Abstract
Information security has become a significant issue in encryption due to the rapid progress of internet and network. Therefore, the development of the encryption algorithm is a growing and significant problem. In this study, a new color image encryption was introduced based on DNA complementary rules and pair coupled chaotic maps. At first, the plain color image was divided into three components (R, G, B) being converted into three DNA matrices using DNA encoding rules. Secondly, DNA addition for R, G and B components was implemented and scrambled the elements position of three DNA sequence via the pair coupled chaotic maps. Three gray coded images obtained and RGB encrypted image was achieved by restructuring R, G, B components. The simulation of experimental result and security analysis showed that this algorithm had larger secret key space and strong secret key sensitivity and it had excellent ability to resist against statistical and differential attacks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In recent years, with the rapid development of the Internet and computer networks and communications, large amounts of digital information and multimedia content, for example images, sounds and videos, are commonly stored and transmitted over the Internet. Most of this digital data available on the Internet and public networks are sensitive and private, and have caused it to be used for malicious purposes. Hence, security and encryption of digital image is receiving wide attention due to the widespread transmission through various communication networks and Internet to guarantee and secure confidentiality and stop unauthorized access to the digital content. One of the most effective ways to secure enough confidential image information against illegal usage, unauthorized users is image encryption. Various methods have been presented to scrambled and encrypted image. Traditional symmetric ciphers such as Advanced Encryption Standard (AES), Data Encryption Standard (DES) and International Data Encryption Algorithm (IDEA) are no more appropriate for protecting of digital images. There are many methods to encrypt images, one of which is based on the theory of chaos and has unique features. Chaotic systems have non-linear dynamic behavior [2, 10] and are used in communication, optimization and process of image, in addition to encryption. The chaos system process includes a lot of excellent features, such as high sensitivity to control parameters and initial values, pseudo-random, ergodicity, deterministic nature and so on, which make them to be suitable for designing image encryption scheme [2, 4, 9, 11, 13, 25]. Image encryption algorithms based on chaotic mappings should consist of iteration of two stages: permutation between the pixels of the image and diffusion between them to provide higher computational security [3, 10, 18]. Logistic map can be considered as a simplest model of chaotic map. Researchers have demonstrated that the Logistic map cannot meet the requirement of being a good random numbers generators, and the generic symmetric non-linearly coupled map is better than it [2]. 1D and 2D chaotic system was used to encrypt image for high security several time [11, 13, 25].
Nowadays, DNA computing is used in cryptography because of Massive parallelism, huge storage and ultra-low power characteristics of DNA calculation. In fact, there is no relationship between encryption and molecular biology. The greatest achievement in this field has implemented by Adelman in 1994, Hamiltonian path solution with building DNA strands, which has caused progress in biology, mathematics and computer [1]. Currently, DNA calculations has developed a new field of encryption. DNA encrypted code uses DNA as data carrier and modern biology techniques for encryption. Due to DNA encoding is not secure and confident to encrypt images, we combine DNA en-coding with chaotic maps to enhance the efficiency and security of image encryption algorithms. Liu and Zhang have demonstrated RGB image encryption method based on DNA sequences operation and chaotic Logistic map in 2012 [20] where the randomness of the key stream produced from Logistic map is not yet high enough. In [8], A novel text and image encryption method with good performance was suggested based on chaos theory and DNA computing, where Image encryption was performed using DNA complementary rule and chaotic maps in [19]. Zhen applied Logistic map and DNA to generate pseudo-random matrix which was encoded by DNA. Then, DNA addition between pseudo-random DNA matrix and plain image are used to obtain a new DNA matrix. The resulting matrix was permutated by the sequence was obtained from the plain image information entropy and the spatiotemporal chaotic system to get the cipher image [32]. Rehman et al. proposed robust dual diffusion and confusion encryption technique for color image according to Chaos, DNA and SHA-2 to protect chosen image against attacks. they used a SHA-256 hash function for modification of the initial conditions for 2-Dimensional Logistic map [23]. Recently, a new encoding scheme for color images using DNA sequences and Chen’s hyper-chaos system was proposed by Amani et al. [7]. Experiment result confirms that the encrypted image is highly disturbed by this proposed algorithm and shows resistance to several security analyses. In addition, Ref. [26] introduced a novel color image encryption scheme using DNA permutation based on the Lorenz system where DNA permutation and addition/subtraction operations can break the bit planes of the plaintext image entirely.
In order to improve the security performance of color image encoding based on chaos and DNA computing, we propose a new scheme to encrypt color image by using pair coupled chaotic maps and DNA coding rules. The results of experiment indicate that our proposed algorithm is very convenient and efficient for RGB image encryption due to resistant against types of attacks. The remainder of this article is organized as follows: In second part, some required theories for suggested algorithm is introduced. The details of encryption algorithm was presented in Section 3. Section 4 is results of simulation and explains information security and comparison with presented algorithms.
2 Basic theory of the proposed algorithm
2.1 DNA encoding and decoding for image
Single- strand DNA (Deoxyribonucleic Acid) contains four nucleic acid bases: A (ade-nine), C (cytosine), G (Guanine), T (Thymine), where A and T are complementary, G and C are complementary. In the theory of binary system, 0 and 1 are complementary, so 00 and 11 are complementary, as well as 01 and 10 are also complementary. Using 00, 01, 10, 11 to express four bases A, C, G, T, there are 24 kinds of encoding rules. Due to the Watson-Crick complement rule [27] between DNA bases, there are acceptable only eight kinds of encoding rules, as listed in Table 1. For example, if the grayscale value of the pixel is 156, its corresponding binary values is 10011100 and using the first DNA encoding Rule the corresponding DNA sequence is CGTA.
Addition and subtraction operation for DNA sequences are defined based on traditional binary addition and subtraction. There are eight kinds of DNA addition rules and subtraction rules based on eight kinds of DNA encoding rules. For example, according to DNA encoding Rule 1, the addition DNA operation is shown in Table 2
2.2 Pair-coupled chaotic maps
Coupled chaotic maps lattices is a non-linear dynamics system with the unit interval, or discrete space and time. In this study, we use pair coupled chaotic maps can be described as a two-dimensional dynamic maps of the form given by (2). Most of linearly or non-linearly symmetric pair-coupled chaotic maps can be described as a symmetric two-dimensional maps as follows [2, 10]:
An example of symmetric non-linearly coupled chaotic maps is defined as follows:
Where 𝜖 ∈ (0,1) is the strength of the coupling and we choose p as an arbitrary integer to have a two-dimensional dynamical system with the property of having an invariant measure at synchronized state. The functions f1(x), f2(y) are two arbitrary one-dimensional maps of the interval [0,1] with an invariant measure which can be described by [2, 18]:
For example, some of these maps is as follows:
The state variables are f2(x,a),f3(x,b) ∈ [0,1] and the system is chaotic when the system parameters a and b are within the range of described below:
The chaotic sequence in the proposed algorithm has produced in the following ways:
(1) The real value of the sequence is produced using non-linearly symmetric pair-coupled chaotic maps with initial values that there are x(0), y(0), p, a, b, 𝜖.
(2) Binary sequences: We transform the real value sequence to the binary sequence by defining a threshold function g(x) is as follows:
Using the above equation, we can convert the chaotic sequence of real value to chaotic sequence of binary.
3 Image encryption and decryption algorithm
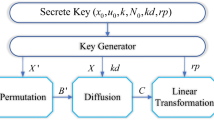
In this section, firstly the color image is divided into R, G and B components. And then, converted the R, G and B components into DNA codes, and get three DNA sequence matrices. Addition operation is carried out for three DNA sequence matrices based on the DNA encoding rules to disturb the correlation between the pixels in spatial domain. Secondly, the three encoded gray images (R, G, B) are permuted using three pair coupled chaotic maps with the initial conditions and the parameters assigned to each individually. Lastly, combine R, G and B components to obtain encrypted RGB images. Block diagram of the algorithm is shown in Fig. 1.
The encryption processes of the algorithm as follows:
Step1: Suppose the size of RGB image is m × n, where m, n are the dimensions of the rows and columns of the image respectively.
Step2: Split the RGB image into R, G and B components and convert the decomposed matrices of R, G and B to binary matrices R(m,n × 8), G(m,n × 8) and B(m,n × 8), then encode respectively using one kind of the DNA encoding rules (key1 is a random key and key1 ∈ [1,8]), and obtain three DNA sequence matrices Ar(m,n × 4), Ag(m,n × 4) and Ab(m,n × 4).
Step3: The additional rule for the encoded sequence matrices Ar(m,n × 4),Ag(m,n × 4) and Ab(m,n × 4) obtained from step 2 is based on the following relations:
Where + is the addition operation for DNA sequences and is performed according to traditional binary addition. There are eight kinds of DNA addition rules corresponding to 8 kinds of DNA encoding rules are described in Section 2. For example, the DNA addition rule 1 according to DNA encoding rule 1 is indicated in Table 2. Perform the additional rule above by using one kind of DNA addition rules (key2 is a random key and key2 ∈ [1,8])) to obtain three (m,n × 4) confused sequence matrices.
Step4: Three chaotic sequences X1, X2 and X3 whose length are l = m × n × 4 are generated using pair coupled chaotic maps with three categories of initial conditions and discrete parameters that is assigned for each of the chaotic sequences individually. Then, we use threshold function g(x) according to (6) to obtain three binary sequences Z(1,l) and transform them into Z1(m,n × 4), Z2(m,n × 4) and Z3(m,n × 4) matrices. The initial conditions and the parameters are given as follow:
Step5: The bases of the corresponding locations of the \(Ar^{\prime }(i,j)\), \(Ag^{\prime }(i,j)\) and \(Ab^{\prime }(i,j)\) DNA matrices are changed to its complement respectively when Z1(i,j), Z2(i,j) and Z3(i,j) is one.
According to Watson-crick base pairing rules, the base of A and T in DNA sequence are complement to each other, as well as the base of G and C in DNA sequence. For example: If \(Ar^{\prime }(i,j)\) is A and z(i,j) = 1 then the base of \(Ar^{\prime }(i,j)\) is changed to T to disrupt the location of the \(Ar^{\prime }(i,j)\), \(Ag^{\prime }(i,j)\) and \(Ab^{\prime }(i,j)\) DNA matrices.
Step6: The result of step 5 to binary matrices is decoded by a kind of DNA decoding rules (the index of this DNA decoding rule serves as secret key3 ∈ [1,8]), then we will get binary matrices \(R^{\prime }(m,n\times 8)\), \(G^{\prime }(m,n\times 8)\) and \(B^{\prime }(m,n\times 8)\).
Step7: Three chaotic sequences C1, C2 and C3 whose lengths are l = (m,n × 8) are produced by using pair coupled chaotic maps system with three categories of initial conditions and discrete parameters, x(0),y(0),p,a,b,𝜖, that is assigned for each of the chaotic sequences individually. By applying threshold function g(x) according to (6) get three binary sequences S(1,l) and transform them into S1(m,n × 8), S2(m,n × 8) and S3(m,n × 8) matrices.
Step8: Exclusive XOR operations are carried out for chaotic binary matric, S1(m,n × 8) with \(R^{\prime }\), to obtain new ciphered matric that is \(R^{\prime \prime }\). We also use this XOR operators for matrices S2(m,n × 8) and S3(m,n × 8) with \(G^{\prime }(m,n\times 8)\) and \(B^{\prime }(m,n\times 8)\), respectively, to obtain new matrices \(G^{\prime \prime }\) and \(B^{\prime \prime }\).
Step9: Finally, RGB image is recovered to obtain the encrypted color image by mixing the three encrypted matrices.
From the analysis of the above algorithm, the attacker must possess 33 keys which include pair coupled chaotic maps parameters and initial values that are: 6 × (𝜖,a,b,x(0),y(0)) and keys of encoding rules key1 and decoding rules key3 and also keys of DNA addition rules key2. In this algorithm, for each of the matrices R, G and B obtained from the original image, we used pair coupled chaotic maps with different initial conditions and parameters and obtained different sequences. We change the values of the matrices R, G, and B, respectively by using these different sequences. The sequence used for the R matric to perform operations on it is different from the sequence used for G or B, and also for R, B matrices. The security of images with the above encryption algorithm is greatly improved. The process of decryption algorithm is the simple reversion of the process of encryption. In decryption algorithm, we should have 33 secret keys: 6 × (𝜖,a,b,x(0),y(0)), key1, key2 and key3. The decryption processes of the algorithm as follows:
Step1: First read the encrypted color image and then separate it to component \(R^{\prime \prime }\), \(G^{\prime \prime }\), \(B^{\prime \prime }\).
Step2: Use three time pair coupled chaotic maps with three different initial values and parameters, 𝜖,p,a,b,x(0) and y(0), for producing three sequences C1,C2 and C3 whose length is (m,n × 8). then prepare three binary sequences S(1,l) by applying threshold function g(x) according to (6) and restructure them into S1(m,n × 8), S2(m,n × 8) and S3(m,n × 8) matrices. Based on XOR operation, we invert the step 8 of the encryption algorithm in the form of:
Step3: Encode the permutated matrices according to DNA rule selected by key3.
Step4: Produce three chaotic sequences X1,X2 and X3 whose length are (m,n × 4) by using pair three time coupled chaotic maps system with three initial different initial values and parameters that they are x(0),y(0),p,a,b,𝜖 and we perform complement operations as in step 5 of the encryption algorithm.
Step5: We do subtract operations using one kind of DNA subtraction rules key2 for DNA sequence matrices according to Table 3 as follow:
Step6: Three DNA sequence matrixes according to DNA rule are decoded as selected by key1 to get the R, G and B component of the plain image.
Step7: Finally, the plain color image is recovered and that is the decrypted image.
4 Experimental results and performance analysis
In this study, the experimental simulation of the proposed algorithm was implements in MATLAB 2014 environment. This study employs the 256 × 256 color image of Lena is shown in Fig. 2a as the plain text image. The initial values and parameters of pair coupled chaotic maps are chosen arbitrarily as follow:
x(1) = 0.2,y(1) = 0,a = 0.7,b = 1.569,𝜖 = 0.342 and p = 100 for R component of color image of proposed algorithm in step 4,
\(x^{\prime }(1)=0.7, y^{\prime }(1)=0.4, a^{\prime }=0.33, b^{\prime }=0.7, \epsilon ^{\prime }=0.5\) and p = 100 for R component of color image of proposed algorithm in step 8,
x(1) = 0.67,y(1) = 0.175,a = 0.6,b = 1.9123,𝜖 = 0.3585 and p = 100 for G component of color image of proposed algorithm in step 4,
\(x^{\prime }(1)=0.2, y^{\prime }(1)=0.7, a^{\prime }=0.3, b^{\prime }=0.8, \epsilon ^{\prime }=0.5\) and p = 100 for G component of color image of proposed algorithm in step 8,
x(1) = 0.2,y(1) = 0.3,a = 0.7,b = 1.7,𝜖 = 0.33 and p = 100 for B component of color image of proposed algorithm in step 4,
\(x^{\prime }(1)=0.7, y^{\prime }(1)=0.2, a^{\prime }=0.2, b^{\prime }=0.5, \epsilon ^{\prime }=0.5\) and p = 100 for B component of color image of proposed algorithm in step 8,
and also key1 = 1 key of encoding rule, key2 = 1 key of DNA addition rule, key3 = 8 key of decoding rule.
In Fig. 2b and c, the encrypted Lena image and decrypted Lena image with above mentioned keys are depicted respectively. The decrypted image is exactly the same as the original one and it is obvious from images there are no relationship between the original Lena image and the ciphered Lena image.
A good image encryption scheme should withstand all types of attacks such as exhaustive attacks, differential attacks and statistical attacks, etc.. Here, we will represent that this encryption system is sufficiently secure against various cryptographical attacks by analyzing the security performance of the proposed encryption scheme.
4.1 Key space analysis
A good image encryption scheme should have a large key space to make any brute-force attack ineffective. Although, the speed of encryption may reduce when the secret key space is very large. From the cryptographical point of view, in order to provide a high security against brute-force attacks, the key space size should be larger than 2100 [6, 21]. In the image encryption algorithm, the secret keys include: (i) The initial values x(1) and y(1) of pair coupled chaotic maps, which are used individually for each component of color image in step 4 of proposed algorithm; (ii) The initial values \(x^{\prime }(1)\) and \(y^{\prime }(1)\) of pair coupled chaotic maps, which are used individually for each component of color image in step 8 of proposed algorithm; (iii) a, b and 𝜖 are parameters of pair coupled chaotic maps in step 4 of proposed algorithm, which are used for each component of color image separately; (iv) \(a^{\prime }\), \(b^{\prime }\) and \(\epsilon ^{\prime }\) are the parameters of pair coupled chaotic maps in the step 8 of proposed algorithm, which are used for each component of color image separately; (v) Key 1 is the index of DNA encoding rule (8 kinds), key 2 is the index of DNA addition rule (8 kinds) and key 3 is the index of DNA decoding rule (8 kinds). Thus, in general we have three initial values for x(1), y(1), \(x^{\prime }\), \(y^{\prime }\) and three values for a, b, 𝜖, \(a^{\prime }\), \(b^{\prime }\), \(\epsilon ^{\prime }\) parameters. The precision of 64-bit double data is 1015, therefore, the key space size will be (1015)3×(1015)3×(1015)3×(1015)3×(1015)3×(1015)3×(1015)3×(1015)3×(1015)3×(1015)3 = (1015)30 = 10450, being large enough to resist the brute-force attack in comparison with the references given in Table 3.
Key space size is the total number of different keys which can be used in the encryption algorithm to provide a good encryption algorithm with high security, should have a large key space to make any brute force attack ineffective. The encryption algorithm consists of 33 key space in this study, they are 6 × (e,a,b,x(0),y(0)), key1, key2 and key3, where \(a, b \in (0, \frac {1}{N})\) for chaotic system, key1, key2 and key3 are random number.
Chaotic system has high sensitivity to initial value and rapid diffusibility. In implementation of the algorithm, a slight change of arbitrary parameter in secret key will change the results of encryption and decryption. In order to test the sensitivity of the secret key slight difference keys are used to decryption. The proposed algorithm has the secret key sensitivity, and it can resist exhaustive attack effectively.
4.2 Sensitivity analysis
Key sensitivity is a critical feature for an effective encryption scheme. That is, a slight change in the key would cause a greatly significant change in output and it can be considered from two aspect: (i) In the image encryption process, when a slightly different key is used to encrypt the same Lena image, a completely different encrypted image is generated; (ii) In the image encryption process, when only a tiny difference exists between the encryption key and decryption keys the encrypted Lena image cannot be decrypted successfully. Pair coupled chaotic maps is extremely sensitive to its initial condition and parameter with 1015. At first, key sensitivity test is carry out by using a key that is just slightly different from the original key to encrypt the same Lena image. The plain image of Lena which is shown in Fig. 3a and its encrypted image with \(b^{\prime }=0.5\), control parameter of B component of color image in step 8 of proposed algorithm, is shown in Fig. 3b and its encrypted image using a tiny change in key \(b^{\prime }=0.5000000000000002\), control parameter of B component of color image in step 8 of proposed algorithm, and other keys are unchanged is displayed in Fig. 3c. The result of chaotic sequence will be totally different by the change of lb parameter. The results of the difference between Fig. 3b and c are shown in Fig. 3d. Therefore, the parameter lb is sensitive and for further investigation of key sensitivity, comparison would be change another key of key space slightly and keep others unchanged to encrypt Lena image. Figure 3 indicates that our proposed algorithm is highly sensitive to the secret key. The pixel-by-pixel difference of two encrypted images that are generated using \(b^{\prime }=0.5\) and \(b^{\prime }=0.5000000000000002\) is measured and the rate of difference is 99.9053 as illustrated in Table 4 for Lena image. Similarly, the key sensitivity of the decryption scheme is performed. The decrypted result for the ciphered image with wrong key is shown in Fig. 4.
The results of key sensitivity compared to other schemes are presented in Table 6 and the proposed cipher algorithm has clear advantage over others (Fig. 5).
4.3 Resistance to statistical attack
4.3.1 The gray histogram analysis
The act of distribution of the image is an important issue. The histograms of the two plain images indicate that the image pixels values are concentrated on some values; however, the histograms of ciphered image are fairly uniformly distributed, which are being significantly different from that in the plain text images. The histogram of an image usually defined as a graph representing the distribution information of pixel values of an image. The histograms of R, G and B three components of the plain image and ciphered image of Lena are displayed in Fig. 3a and b. Through the comparison of histogram, it can be observed that the proposed algorithm results are a very flat in ciphered image distribution and that statistical attack is not effective to our proposed algorithm. It should be flat enough to be safe and secure and certain amount of information cannot be analyzed by the statistical attack opponent. Thus, a flat distribution is appropriate in cryptography. In order to analyze the distribution of pixel values, the uniformity of the encrypted images is measured by computing the variance of histograms. When the variances are lower, the images have higher uniformity and The value of the variance for an encrypted image of dimension n × n is given by the following formula:
where Z is the vector of the histogram values Z = z1,z2,z3,...,z256 and zi and zj is the number of pixels where gray values are equal to i and j, respectively. Table 5 presents the variances of the histograms of the plain image for Lena and of the encrypted image for Lena. It can be observed that from Table 5 the variance of the original image of Lena is 6405 and for the encrypted image produced by our proposed algorithm is 3429. Thus, it indicates that the proposed algorithm has a better ability of resisting statistical attacks.
4.3.2 Correlation coefficient analysis
The less correlation of two adjacent pixels is shown the stronger ability of resisting statistical attack. A number of 1000 pairs of adjacent pixels were randomly selected in vertical, horizontal and diagonal directions from the original image of Lena and its encrypted image of Lena respectively, and calculate the correlation coefficients of two adjacent pixels according to the following formula:
Where x, y represent gray between adjacent pixels in the image, and N represents the number of pixels are chosen from the image, and E(x) and D(x) display mathematical expectation and variance of x, respectively. Figures 6, 7 and 8 respectively shows the correlation distribution of adjacent pixels of R, G and B layer of Lena as the plain image along horizontal and vertical and diagonal directions. Table 6 shows the computation results of correlation coefficients of the original Lena image and its ciphered image. From this table, it can be found that the correlation coefficients of adjacent pixels in original Lena image in each direction are close to 1, while the correlation coefficients of adjacent pixels in encrypted Lena image in each direction are close to 0. It is obvious From the result in Table 6, our encryption scheme has low correlation as compared to original image and is better than other algorithms. So our proposed algorithm is efficient.
Correlation analysis of two adjacent pixels of R layer of Lena. a Horizontal distribution of plain image; b Horizontal distribution of ciphered image; c Vertical distribution of plain image; d Vertical distribution of ciphered image; e Diagonal distribution of plain image; f Diagonal distribution of ciphered image
Correlation analysis of two adjacent pixels of G layer of Lena. a Horizontal distribution of plain image; b Horizontal distribution of ciphered image; c Vertical distribution of plain image; d Vertical distribution of ciphered image; e Diagonal distribution of plain image; f Diagonal distribution of ciphered image.
Correlation analysis of two adjacent pixels of B layer of Lena. a Horizontal distribution of plain image; b Horizontal distribution of ciphered image; c Vertical distribution of plain image; d Vertical distribution of ciphered image; e Diagonal distribution of plain image; f Diagonal distribution of ciphered image
4.3.3 Information entropy
The information entropy is an important feature which is used to represent randomness of the system and measure the gray pixel value distribution in the image. It is well known that the information entropy H(m) of plaintext images can be described as follows:
Where, m represents the information source and P(m) denotes the probability of symbol m. Because there are 28 state of the information source for the 256 gray image with the same probability, according to (11), the information is completely random when the entropy image is 8. The information entropy of original Lena image and its encrypted Lena image obtained by the proposed algorithm are calculated and listed in Table 7. The information entropy of encrypted image should be close to eight to be chaotic as can be observed in Table 7, the proposed image encryption algorithm have good performance in information entropy.
4.4 performance
Finally, the time complexity of the proposed algorithm for encryption and decryption of lena image is evaluated. All Experimental results are based on color Lena image of size 256 256. The time complexity analysis is measured on Intel(R) Pentium(R) Core(i5) processor 2.3 GHz with 2048 MB RAM personal computer. The algorithm is developed in Matlab R2014a and compiled on Windows 10. The proposed image encryption scheme indicate good performances in term of execution time and statistical data. For color image Lena of size 256256, the average running speed is 1.28MB/s for encryption and 1.31 MB/s for decryption. However, In this algorithm the operation speed may be slower but this could be useful in domains like the military where the robustness of top secret data is privileged at their encryption speed. Table 8 displays the Comparison of our proposed method with other algorithms.
5 Conclusions
This study proposed a new color image encryption using the pair coupled chaotic maps and DNA encoding to enhance the security of image encryption. Based on the above discussion, the original color image is encoded using one kind of DNA rules and then scrambled by the chaotic sequences is achieved separately from pair coupled chaotic maps for R, G and B components of RGB image. we used pair coupled chaotic maps with different initial conditions and parameters and obtained different sequences. We change the values of the matrices R, G, and B, respectively by using these different sequences. The security of images with the above encryption algorithm is greatly improved. The process of decryption algorithm is the simple reversion of the process of encryption. The proposed scheme will be high-speed image encryption and decryption when used in the future DNA computer. Therefore, the proposed image scheme can be used as a candidate in the future DNA computer. Experimental simulation and security indicated that the proposed algorithm can resist against most known attacks, such as exhaustive attacks and statistical attacks and so on. This make it suitable for transmission of data via Internet and network in the secure communication.
References
Adleman LM (1994) Molecular computation of solutions of combinatiorial problems. Science 266:1021–1024
Ahadpour S, Behnia S, Jafarizadeh MA (2009) Synchronization in pair-coupled maps with invariant measure. Commun Nonlinear Sci Numer Simulat 14:2916–2922
Ahadpour S, Sadra Y (2012) Randomness criteria in binary visibility graph and complex network perspective. J Inf Sci 197:161–176
Ahadpour S, Majidpour M (2013) Image steganography using discrete cross-coupled chaotic maps. Watam Press Dynamics of Continuous. Discrete and Impulsive Systems Series B: Applications Algorithms 20:283–292
Akhavan A, Samsudin A, Akhshani A (2017) Cryptanalysis of an image encryption algorithm based on DNA encoding. Opt Laser Technol 95:94–99
Alvarez G, Li S (2006) Some basic cryptographic requirements for chaos-based cryptosystems. Int J Bifurcation Chaos 16:2129–2151
Amani H, Yaghoobi M (2019) A new approach in adaptive encryption algorithm for color images based on DNA sequence operation and hyper-chaotic system. Multimed Tools Appl 78:21537–21556
Babaei M (2013) A novel text and image encryption method based on chaos theory and DNA computing. Natural Comput 12:101–107
Behnia S, Akhshani A, Ahadpour S (2011) An analytical invariant measure for random maps with position dependent. Dynamics of Continuous, Discrete and Impulsive Systems Series A: Mathematical Analysis 18:245–255
Behnia S, Akhshani A, Ahadpour S, Mahmodi H, Akhavan A (2007) A fast chaotic encryption scheme based on piecewise nonlinear chaotic maps. Phys Lett A 366:391–396
Belazi A, Abd AA, El-Latif SB (2016) A novel image encryption scheme based on substitution-permutation network and chaos. Signal Procss 128:155–170
Chai X, Chen Y, Broyde L (2017) A novel chaos-based image encryption algorithm using DNA sequence operations. Opt Lasers Eng 88:197–213
Chai XL, Yang K, Gan ZH (2017) A new chaos-based image encryption algorithm with dynamic key selection mechanisms. Multimed Tools Appl 76:9907–9927
Chen J, Zhu Z, Zhang L, Zhang Y, Yang B (2018) Exploiting self-adaptive permutation-diffusion and DNA random encoding for secure and efficient image encryption. Signal Process 142:340–353
Gan Z, Chai X, Yuan K, Lu Y (2018) A novel image encryption algorithm based on LFT based S-boxes and chaos. Multimed Tools Appl 77:8759–8783
Guesmi R, Farah MAB, Kachouri A, Samet M (2016) A novel chaos-based image encryption using DNA sequence operation and Secure Hash Algorithm SHA-2. Nonlinear Dyn 83:1123–1136
Hu T, Liu Y, Gong L, Guo S, Yuan H (2017) Chaotic image cryptosystem using DNA deletion and DNA insertion. Signal Process 134:234–243
Jafarizadeh A, Behnia S, Khorram S, Nagshara H (2001) Hierarchy of chaotic maps with an invariant measure. J Statistical Phys 104:1013–1028
Liu H, Wang X, Kadir A (2012) Image encryption using DNA complementary rule and chaotic maps. Appl Soft Comput 12:1457–1466
Liu L, Zhang Q, Wei X (2012) A RGB image encryption algorithm based on DNA encoding and chaos map. Comput Electr Eng 38:1240–1248
Norouzi B, Seyedzadeh SM, Kuchaki SM, Mosavi MR (2014) A novel image encryption based on hash function with only two-round diffiusion process. J Multimed Syst 20:45–64
Rehman A, et al. (2018) An efficient mixed inter-intra pixels substitution at 2bits-level for image encryption technique using DNA and chaos. Optik 153:117–134
Rehman A, Liao X (2019) A novel robust dual diffusion/confusion encryption technique for color image based on Chaos, DNA and SHA-2. Multimed Tools Appl 78:2105–2133
Rehman A, Liao X, Ashraf R, Ullah S, Wang H (2018) A color image encryption technique using exclusive- OR with DNA complementary rules based on Chaos theory and SHA-2. Optik 159:348–367
Wang X, Teng L, Xue Q (2012) A novel colour image encryption algorithm based on chaos. Signal Process 92:1101–1108
Wang X, Li P, Qian Y, Liu L, Zhang H, Wang X (2018) A novel color image encryption scheme using DNA permutation based on the Lorenz system. Multimed Tools Appl 77:6243–6265
Watson JD, Crick FHC (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171:737–738
Wu X, Kan H, Kurths J (2015) A new color image encryption scheme based on DNA sequences and multiple improved 1D chaotic maps. Appl Soft Comput 37:24–39
Yaghouti Niyat A, Moattar MH, Niazi Torshiz M (2017) Color image encryption based on hybrid hyper-chaotic system and cellular automata. Opt Lasers Eng 90:225–237
Zeng L, Liu R (2015) Cryptanalyzing a novel couple images encryption algorithm based on DNA subsequence operation and chaoticsystem. Optik 126:5022–5025
Zhang Y, Li Y, Wen W, Wu Y, Chen J (2015) Deciphering an image cipher based on 3-cell chaotic map and biological operations. Nonlinear Dyn 82:1831–1837
Zhen P, Zhao G, Min L, Jin X (2016) Chaos-based image encryption scheme combining DNA coding and entropy. Multimed Tools Appl 75:6303–6319
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Azimi, Z., Ahadpour, S. Color image encryption based on DNA encoding and pair coupled chaotic maps. Multimed Tools Appl 79, 1727–1744 (2020). https://doi.org/10.1007/s11042-019-08375-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-08375-6