Abstract
The goal of this paper is to propose a modified maximally stable extremal region (MSER) based method for the segmentation of ultrasound liver images. Firstly, the feature regions including liver lesions are extracted using the modified MSER detector. Unlike the MSER algorithm, the improved MSER detector merely needs dozens of gray levels rather than 256 possible gray levels ranging from 0 to 255. Next, the edges of the liver lesions are detected from the binary images, and a merging strategy is designed to refine the contour of the liver lesion. The last step is the segmentation of the liver lesion according to the refined contour. The segmentation results of ultrasound liver images demonstrate that there is a significant correlation between the liver lesions selected by a medical expert and the liver lesions segmented by the proposed method. A comparison of the proposed method and other segmented methods shows that the proposed method can detect a more accurate contour of liver lesion images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Semi-automatic or fully automatic segmentation methods for liver CT images have been attracted many researchers’ attention in the past 10 years [6,17,31,33]. Compared to CT images, ultrasonic images have lower quality due to the inherent presence of speckle noise. This makes the segmentation of the object and the background in ultrasonic image is more difficult. In particular, effective and robust algorithms for correct segmentation of ultrasound liver images are hard to find.
For years researchers have focused on the segmentation techniques of ultrasound images [1–5,8,11,13,15,18–21,23,24,26–30,34–39]. Traditional segmentation methods in digital image processing were applied to ultrasound images in the reported literature. Liver tumor boundaries extracted using snake model algorithms [2–4,8] were used to compare tumor size and shape, before and after treatment. Liu [23] presented an improved GVF snake model to segment breast boundaries of digital mammograms. Li [21] proposed a radiating GVF snake model to segment cytoplasm and nucleus in cervical smear images. Rodtook [35] studied an adaptive segmentation of the ultrasound images of breast cancer using multi-feature GVF snakes model. Wu [39] developed an accelerated GVF algorithm for CT images. In addition, K-nearest neighbor [26] and closest neighbor approach [34] were also performed in the segmentation of ultrasonic liver images. Fractal feature vector [19], level set method [36] and hidden Markov model [15] have been employed to 2D or 3D ultrasound liver image segmentation.
Intelligent optimization methods were also utilized in liver medical image processing. Support vector machines [1,18] were adopted to extract and optimize the liver boundary of ultrasound images. An optimization method based on ensemble [13] was proposed to detect the liver shape in ultrasound image. Masoumi [24] combined an iterative watershed algorithm with artificial neural network to segment automatically liver region in MRI images. Moraru [28] developed an optimization of breast lesion segmentation by combining the snake evolution techniques with statistical textures information of ultrasound image. Torbati [38] improved a self-organizing map network to segment medical images including ultrasound image and MRI images. There are a few disadvantages of these methods for the segmentation of ultrasound liver image. The first drawback is that the segmented result heavily depended on the initialization of boundary. The second is that these segmentation methods run time consuming.
The maximally stable extremal region (MSER) algorithm is a well-known region detector used in wide-baseline stereo matching [25]. The set of stable feature regions in the MSER algorithm has the following advantages: (1) it is closed under continuous geometric transformations; (2) it is invariant to affine intensify changes; (3) it is detected at different scales. The MSER algorithm has successfully been used in applications such as the automatic 3D-reconstruction from a set of images [9,25], object recognition [10] and SAR image segmentation [14].
However, the MSER algorithm is few applied to segment ultrasound liver image in the reported literature. Because the liver lesions lead to connected shadow area in ultrasound image, it is nature to explore an idea of applying the MSER algorithm in ultrasound image segmentation. This paper develops an improved segmentation method for ultrasound image of the liver lesions in terms of the extracted MSERs. The improvement of our method has: (1) Merely need part of gray levels instead of the total ordered gray levels from 0 to 255, (2) Smooth the region of interest (ROI) image using anisotropic diffusion filter with a triangle kernel function, and (3) Merge the edge images corresponding the feature regions.
This paper is organized as follows. Section 2 presents the generalized MSER segmentation algorithm for ultrasonic liver image. Performance evaluation indicators are introduced to assess the proposed method in Section 3. The tested results of ultrasonic liver image and the comparison of several segmentation methods and the improved method are shown in Section 4. Section 5 concludes the paper with a discussion.
2 The generalized MSER segmentation algorithm
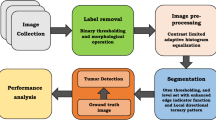
Inspired by the MSERs algorithm [25], this section presents a generalized MSER extracting method to segment liver tumor, liver cyst, or liver hemangioma in B-mode ultrasound images. There are two main steps in the generalized MSER segmentation algorithm. First, the proposed algorithm uses the generalized MSER detector to obtain the feature regions of the liver lesions. Then an edge merging strategy is employed to refine the edge map of liver lesions. Figure 1 shows a flow chart of the proposed method for the segmentation of ultrasonic liver lesions. Details of the presented segmentation method are described in the following text.
2.1 Image preprocessing
Although the MSER detector can extract stable feature regions for nature image, the speckle noise inherent in ultrasonic liver image makes the MSER algorithm difficult perfectly to segment the liver lesions in ultrasound images. In order to obtain a better MSER of liver lesions in ultrasound image, anisotropic diffusion filtering algorithm and mathematical morphology algorithm are used to remove noise and to enhance image.
The main effect of the anisotropic diffusion filtering algorithm is to remove speckles while preserving the edges in the image [11,12,32]. In the Perona [32], the formulation of an anisotropic diffusion filter is expressed by
where I is the image intensity, div(⋅) is the divergence operator, c(⋅) is the diffusion coefficient, ∇ denotes the gradient and G 0 ⊗ I is given as a convolution between the image I and a Gaussian kernel G 0 at the time t. Here, we selected a triangle window function instead of the Gaussian kernel function because of the time consumption using the Gaussian kernel in Eq. (1).
The main effect of mathematical morphology algorithm is to eliminate details or isolated pixels from the binary images and smooth boundaries. The openings or closings in mathematical morphology [7] are utilized many times in the implementation process of the proposed method.
2.2 Generalized MSER detector
The standard MSER algorithm [25] includes four major parts: (1) A sequence of binarization images are obtained from threshold 0 to threshold 255. (2) A representation of maximal intensity regions at each threshold is created from the set of all connected components of all frames of the sequence. (3) MSER detectors of all regions are tracked and the growth rates are monitored for local minimums. (4) All pixels belonging to the same detected MSER are identified. The generalized MSER detector in the proposed method merely needs part of gray levels instead of the total ordered gray levels from 0 to 255, and it is faster than the standard MSER detector.
The generalized MSER detector is performed for the ROI of the input image according to the next steps.
-
(1)
Calculate a gray threshold M of the ROI including the liver lesions using the Otsu’s adaptive algorithm [30].
-
(2)
Take M as a mid-value and obtaining an interval [M − δ, M + δ], δ is a positive number indicating the threshold range.
-
(3)
Select one threshold value in the interval [M − δ, M + δ] for the ROI and build a set of local binary images Q = {Q M − δ , ⋯, Q j , ⋯ Q i , ⋯, Q M + δ }.
-
(4)
Remove the noise or isolated pixels from the binary image set Q = {Q M − δ , ⋯, Q j , ⋯ Q i , ⋯, Q M + δ }, and each binary image in Q is a connected region.
-
(5)
Compute the area A(i) of all connected binary regions in Q = {Q M − δ , ⋯, Q j , ⋯ Q i , ⋯, Q M + δ }.
-
(6)
Analyze the area function A(i) for the potential regions. Suppose there have three consecutive binary regions {Q i − 1, Q i , Q i + 1} in set Q, the corresponding area is A(i − 1), A(i), and A(i + 1) respectively. The changed rates ΔA(i) of the area A(i) is estimated by Eq. (2)
If ΔA(i) persists with similar value over a range of thresholds i ∈ [M − δ, M + δ], the connected binary regions are named the generalized MSER. Generally speaking, several possible MSERs can be acquired for an image using the generalized MSER extraction algorithm.
In Fig. 1, the gray mean M of the ROI image is 105 and the threshold range δ is set as 5. Eleven binary images with threshold values from 100 to 110 are gotten. The change rate ΔA of the segmented region area A calculated by Eq. (2) is illustrated in Fig. 2. It can be seen the ΔA reaches its local minimum at the threshold value 106. Therefore, we can detect maximally stable extremal regions from a dozen binary images without needing all binary images in terms of S = (0, 1, 2, ⋯, 255).
2.3 Edge merging strategy
The edge merging strategy is implemented for a few stable binary regions of the object in the ROI. The edges of the generalized MSERs may be detected by canny operator for the stable binary regions. These edges represent the contour of the object in the ultrasound image. Which may be the true edge of the object? This step will refine the contour of the object through merging these edges. Let the edges E corresponding the generalized MSER be
where N is the number of the generalized MSER, and e i is an edge of the object corresponding each generalized MSER. Each edge e i is a matrix which has the same size as the ROI image. We may merge the edge through searching the edge points in the adjacent matrix. If three matrices adjacent to each other have edge point at the same positions, the value of the element in the merged matrix is one, otherwise, it is zero. That is to say, the merged edge may be refined from N edge images through Eq. (4)
where e m (x, y) is the merged edge image.
According to the merged edge, the ROI is segmented into two parts. The first part is background, and the second part is the edge of the object. Namely, when a pixel is inside the merged edge, the pixel is set as zero. Otherwise, the pixel remains the image background.
3 Performance evaluation indicators
In order to analysis quantitatively the performance of the proposed method, Hausdorff distance (HD) [38], mean absolute distance (MD) [38], Dice’s similarity index [16] and a relative error of the extracted area are estimated to describe the segmentation accuracy. HD and MD are two boundary error metrics to analyze the difference between the manual extracted edge and the automatic extracted edge. Similar to [38], the vector Q = (q 1j , q 2j ) n × 2 and R = (r 1i , r 2i ) m × 2 are the manual extracted edge points and the automatic extracted edge points, respectively. The shortest Euclidean distance between each point of R and all points of Q is defined as
where m and n are the number of edge pixels in R and Q,respectively.
HD and MD are written as
HD and MD stand for the longest and average distance between the two boundaries. The smaller are HD and MD, the closer are the detected edges to the truth.
Next, Dice’s similarity index is used to find the overlapping similarity between the manual segmented area A MS and the automatic segmented areas A AS . Dice’s similarity index is computed by Eq. (8).
where |A MS ∩ A AS | represents the overlapping area between the manual segmented area and the automatic segmented areas. Dice’s similarity index approaching to 1 indicates that the segmented result is closer to the manual segmented area. The relative error of the extracted area is defined as follows:
where A MS indicates the manual segmented area, and A AS is the automatic segmented areas.
4 Experiment results
We utilize a dataset of 40 ultrasound liver lesion images from Zhangshu people hospital and 48 ultrasound abdominal images from Shijiazhuang forth people hospital to validate the proposed method in the experiments.
4.1 Comparison of standard MSER and generalized MSER detector
A comparison of the standard MSER detector and the generalized MSER detector is presented in this experiment. More than 30 ultrasonic images of the liver lesions are tested. One of experiment results is shown in Fig. 3.
Figure 3a is the ROI image including liver cyst. Figure 3b1 to b8 is eight binary images, and their corresponding threshold values are 1, 5, 10, 40, 70, 120, 250, and 255 respectively. Figure 3c1 to c7 displays part of the feature extractions, and the corresponding threshold values are 59, 61, 63, 65, 67, 69, and 71 respectively. Figure 3b9 is the detected feature MSERs using Matas’s method [25] where a few background regions are also regarded as the MSER extraction. This result shows that Matas’s method needs to obtain the scores or even hundreds of the binary images in order to extract the MSER.
The running time both the standard MSER detector and the generalized MSER detector is estimated for the ROI image Fig. 3a of ultrasound liver cyst image, respectively. PC with 2.93GHz and Matlab7.0 are used in experiment. The estimated running time are shown in Table 1. In the standard MSER detector, the range of threshold is [0, 255] and we use three kinds of threshold steps. For the generalized MSER extraction, 30 threshold levels from 48 to 77 are applied to detect the feature extractions of the ROI image. It can be seen that the generalized MSER extraction is less time-consuming.
In the proposed method, the binary images are extracted using threshold values from 50 to 68 gray levels. The area changed rates ΔA of the feature extractions are estimated by Eq. (2). The mean and standard deviation of ΔA are 0.0095 and 0.0023, respectively. These results indicate that ΔA holds stable over a range of thresholds from 50 to 68. These binary images are considered as the generalized MSER extractions in this paper. Figure 3c8 displays the detected edges of the feature extractions. Figure 3c9 is the refined result according to edge merging strategy, and the broken yellow line in Fig. 3c10 is the segmented result. These figures show that the proposed method can acquire a more accurate segmentation.
4.2 Preprocess experiment of ultrasound image
This experiment tests and verifies the effect of the anisotropic diffusion filtering algorithm and mathematical morphology algorithm. Figure 4 is the preprocessed results of ultrasound images. The first original ultrasound image is from Zhangshu people hospital, and the latter two original images is from Shijiazhuang forth people hospital. Although the third original image is blurry, the contour of the object is still distinct after the anisotropic diffusion filtering. It can be seen that the anisotropic diffusion filtering algorithm preserves the edges of the object from the third column of Fig. 4. The effect of mathematical morphology algorithm to eliminate isolated pixels is very apparent by comparing the forth column with the last column in Fig. 4.
4.3 Experiments on free parameter of the proposed method
In the propose method, there have several free parameters: the average gray value of the ROI and the number of adjacent matrices showing an edge at the same location. This experiment demonstrates the method performance depending on these parameters.
Figure 5 is the experimental results. The first and second columns are applied to verify the dependence on the average gray value of the ROI, and the second and third columns validate the dependence on the number of adjacent matrices showing an edge at the same location. In the first experiment, the area of the ROI in the same original image is 22,932 pixels and 17,666 pixels, respectively. The estimated average gray value of the ROI is 55.89 and 51.62 pixels, respectively. The segmented areas of the object have little difference under the same number of adjacent matrices. These results are shown as the first two rows in Table 2.
The tested results on free parameters in the proposed method. The first row is original image, the second one is ROI, the third one is the merged edge by two adjacent matrices, the fourth one is the merged edge by three adjacent matrices, the fifth one is the merged edge by four adjacent matrices, and the last column is the merged edge by five adjacent matrices
In the latter experiment, we select 2, 3, 4 and 5 adjacent matrices showing an edge at the same location in the ROI of ultrasound liver image and obtain the different segmented result of the object. These results in Fig. 5 and Table 2 are shown that the merged edge points are sparse with increasing of the number of adjacent matrices. It can be seen that the extracted edge is a better while three or four adjacent matrices are applied in the edge merging strategy.
4.4 Segmentation of ultrasound liver image
In this subsection, we first present the segmentation of the liver abscess image and the liver cyst image using the proposed method. Then two comparison experiments are performed to evaluate the segmentation accuracy and the segmentation runtime.
4.4.1 Tested experiment of the proposed method
The tested experiment is the segmentation of the liver abscess and the liver cyst image. Figure 6a and 7a are ultrasound liver cyst image and ultrasound liver abscess image respectively. Figures (b1) in Figs. 6 and 7 are the binary images using Otsu’s algorithm [30], and it is difficult to segment the object from the two binary images. Figures (b2) to (b5) in Figs. 6 and 7 are the generalized MSER extraction using 4 threshold values. Figures 6c1 and 7c1 are the merged edges, and the segmented results are shown in Figs. 6c2 and 7c2. These experimental results show that the proposed method is effective and feasible.
4.4.2 Comparison of the proposed method and other methods
This experiment is the comparison of the proposed method with other segmentation method for ultrasound liver lesion image, which is from Zhangshu people hospital and Shijiazhuang forth people hospital. Figure 8 is the segmentation results of the liver lesions in ultrasound image using the proposed method, traditional snake-GVF algorithm [40], improved snake-GVF algorithm [23], watershed algorithm [24] and level set method [22], where three images from left to right are named as “image 1”, “image 2”, and “image3”.
Comparisons experiments the proposed method, traditional snake-GVF algorithm [40], watershed algorithm [24], Level Set [22] and improved snake-GVF algorithm [23] for liver lesions. The first row is original image, the second row is ROI, the third row is manually delineated result by a medical expert, the forth row is the segmented result by the proposed method, the fifth and sixth row are the segmented results by traditional snake-GVF algorithm with different initial contour, the seventh row is the segmented result by watershed algorithm, the eighth and ninth row are initial contour and the segmented result by Level Set, and the tenth and last row are gradient map and the segmented result by improved snake-GVF algorithm
The segmentation result is highlighted by red line in Fig. 8. The first and second row is the original ultrasound image and the ROI selected by white rectangle. The third row is region of liver lesions manually delineated by a medical expert. The forth row is the segmentation of the proposed method. The fifth and sixth row is the segmented results using traditional snake GVF algorithm [40] with different initial contour. It’s obvious that the segmented results are changed with the initial contour. The seventh row is the segmented results of watershed algorithm [24], which used the eight-connected neighborhood.
The parameters of level set algorithm [22] are selected with different images in Fig. 8 and are listed in Table 3. The initial contour is marked by red rectangle in the eighth row, and the ninth row is the final segmented contour. It can be seen that the segmented results usual include the strong reflection regions of ultrasound images besides the liver lesions.
The tenth and last row are the results using improved snake-GVF algorithm [23], where the tenth row is gradient map in terms of adjusting strategy. Liu [23] presented weights adjusting strategy to improve the edge map in the traditional snake-GVF algorithm. The improved method increased the segmentation accuracy, but it still ran time consuming.
Compared these results, it can be seen that the segmented results using the proposed method is approximate to the result manual delineated by a physician, and even a few details of the liver lesions are segmented. These experimental results show that the proposed method is effective and more precise.
In order to assess these segmented results, we estimated HD, MD, Dice’s similarity index and a relative error of the extracted area for ultrasound images of the liver lesions. The liver lesions in Fig. 8 are segmented manually 10 times by the physician and the average segmented areas are obtained. The automatic segmented areas using the proposed method, traditional snake-GVF algorithm [40], improved snake-GVF algorithm [23], watershed algorithm [24] and Level Set [22] is calculated 30 times for the ROI of ultrasound image in Fig. 8.
Let the boundary in the third row of Fig. 8 be Q and the boundaries in the forth, sixth, seventh, ninth and last row of Fig. 8 be R respectively. The HD, MD and Dice’s similarity index are estimated from Eqs. (6), (7) and (8), and are displayed in Table 4. HD and MD in Table 4 show that the results using the proposed method and improved snake-GVF algorithm are more stable and closer to the manual detected object. And the estimated Dice’s similarity index indicates that the overlapping areas using the proposed method are better in most cases. These results show that the segmented result using the proposed method is close to the manual segmented result.
The estimated relative errors A ERR for three ultrasound images in Fig. 8 are shown in Table 5. In the experiment, the proposed method and improved snake-GVF algorithm [23] are more stable than other methods. Traditional snake-GVF algorithm [40] and Level set algorithm [22] are actually influenced by the initial contour of the object.
We also estimated the runtime performance of these methods. PC with 2.93GHz and Matlab7.0 are used in experiment. The runtime of these methods includes the ROI input, program running, and the segmented result output. For each ultrasound liver image, we carried out 15 experiments using the proposed method, traditional snake-GVF algorithm [40], improved snake-GVF algorithm [23], watershed algorithm [24] and Level set algorithm [22]. The mean of runtime is listed in Table 6. It can be seen that the running time of the proposed method is nearly in concert with watershed algorithm [24] and Level set algorithm [22]. Although improved snake-GVF algorithm [23] may improve the segmented accuracy, it still run time consuming.
In addition, we further estimated the segmentation accuracy and the runtime of the proposed method, Milko [26], Lee [19] and Chen [5]. The ultrasound liver image is from Shijiazhuang forth people hospital. The Dice’s similarity indices are computed to evaluate the segmentation accuracy of these methods in this experiment. The manual segmented area in Fig. 9b is regarded as a reference value and it is 3482 pixels. The Dice’s similarity indices and the segmentation runtime are shown in Table 7. Although the proposed method needs a little long runtime, the segmentation result is more consistent with the manual segmented result from Fig. 9 and Table 7.
Comparisons experiments on our method, Milko’s method [26], Lee’s method [19] and Chen’s method [5] for liver cyst. a Original image, b manually delineated result, c segmented result by our method, d segmented result by Chen’s method, e the texture image and f segmented result by Milko’s method, g the multifractal image and h segmented result by Lee’s method
5 Conclusions
This paper proposed a modified MSER extraction based method for the segmentation of ultrasound image of the liver lesions. The proposed method utilized the improved MSER algorithm to obtain stable feature regions, and saved the running time. Plenty of ultrasound images including the liver lesions were evaluated the performance of the proposed method. The experimental results indicated that there were a better consistency between the proposed method and the manually extracted regions by medical experts. The future work is to integrate the proposed method into an automatic segmentation software system.
References
Akbari H, Fei BW (2012) 3D ultrasound image segmentation using wavelet support vector machines. Med Phys 39(6):2972–2984
Chen CM, Lu HS (2000) An adaptive snake model for ultrasound image segmentation: Modified trimmed mean filter, ramp integration and adaptive weighting parameters. Ultrason Imaging 22(4):214–236
Chen CM, Lu HHS, Hsiao AT (2001) A dual-snake model of high penetrability for ultrasound image boundary extraction. Ultrasound Med Biol 27(12):1651–1665
Chen CM, Lu HHS, Huang YS (2002) Cell-based dual snake model: A new approach to extracting highly winding boundaries in the ultrasound images. Ultrasound Med Biol 28(8):1061–1073
Chen MF, Zhu HS, Zhu HJ (2013) Segmentation of liver in ultrasonic image applying local optimal threshold method. Imaging Sci J 61(7):579–591
Ciecholewski M (2014) Automatic liver segmentation from 2D CT images using an approximate contour model. J Signal Process Syst 74(2):151–174
Crespo J, Maojo V (1998) New results on the theory of morphological filters by reconstruction. Pattern Recogn 31(4):419–429
Cvancarova M, Albregtsen F, Brabrand K, Samset E (2005) Segmentation of ultrasound images of liver tumors applying snake algorithms and GVF. Int Congr Ser 1281:218–223
Donoser M, Bischof H (2006) Efficient maximally stable extremal region (MSER) tracking. In Proc. Conf Comput Vis Pattern Recognit
Donoser M, Bischof H (2006) 3D segmentation by maximally stable volumes (MSVs). 18th Int Conf Pattern Recog (ICPR) 1:63–66
Elamvazuthi I, Muhd Zain M, Begam KM (2013) Despecking of ultrasound images of bone fracture using multiple filtering algorithms. Math Comput Model 57(1–2):152–168
Fabbrini L, Greco M, Pinelli G (2014) Improved edge enhancing diffusion filter for speckle-corrupted images. IEEE Geosci Remote Sens Lett 11(1):99–103
Feng X, Shen X, Wang Q, Kim J, Hao Z, et al (2013) Learning based ensemble segmentation of anatomical structures in liver ultrasound image. Proc SPIE 8669, Med Imaging 2013: Image Process, 866947; doi:10.1117/12.2006758
Gui Y, Zhang X, Shang Y (2012) SAR image segmentation using MSER and improved spectral clustering, EURASIP J Adv Signal Process, 83
Häme Y, Pollari M (2011) Semi-automatic liver tumor segmentation with hidden Markov measure field model and non-parametric distribution estimation. Med Image Anal
http://en.wikipedia.org/wiki/S%C3%B8rensen%E2%80%93Dice_coefficient,2014.
Jiang HY, Cheng, QS (2009) Automatic 3D segmentation of CT images based on active contour models. In D. Thalmann, J.J. Shah, Q.S. Peng (Eds.), Proceedings of IEEE international conference on computer-aided design and computer graphics, pp. 540–543
Kotropoulos C, Pitas I (2003) Segmentation of ultrasonic images using support vector machines. Pattern Recogn Lett 24(4–5):715–727
Lee WL, Chen YC, Chen YC, Hsieh KS (2005) Unsupervised segmentation of ultrasonic liver images by multi-resolution fractal feature vector. Inf Sci 175:177–199
Lee M, Cho W et al (2012) Segmentation of interest region in medical volume images using geometric deformable model. Comput Biol Med 42:523–537
Li K, Lu Z, Liu W, Yin J (2012) Cytoplasm and nucleus segmentation in cervical smear images using Radiating GVF Snake. Pattern Recogn 45(4):1255–1264
Li C, Xu C, Gui C, Fox MD (2005) Level Set Evolution Without Re-initialization: A New Variational Formulation, In Proceedings of CVPR’05, vol. 1, pp. 430–436
Liu C, Tsai C, Tsui T, Yu S (2012) An improved GVF snake based breast region extrapolation scheme for digital mammograms. Expert Syst Appl 39(4):4505–4510
Masoumi H, Behrad A, Pourmina M, Roosta A (2012) Automatic liver segmentation in MRI images using an iterative watershed algorithm and artificial neural network. Biomed Signal Process Control 7(5):429–437
Matas J, Chum O, Urban M, Pajdla T (2004) Robust wide baseline stereo from maximally stable extremal regions. Image Vis Comput 22(10):761–767
Milko S, Samset E, Kadir T (2008) Segmentation of the liver in ultrasound: a dynamic texture approach. Int J Comput Assist Radiol Surg 3:143–150
Min ZF, Song EM, Jin RC, Li GK (2009) B-scan ultrasound image feature extraction for quantitative grading of fatty liver severity. J Comput Aided Des Comp Graph 21(6):752–757 (In Chinese)
Moraru L, Moldovanu S, Biswas A (2014) Optimization of breast lesion segmentation in texture feature space approach. Med Eng Phys 36(1):129–135
Nowatschin S, Markert M, Weber S, Lueth TC (2007) A system for analyzing intra-operative B-mode ultrasound scans of the liver. Annu Int Conf IEEE Eng Med Biol, pp: 1346–1349
Ostu N (1978) Discriminate and least square threshold selection. Proc Int Conf Pattern Recog, pp: 592–596
Pan S, Dawant BM (2006) Automatic 3D segmentation of the liver from abdominal CT images: a level-set approach. Proc SPIE 4322:128–138
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12:629–639
Pohle R, Toennies KD (2001) Segmentation of medical images using adaptive region growing. Proc SPIE Med Imaging 4322:1337–1346
Rani N, Thind T (2013) Segmentation of ultrasound images using closest neighbuor approach. Int J Recent Technol Eng 1(6):101–103
Rodtook A, Makhanov S (2013) Multi-feature gradient vector flow snakes for adaptive segmentation of the ultrasound images of breast cancer. J Vis Commun Image R 24:1414–1430
Smeets D, Loeckx D, Stijnen B, Dobbelaer BD, Vandermeulen D, Suetens P (2010) Semi-automatic level set segmentation of liver tumors combining a spiral-scanning technique with supervised fuzzy pixel classification. Med Image Anal 14:13–20
Tian Y, Duan F, Lu K et al (2013) A Flexible 3D Cerebrovascular Extraction from TOF-MRA Images. Neurocomputing 121:392–400
Torbati N, Ayatollahi A, Kermani A (2014) An efficient neural network based method for medical image segmentation. Comput Biol Med 44:76–87
Wu X, Spencer S (2009) etal., Development of an accelerated GVF semi-automatic contouring algorithm for radiotherapy treatment planning. Comput Biol Med 39:650–656
Xu C, Prince JL (1998) Snakes, shapes, and gradient vector flow. IEEE Trans Image Process 7(3):359–369
Acknowledgments
This work was part supported by the National Natural Science Foundation of China under grant No. 61473025, the Fundamental Research Funds for the Central Universities (YS1404), Beijing University of Chemical Technology Interdisciplinary Funds for “Visual Media Computing” and the open-project grant funded by the State Key Laboratory of Synthetical Automation for Process Industry at the Northeastern University in China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, H., Sheng, J., Zhang, F. et al. Improved maximally stable extremal regions based method for the segmentation of ultrasonic liver images. Multimed Tools Appl 75, 10979–10997 (2016). https://doi.org/10.1007/s11042-015-2822-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-015-2822-z