Score).
The effectiveness of various models with the suggested method is compared in Table 6. It demonstrates that the proposed strategy had the best results in comparison to others. Figure 14 showed the proposed effectiveness when compared to CNN, MobileNet, and BIR in terms of the F1 score, recall, accuracy, and precision.
5 Statistical analysis
To examine the statistical significance of observed performance results, we used a T-test. The significance level for the test was set to α = 0.05.
Among the evaluated models, the proposed approach produces the best results with good efficiency and accuracy. The outcomes of the statistical tests also show that the enhancement of our proposed method is statistically significant. The value of the two models is indicated byρ an amount in each grid cell for the relevant row and column. Comparing the proposed strategy to other approaches, we can observe that the enhancement is statistically significant at the 0.01 level. The t-value is −2.51921. The p value is .008661. The result is significant at p < .05. Figure 14 is the statistical results of paired T-Test. The dark grey indicates <0.01. The light grey indicates 0.01–0.05. The blank space indicates>0.05. Figure 15 represents the statistical test result of paired T-test.
The confusion matrix for the proposed end-to-end trained method is shown in Fig. 16. The primary purpose of the confusion matrix, also known as the error matrix, is to highlight the differences and inconsistencies between the actual class and the predicted class. In this article, the possible number of classes is five, including dyed-lifted-polyps, dyed-resection-margins, esophagitis, polyps, and ulcerative colitis.
6 Conclusion
WCE image analysis is a crucial field in computer vision because of its substantial role in clinical diagnosis. But it is also a difficult task. WCE photos frequently have numerous small areas that are difficult to detect. In this study, a novel Lenet-5-based GI tract classification and detection system are proposed. The effectiveness of this system is demonstrated through in-depth experimental studies on a standard dataset and compared with the existing state-of-the-art approaches. The proposed system utilizes a better segmentation and demonstrates 99.12% of accuracy, 98.79% of recall, 99.05% of precision, and 98.49% of F1- a score that is superior to the existing models.
There will be three key topics covered in future works. The performance of the proposed expert system will first be assessed using various datasets. It will also be important to increase the number of comparative deep learning methods.
Data availability
Not applicable.
Code availability
Not applicable.
References
Al Mamun A, Em PP, Ghosh T, Hossain MM, Hasan MG, Sadeque MG (2021) Bleeding recognition technique in wireless capsule endoscopy images using fuzzy logic and principal component analysis. Int J Electric Comput Eng (2088–8708) 11(3):11
Alam MW, Vedaei SS, Wahid KA (2020) A fluorescence-based wireless capsule endoscopy system for detecting colorectal cancer. Cancers 12(4):890
Alaskar H, Hussain A, Al-Aseem N, Liatsis P, Al-Jumeily D (2019) Application of convolutional neural networks for automated ulcer detection in wireless capsule endoscopy images. Sensors 19(6):1265
Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T (2019) Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 89(2):357–363
Fan S, Xu L, Fan Y, Wei K, Li L (2018) Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol 63(16):165001
Gao Y, Lu W, Si X, Lan Y (2020) Deep model-based semi-supervised learning way for outlier detection in wireless capsule endoscopy images. IEEE Access 8:81621–81632
Ghosh T, Fattah SA, Wahid KA (2018) CHOBS: color histogram of block statistics for automatic bleeding detection in wireless capsule endoscopy video. IEEE J Transl Eng Health Med 6:1–12
He JY, Wu X, Jiang YG, Peng Q, Jain R (2018) Hookworm detection in wireless capsule endoscopy images with deep learning. IEEE Trans Image Process 27(5):2379–2392
Jain S, Seal A, Ojha A, Krejcar O, Bureš J, Tachecí I, Yazidi A (2020) Detection of abnormality in wireless capsule endoscopy images using fractal features. Comput Biol Med 127:104094
Jani KK, Srivastava S, Srivastava R (2019) Computer aided diagnosis system for ulcer detection in capsule endoscopy using optimized feature set. J Intell Fuzzy Syst 37(1):1491–1498
Jani KK, Srivastava S, Srivastava R (2021) Framework for the restoration of capsule endoscopy images using partial differential equations-based filter. IETE J Res, 1-11
Khan MA, Kadry S, Alhaisoni M, Nam Y, Zhang Y, Rajinikanth V, Sarfraz MS (2020) Computer-aided gastrointestinal diseases analysis from wireless capsule endoscopy: a framework of best features selection. IEEE Access 8:132850–132859
Lu F, Li W, Lin S, Peng C, Wang Z, Qian B, Ranjan R, Jin H, Zomaya AY (2021) Multi-scale features fusion for the detection of tiny bleeding in wireless capsule endoscopy images. ACM Transact Internet Things 3(1):1–19
Oleksy P, Januszkiewicz Ł (2020) Wireless capsule endoscope localization with phase detection algorithm and simplified human body model. Int J Electron Telecommun 66(1):45–51
Pogorelov K, Suman S, Azmadi Hussin F, Saeed Malik A, Ostroukhova O, Riegler M, Halvorsen P, Hooi Ho S, Goh KL (2019) Bleeding detection in wireless capsule endoscopy videos—color versus texture features. J Appl Clin Med Phys 20(8):141–154
Ponnusamy R (2020) Wireless capsule endoscopy image classification model to detect gastro intestinal tract diseases using visual words based on feature fusion. Int J Future Gener Commun Netw 13(1):985–998
Prasath VB, Thanh DN, Thanh LT, San NQ, Dvoenko S (2020) Human visual system consistent model for wireless capsule endoscopy image enhancement and applications. Pattern Recognition Image Anal 30(3):280–287
Rathnamala S, Jenicka S (2021) Automated bleeding detection in wireless capsule endoscopy images based on color feature extraction from Gaussian mixture model superpixels. Med Biol Eng Comput 59(4):969–987
Rustam F, Siddique MA, Siddiqui HUR, Ullah S, Mehmood A, Ashraf I, Choi GS (2021) Wireless capsule endoscopy bleeding images classification using CNN based model. IEEE Access 9:33675–33688
Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T (2020) Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 92(1):144–151
Shrivastava A, Chaudhary A, Kulshreshtha D, Singh VP, Srivastava R (2017) Automated digital mammogram segmentation using dispersed region growing and sliding window algorithm. In: 2017 2nd international conference on image, vision and computing (ICIVC). IEEE, pp 366–370
Singh NP, Singh VP (2020) Efficient segmentation and registration of retinal image using Gumble probability distribution and BRISK feature. Traitement du Signal 37(5):855–864
Sivakumar P, Kumar BM (2019) A novel method to detect bleeding frame and region in wireless capsule endoscopy video. Clust Comput 22(5):12219–12225
Sornapudi S, Meng F, Yi S (2019) Region-based automated localization of colonoscopy and wireless capsule endoscopy polyps. Appl Sci 9(12):2404
Souaidi M, Abdelouahed AA, El Ansari M (2019) Multi-scale completed local binary patterns for ulcer detection in wireless capsule endoscopy images. Multimed Tools Appl 78(10):13091–13108
Wang S, Xing Y, Zhang L, Gao H, Zhang H (2019) A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys Med Biol 64(23):235014
Wang S, Xing Y, Zhang L, Gao H, Zhang H (2019) Deep convolutional neural network for ulcer recognition in wireless capsule endoscopy: experimental feasibility and optimization. Computation Math Methods Med 2019:1–14
Acknowledgements
We declare that this manuscript is original, has not been published before, and is not currently being considered for publication elsewhere.
Author information
Authors and Affiliations
Contributions
The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Corresponding authors
Ethics declarations
Ethics approval
This material is the authors’ own original work, which has not been previously published elsewhere. The paper reflects the authors’ own research and analysis in a truthful and complete manner.
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Padmavathi, P., Harikiran, J. & Vijaya, J. Effective deep learning based segmentation and classification in wireless capsule endoscopy images. Multimed Tools Appl 82, 47109–47133 (2023). https://doi.org/10.1007/s11042-023-14621-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14621-9