Abstract
Introduction:
Epigenetics studies gene expression changes influenced by environmental and lifestyle factors, linked to health conditions like reproductive aging. Male reproductive aging causes sperm decline, conceiving difficulties, and increased genetic abnormalities. Recent research focuses on epigenetics' role in male reproductive aging. Objectives: This review explores epigenetics and male reproductive aging, focusing on sperm quality, environmental and lifestyle factors' impact, and potential health implications for offspring.
Methods:
An extensive search of the literature was performed applying multiple databases, such as PubMed and Google Scholar. The search phrases employed were: epigenetics, male reproductive ageing, sperm quality, sperm quantity, environmental influences, lifestyle factors, and offspring health. This review only included articles that were published in English and had undergone a peer-review process. The literature evaluation uncovered that epigenetic alterations have a substantial influence on the process of male reproductive ageing.
Result:
Research has demonstrated that variations in the quality and quantity of sperm that occur with ageing are linked to adjustments in DNA methylation and histone. Moreover, there is evidence linking epigenetic alterations in sperm to environmental and lifestyle factors, including smoking, alcohol intake, and exposure to contaminants. These alterations can have enduring impacts on the well-being of descendants, since they can shape the activation of genes and potentially elevate the likelihood of genetic disorders. In conclusion, epigenetics significantly influences male reproductive aging, with sperm quality and quantity influenced by environmental and lifestyle factors.
Conclusion:
This underscores the need for comprehensive approaches to managing male reproductive health, and underscores the importance of considering epigenetics in diagnosis and treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As global life expectancies increase, understanding the complexities of aging is critical for addressing a variety of health concerns, including reproductive health. While female reproductive aging and its effects on fertility and pregnancy have been extensively studied, there is growing recognition of the significance of male reproductive aging as a major factor contributing to declining fertility and reproductive health outcomes [1,2,3].
As men age, their reproductive capacity gradually declines, a process known as male reproductive aging. Unlike the clearly defined menopause experienced by women, men experience a steady decrease in fertility over time. This decline significantly impacts natural conception and assisted reproductive technologies like in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) [4]. However, the consequences of male reproductive aging extend beyond fertility. Advanced paternal age has been linked to an increased risk of adverse health outcomes in children, including neurodevelopmental disorders, mental health issues, and specific genetic conditions [5, 6]. Understanding the complex interplay between aging and male reproductive function, particularly the epigenetic mechanisms involved, is crucial for developing innovative solutions to fertility challenges and safeguarding the well-being of future generations.
Epigenetics explores how heritable changes in gene expression occur without altering the DNA sequence itself [7]. These changes, mediated by mechanisms such as DNA methylation, histone modifications, and non-coding RNAs, have a profound impact on gene regulation and cellular functions [8]. In the context of male reproductive ageing, epigenetics plays a crucial role in regulating spermatogenesis, sperm function, and sperm quality [9]. Dysregulation of epigenetic processes can lead to abnormal gene expression patterns in sperm, affecting both fertility and the health of future generations. Understanding the role of epigenetic alterations in male reproductive ageing is therefore essential for unraveling the underlying mechanisms and developing strategies to mitigate their detrimental consequences.
This review delves into the intricate role of epigenetics in the aging male reproductive system, exploring how physiological changes impact fertility. It examines key epigenetic mechanisms – DNA methylation, histone modifications, and non-coding RNAs – and their influence on gene expression within the male germline. The review investigates how disruptions in these epigenetic processes affect sperm quality, fertility potential, and the well-being of offspring. By exploring the link between age-related epigenetic alterations and declining reproductive function, the study seeks to identify potential therapeutic interventions and preventative strategies targeting specific epigenetic pathways. Ultimately, the goal is to pave the way for innovative research and clinical therapies that improve reproductive health in aging men.
Search strategy
The search technique involved identifying keywords related to epigenetics, male reproductive ageing, and correlated terms like spermatogenesis and DNA methylation. Boolean operators were used to merge these keywords, and databases like PubMed, Google Scholar, Web of Science, Scopus, and Embase were used to gather relevant literature. The search was enhanced by adding more precise phrases like “histone modifications” and “sperm quality”. Filters and constraints were implemented to refine the search results. Snowballing was used to find further sources. The search strategy was iterative, with adjustments made based on initial findings. Proper documentation was maintained for transparency and reproducibility.
Epigenetics mechanism in male reproductive ageing
The main epigenetic processes that control gene expression include DNA methylation, modifications to histones, and the presence of short, non-coding RNAs.
DNA methylation
In mammals, DNA methylation primarily occurs at the 5’ position of cytosine residues, most commonly within CpG dinucleotides. Nasrullah et al. [10] found that approximately 60–80% of CpG dinucleotides in gene promoters are methylated. This methylation in promoter regions suppresses transcription by altering chromatin condensation. DNA methyltransferases (DNMTs) are the enzymes responsible for this process. During early embryonic development, ‘de novo’ DNMTs establish methylation patterns at specific chromosomal locations. Subsequently, maintenance methyltransferases (DNMT1) ensure the accurate replication of these methylation patterns during each DNA replication cycle [11]. Gametogenesis is intricately linked to DNA methylation. Primordial germ cells (PGCs) migrate into the developing gonad with a notable reduction in DNA methylation. The drop is reversed throughout male embryonic development and female postnatal folliculogenesis. Cytosine methylation, although typically associated with CpG sites, also occurs at non-CpG sites, including CpA, CpT, and CpC. The significance of these non-CpG methylations remains unclear. Research by Ross et al. [12] suggests that non-CpG methylation is prevalent in brain, embryonic stem cells (ESCs), induced pluripotent stem cells, and oocytes. Ichiyanagi et al. [13] reported its presence in male germ cells, where it is particularly pronounced in pro spermatogonia and decreases during mitosis [14].
5-hydroxymethylcytosine (5hmC) is a crucial DNA modification that plays a key role in regulating gene expression and the removal of methyl groups from DNA. This process, known as demethylation, is essential for various cellular functions, including the development of mammals and the suppression of tumor formation. Research by Zheng et al. [15] revealed the critical role of 5hmC in the formation of spermatogenic cells in mice. A subsequent study by Wang et al. compared 5hmC patterns in normal sperm, aberrant sperm, and globozoospermia spermatozoa. This analysis identified a significant number of genes (6664, 9029, and 6318, respectively) containing 5hmC in each group. The presence of 5hmC in genes linked to sperm production, motility, and morphology suggests a potential influence of this molecule on sperm characteristics [16, 17].
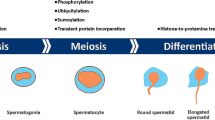
Next-generation sequencing (NGS) is a powerful tool for studying DNA methylation in humans. The NGS DNA methylation workflow involves digesting genomic DNA with methylation-sensitive restriction enzymes, enriching for methylated fragments using affinity-based methods, and chemically modifying the fragments [18]. Whole-genome bisulfite sequencing (WGBS) allows for the examination of methylation patterns across the entire genome, down to individual DNA bases. Recent WGBS studies have elucidated the methylome profiles of human embryonic stem cells and induced pluripotent stem cells [19]. The epigenetic changes associated with spermatogenesis are summarized in the diagram provided in Fig. 1.
During spermatogenesis, many epigenetic modifications, together with DNA methylation and histone moderation, occur at different stages. (1) Primordial germ cells (PGCs) undergo a demethylation process characterised by the elimination of DNA methylation, which leads to the deletion of genomic imprinting. Furthermore, this mechanism specifically impacts certain histones, particularly the K4 and K9 residues of H3. Furthermore, there is a procedure known as H4 deacetylation. Presently, the genes DNMT3A, DNMT3B, and DNMT3L are undergoing expression. (2) Spermatogonia experience a progressive DNA methylation process, leading to the formation of paternal methylation. (3) The presence of methylation on H3K9 and H3K4 is observed in spermatocytes. (4) Throughout the maturation of round spermatids, the protein H4 experiences hyperacetylation, the presence of DNMT1 is detected, and there is a shift from histones to transition proteins (TPs). (5) Lengthened spermatids demonstrate the retention of DNA methylation, together with the elimination of H3K9 methylation. At this stage, the conversion from transition proteins (TPs) to protamines takes place. (6) Sperm maintain genomic imprinting.
Histone modifications
Histone tail changes are another epigenetic chromatin marker that regulates gene expression. Histone tail post-translational changes include methylation, acetylation, phosphorylation, ubiquitination, ribosylation, and sumoylation. Histone marks are active because many enzymes can make and remove them. Acetylation of amino-terminal tail lysine residues is the principal histone modification. The quantities of this alteration are regulated by HAT and HDAC. Acetylation reduces histone-DNA binding strength, activating genes. Conversely, histone deacetylation compacts chromatin, inactivating genes transcriptionally. However, histone methylation controls transcription activation and inactivation. Lysine 4 on histone H3 (H3–K4) is linked to gene expression, but di- and tri-methylation of H3K9 and H3K27 is linked to gene silent [20]. Only animal testes contain histone H3T. Tachiwana et al. showed that Nap2 chaperone helps H3T nucleosomes assemble. Also, these nucleosomes are much less stable than H3.1-containing ones. These authors also suggested that H3T’s unstable nucleosome structure may affect spermatogenesis’ chromatin reconfiguration [20]. The first study of histone alterations used Western blot analysis with antibodies that target altered histones. Today, mass-spectrometry-based proteomic tools are used to analyze these alterations. Recently developed NGS systems can analyse histone alterations across the genome. The methods use chromatin immunoprecipitation and sequencing to map the genome-wide binding pattern of chromatin-associated proteins, including modified histones (ChIP–seq) [21].
Small non-coding RNAs
Small RNAs and long intergenic non-coding RNAs (lincRNAs) represent a third key mechanism for regulating gene activity through epigenetic modification. Sperm cells employ short non-coding RNAs to control epigenetic processes, ensuring the preservation of histone-bound DNA during the transition to protamine, a critical step in spermatogenesis and early embryonic development. LincRNAs, on the other hand, engage with chromatin to recruit histone-modifying enzymes. This interaction is exemplified by the lincRNA HOTAIR, which facilitates the modulation of H3 histone lysine 27 and 4 methylation by the PRC2 and LSD1 enzymes. The presence and activity of small non-coding RNAs can be detected using techniques such as RT-PCR, in situ hybridization, and small RNA sequencing. Moreover, microarray technologies have enabled the investigation of spermatozoa microRNAs (miRNAs), revealing distinct expression patterns in individuals with varying fertility levels.
Epigenetics changes in male’s reproductive ageing
Male reproductive health is significantly affected by the ageing process, resulting in decreased fertility and a higher likelihood of negative reproductive outcomes. Recent research indicates that epigenetic modifications are highly influential in facilitating the age-related alterations in male reproductive function [24, 25]. Male reproductive ageing is marked by a progressive decrease in reproductive capacity and fertility as one gets older. Physiological changes including variations in hormone levels, diminished semen quality, and modifications in testicular structure. The physiological changes associated with ageing lead to reductions in the production of sperm, the ability of sperm to move, and the integrity of sperm DNA. These changes eventually have a negative impact on fertility outcomes [26]
DNA methylation and histone modification and non coding RNA expression
Epigenetic processes are essential for controlling gene expression patterns without modifying the DNA sequence itself [27]. During male reproductive ageing, there are changes in epigenetic modifications that play a role in the decline of sperm quality, fertility, and the health of offspring associated with ageing. This study examines the effects of ageing on DNA methylation, histone modification, and non-coding RNA expression in the male germline, providing insights into their involvement in reproductive ageing [27].
DNA methylation is the process of adding a methyl group to cytosine residues, mainly found inside CpG dinucleotides. This epigenetic modification has been extensively researched and has a significant impact on gene expression patterns. Studies have shown that as men age, there are changes in DNA methylation patterns both globally and at specific locations [24, 28].
Research has indicated that as individuals age, the levels of DNA methylation in sperm decrease, indicating a decrease in the stability of epigenetic factors. The phenomenon of global hypomethylation in sperm from older guys may lead to genomic instability and abnormal gene expression patterns [29,30,31]. Epigenetic mechanisms play a crucial role in regulating gene expression patterns without altering the DNA sequence itself [32]. Epigenetic mutations during male reproductive ageing contribute to the deterioration of sperm quality, fertility, and the health of offspring. This work investigates the impact of ageing on DNA methylation, histone modification, and non-coding RNA expression in the male germline. It offers valuable insights into how these factors contribute to reproductive ageing [25, 29].
DNA methylation is the enzymatic addition of a methyl group to cytosine residues, primarily located inside CpG dinucleotides. The investigation of this epigenetic alteration has been thoroughly studied and has a substantial influence on the patterns of gene expression. Research has demonstrated that as males get older, there are alterations in the patterns of DNA methylation, both on a global scale and at particular sites [24, 33, 34].
Aside from worldwide modifications, there are also age-related modifications in DNA methylation that take place at particular locations in the genome, specifically in genes that play a role in spermatogenesis, hormone signalling, and fertility. For instance, it has been discovered that the promoter regions of genes that are crucial for sperm motility and fertilization experience hypermethylation, which may hinder their production and function in the ageing structure of sperm chromatin and gene expression. Changes in histone modifications during male reproductive ageing affect the organization of chromatin and the control of genes in sperm [35,36,37].
Research has shown that there are changes in the patterns of histone acetylation and methylation in sperm as male age [29]. These changes are specifically related to the compaction of chromatin and the silencing of genes, and they result in altered amounts of certain histone marks in older males. These alterations have the potential to influence the structural integrity of sperm chromatin and the patterns of gene expression, ultimately leading to consequences on fertility and the development of embryos [29].
Non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have important functions in regulating gene expression after transcription and modifying the structure of chromatin. Dysregulation of non-coding RNA expression in male reproductive ageing leads to changes in sperm function and fertility outcomes [38].
Changes in the expression profiles of microRNAs (miRNAs) have been reported in sperm as a result of ageing. These changes involve changed levels of certain miRNAs that play a role in regulating genes associated with spermatogenesis, sperm motility, and fertilization. Abnormal miRNA expression can interfere with regular sperm function and have a role in the reduction of fertility in older male [39].
Recent research suggests that long non-coding RNAs (lncRNAs) may have a substantial impact on male reproductive ageing. The expression patterns of these lncRNAs in the sperm of older boys are unique and distinguishable. Long non-coding RNAs (lncRNAs) has the capacity to regulate gene expression through many mechanisms, including chromatin modification and transcription regulation [40]. Consequently, they have the potential to influence the quality of sperm and the results of fertility. This has been evidenced in research undertaken by Zhou et al. [41].
The involvement of long non-coding RNAs (lncRNAs) in the process of male reproductive ageing underscores their many methods of impacting gene expression. The impact of long non-coding RNAs (lncRNAs) on the quality of sperm and reproductive outcomes in older males can be ascribed to many processes, including chromatin remodelling and transcriptional regulation [42].
Particular long non-coding RNAs (lncRNAs) have been found to interact with chromatin-modifying complexes, such as polycomb repressive complexes (PRCs) and lysine-specific demethylases (KDMs), to regulate chromatin structure and gene expression [43]. LncRNAs have the ability to affect the quality of sperm and the consequences of fertility by attracting complexes to certain locations in the genome. This, in turn, modifies the accessibility of chromatin and controls the activity of transcription [44].
lncRNAs can function as transcriptional regulators by directly attaching to DNA or RNA molecules. This enables them to exert influence over the function of transcription factors and RNA polymerase complexes. Through these interactions, long non-coding RNAs (lncRNAs) can precisely regulate the expression of genes involved in spermatogenesis, sperm maturation, and fertilisation. Consequently, they exert a direct impact on the reproductive outcomes of older males.
The dysregulation of long non-coding RNA (lncRNA) expression in elderly males can have profound implications for sperm quality and reproductive outcomes [45]. The altered configurations of certain long non-coding RNAs (lncRNAs) can disrupt the meticulously synchronised processes of spermatogenesis, sperm motility, and fertilization [46]. The disturbance mentioned can lead to impaired sperm function and reduced fertility in older males.
Impact of epigenetics changes on male reproductive function
Epigenetics and spermatogenesis
Sperm possess tightly packed and organized DNA, a result of a crucial transformation during their development known as spermiogenesis. This process involves the replacement of nearly all histone proteins with protamines, creating a highly condensed chromatin structure [48]. This conversion, marked by the formation of disulfide bonds, stabilizes the sperm nucleus and confers several advantages. Sperm motility is enhanced, providing the necessary propulsion for fertilization. The condensed nucleus also offers protection against harmful oxidative stress and toxic chemicals within the female reproductive system. Moreover, protamine-mediated DNA compaction inhibits transcription, ensuring the preservation of the paternal genetic blueprint. The transition from histones to protamines is a meticulously orchestrated multi-step process. Early stages involve the replacement of histones with transition proteins (TP) through histone hyperacetylation. Subsequently, the spermatids elongate, and TP1 and TP2 are replaced by protamines. These protamines, particularly P1 and the P2 family (comprising P2, P3, and P4), contribute to the compact nucleus, safeguarding genetic information and epigenetic imprints. The balance between P1 and P2 protamines appears to be critical for male fertility [48]. A P1/P2 ratio of 0.8–1.2 is associated with healthy fertility. Ratios below 0.8, however, indicate insufficient DNA condensation, leading to reduced sperm motility, count, and morphology. Furthermore, research suggests that low P1/P2 ratios increase DNA fragmentation. Reduced total levels of P1 and P2 protamines may also compromise DNA protection. Disruptions in the production of mature protamine P2 have been linked to subfertility, highlighting the importance of this protein in male reproductive health [48].
DNA methylation and histone modifications during spermatogenesis
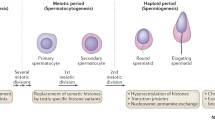
Male gametes need epigenetic alterations to mature properly. Epigenetic events begin with DNA demethylation and remethylation before meiosis. DNMT3A, DNMT3B, and DNMT3L regulate meiotic de novo DNA methylation. Pachytene spermatocytes finish this process after delivery [49, 50]. The methylation profile is maintained by DNMT1. Histone methylation and acetylation also modify DNA accessibility to transcription factors (Fig. 1). Histone methyltransferases (HMT) and demethylases (HDM) regulate the methylation of H3–K9 and H3–K4. Meiosis has high histone H3–K9 methylation, but it is removed to activate genes. However, meiosis decreases histone H3–K4 methylation and is associated to DNA silencing. During spermatogenesis, numerous enzymes, including HAT and HDAC, govern H3 and H4 lysine residue acetylation and deacetylation. During spermiogenesis, H4 hyperacetylation is necessary for histone to protamine transfer and nucleosome disintegration in elongating spermatids [50].
Environmental agents inducing epigenetic modifications
Various environmental and lifestyle factors, like s stress, physical activity, alcohol use, smoking, and shift work, have been found to impact both male and female fertility [51]. Additionally, these factors have been demonstrated to have an effect on the occurrence of epigenetic alterations, which can have implications for human diseases.
Numerous studies have highlighted the profound impact of environmental and lifestyle factors on male and female fertility through epigenetic modifications. For example, a study by Sciorio et al. [52] demonstrated that exposure to air pollution in urban areas can disrupt DNA methylation patterns in sperm, leading to decreased sperm quality and fertility [52]. Similarly, a study by Huang et al. [25] found that smoking in women alters the epigenetic profile of ova, potentially compromising embryo development and implantation success [25]. These findings emphasize the critical role of environmental factors in shaping reproductive health.
Studies conducted on several animal models have provided evidence for the existence of environmental epigenetic inheritance through gametes [53]. Several studies have indicated that both food and physical activity can have an impact on histone modifications and miRNA expression. Hudlikar et al. showed that consuming cruciferous vegetables once can reduce HDAC activity in mononuclear cells of peripheral blood, leading to increased acetylation of H3 and H4 [54]. Conversely, other studies have indicated that exposure to cigarette smoke can decrease the expression of mir-34b, mir-421, mir450-b, mir-466, and mir-469 [55]. Multiple studies have demonstrated that both diet and exercise can influence histone modifications and miRNA expression. Dashwood et al. [56] demonstrated that the consumption of cruciferous vegetables has the ability to decrease HDAC activity in mononuclear cells found in peripheral blood. This reduction in HDAC activity results in an increase in the acetylation of H3 and H4 proteins [54]. In contrast, alternative research has suggested that being exposed to cigarette smoke can reduce the manifestation of mir-34b, mir-421, mir450-b, mir-466, and mir-469 [55]. However, the most substantial amount of information is derived from research that examines the impact of the environment on DNA methylation. Evidence has shown those hazardous substances, excessive alcohol consumption, the mother’s nutrition, and smoking during foetal life can cause changes in certain regions of the genome. Additional insights in this domain have been furnished by research examining the impact of paternal exposure to different contaminants and lifestyle-related factors on the health of the child and subsequent generations [57].
Lifestyle and environmental factors like as nutrition, smoking, radiation, and alcohol usage can cause epigenetic changes that significantly impact sperm function. Initially, these mutations can cause changes in sperm that result in a decline in male fertility. When fertilisation happens, either spontaneously or by assisted reproductive technology (ART), there can be detected transgenerational epigenetic effects (as shown in Fig. 2). These effects can result in specific changes, including (1) alterations in the development of the embryo, (2) the occurrence of congenital disorders at birth, and (3) the beginning of diseases later in adult life, such as obesity, hypertension, diabetes, and so on.
Paternal exposure to toxins or ionizing radiation
Many studies have examined the effects of paternal lifestyle choices, environmental pollutants, and low ionising radiation. Previous study has repeatedly linked parental occupational toxicity to child health issues. Bond et al. [57] found considerable exhaust fume exposure in paternal grandmothers of acute lymphoblastic leukaemia patients. However, abnormalities in sperm DNA may cause several of these illnesses, suggesting a genetic rather than epigenetic process. Is there empirical evidence that epigenetic mechanisms drive transgenerational effects from paternal chemical exposure? Animal models show that male exposure to pesticides or other hazardous chemicals can cause gamete abnormalities and aberrant offspring development. This is mostly attributable to germ line DNA methylation alterations [58]. Anway et al. [59] found that exposing embryos to vinclozolin during gonadal sex determination in rats caused prostate, kidney, immune system, and testis diseases and cancers in subsequent generations. This suggests transgenerational effects. Soubry et al. [60] reported that brief vinclozolin exposure of pregnant female F0 mice during gonadal sex caused adult-onset illnesses in F3 male and female mice. Recent data suggests that ionising radiations may enhance DNA methylation alterations, which might cause genetic disorders. Radiations cause genotoxic biological processes like DNA breakage. The mechanism behind transgenerational effects is still unknown. Leung et al. [61] postulated that DNA methylation influences DNA repair pathways and transmits the radiation-exposure signal through sperm. The continuing instability in the reproductive cells of unexposed descendants of irradiation mice may generate mosaicism in germ cells, a well-known phenomenon in human genetic illnesses [58]. Recently, the Piwi-interacting RNAs (piRNA) pathway may play a major role in transgenerational radiation impacts, including genomic and epigenomic instability. DNA methylation of transposable elements helps maintain genomic integrity. It also affects cellular regulation by additional epigenetic alterations. More animal model data supports transgenerational epigenetic changes caused by parental exposure to genotoxic stressors such radiation, nutrition, and anti-androgens. Research has showed that female mice given vinclozolin have epigenetic alterations in their offspring’s sperm compared to a control group [62].
Paternal diet
The paternal diet’s effect on gametogenesis is an attractive germ line epigenetic topic. This link was first identified in animal models. Carone et al. [63] showed that male mice fed a protein-deficient diet generated progeny with higher lipid and cholesterol gene expression than those fed a normal diet. Cytosine methylation patterns were similar in all three diets, showing that these diets mainly do not affect the sperm epigenome. They also observed that modest changes in a few sites could affect organism development [64]. However, Radford et al. [65] found that poor pregnancy nutrition impairs adult sperm methylation. This shows that reproductive cell methylation may alter chromatin structure, transcriptional network differentiation, or tissue organisation. Changes may lead to environmental disease transmission across generations. In female progeny of male mice fed a high-fat diet, Ng et al. [66] found that insulin secretion and glucose tolerance decreased over time. The observed impact was aided by adult female offspring expressing 642 pancreatic islet genes differently. The 13 functional clusters of these genes include cation and ATP binding, cytoskeleton, and intracellular transport [67]. Bodden et al. [68] found that high-fat-fed male mice have altered testis transcription and global methylation in mature sperm. Changes affect future generations’ metabolism. Human evidence on diet’s effects on epigenetic alterations and transgenerational effects is available with animal model data. In 1944–45, the Germans banned all food and fuel transports to the Netherlands as retaliation for the Dutch government-in-exile’s efforts to hamper German reinforcements and troops. The official daily rations for adults dropped from 1800 cal in December 1943 to 800 cal in April 1945, causing a severe famine. After the liberation of the Netherlands in May 1945, the situation improved, and by June 1945, daily rations exceeded 2000 cal. The starvation killed many Amsterdamers, yet many babies were born. Several decades’ later, other research analysed the health of Amsterdam residents born during the famine to determine how starvation affected their descendants. For the first time, the studies linked chronic diseases in adulthood (such as coronary heart disease, atherogenic lipid profile, obesity, elevated plasma fibrinogen, and decreased factor VII) to famine during certain gestational stages. This connection was substantial in children [67]. Molecular analysis of epidemiological data showed that the Dutch famine families study provided the first direct proof of epigenetic programming caused by pregnant starvation. Research has showed that starvation during conception decreases methylation in a mother-inherited region of the IGF2 gene due to a lack of methyl donors. This implies early starvation can cause long-term epigenetic changes. IGF2 methylation did not change in late-pregnant famine victims. Hunger during embryonic development often causes long-term DNA methylation changes, according to additional studies. When exposed, gender and pregnancy stage can affect these changes [69]. Recent investigations revealed that prenatal malnutrition-related differentially methylated regions (P-DMRs) are typically found in the regulatory domains of genes with variable expression throughout early development [69]. All of the above studies show that under nutrition affects the foetus early on. Should we not think that the hunger may target other gametes besides embryos? Animal studies and other evidence clearly suggest that the father’s diet affects his children’s health. Bygren et al. [70] studied how grandparents’ nutrition affects grandchildren’s growth in humans. They observed that greater food availability during the paternal grandfather’s childhood decreased offspring survival. Later, the same research found that childhood cardiovascular disease risk was reduced when the father had limited food access before puberty. However, if the paternal grandfather ate too much during the same era, the child was at risk of diabetes death. This suggests epigenetic inheritance may explain these events. Children with fat parents had altered methylation at imprint regulatory locations, according to Soubry et al. [60] The lifestyle and nutrition of parents before conception may alter gametogenesis imprint marks. The substantial link between father obesity and kid methylation patterns suggests that developing sperm is susceptible to environmental variables. Recent research suggests that paternal grandmothers’ early dietary supply may affect female grandchildren’s cardiovascular mortality. This suggests an X-linked epigenetic inheritance through spermatozoa [60]. However, a complete study of diet-related transgenerational inheritance should focus on the third generation. The real first “unexposed” generation is directly exposed to the maternal food during gestation, while its second generation is formed from in utero gametes. Although there are limitations, transgenerational effects connected with paternal nutrition’s epigenetic impact are intriguing [71].
Therapeutic Interventions Targeting Epigenetic Changes in male’s reproductive aging
There is increasing interest in creating therapeutic therapies that specifically target these changes because of the significance that epigenetic modifications play in male reproductive aging. The aging-related characteristics of reproductive abnormalities have been addressed using pharmaceutical, nutritional, and environmental therapies [72]. Upholding a healthy lifestyle is highly correlated with a decreased likelihood of experiencing age-related epigenetic modifications in male reproductive outcomes, such as infertility. However, to prevent or treat epigenetic alterations in male reproductive aging, pharmaceutical treatments might be required. Human testing has been done on or is now being done on some of the therapies that had positive results in preclinical trials.
These potential therapeutic strategies include:
Epigenetic Modulators: Preclinical studies have demonstrated the promise of small molecules or natural compounds that can modulate epigenetic modifications [73], including antioxidant supplement and miRNA mimics or inhibitors, spermidine, NAD + precursors, sirtuin-activating compounds, metformin, mTOR inhibitor, and SGLT2-inhibitors. MicroRNA mimics and inhibitors of caloric restriction have also been studied. These substances may be able to counteract or lessen the negative effects of aging on the health of male reproduction.
DNMTi: DNMT inhibitors are medications that prevent DNMT enzyme activity, causing DNA to become hypomethylated [74]. DNMT inhibitors have the ability to restore gene expression profiles and undo age-related epigenetic alterations by modifying DNA methylation patterns [75, 76]. DNMTi medications, which include decitabine and 5-azacytidine, function by preventing DNA methyltransferases—the enzymes that add methyl groups to DNA—from doing their job [77]. This lowers DNA methylation, which modifies the expression of some genes. DNMT inhibitors have shown promise as therapeutic interventions for male reproductive aging, improving sperm quality, fertility, and testicular function [78]. Clinical trials have evaluated their safety and efficacy, with mixed results [79]. While generally well-tolerated, side effects like fatigue, nausea, and vomiting have been reported. However, the long-term safety of DNMT inhibitors in males remains to be fully established [80].
HDACi: HDACis have emerged as promising therapeutic interventions for targeting epigenetic changes in male reproductive aging. By inhibiting the activity of HDACs, HDACis can promote chromatin relaxation and the activation of genes that are essential for spermatogenesis and testicular function [81, 82]. HDACis have antioxidant and anti-inflammatory properties, which can protect against testicular damage and improve testicular function [83, 84]. HDACis, such as vorinostat and romidepsin, induce epigenetic reprogramming, restoring youthful epigenetic landscape and rejuvenating testicular function [85, 86]. These drugs inhibit histone deacetylases, increasing histone acetylation and gene expression. Epigenetic drugs can improve male fertility by targeting specific epigenetic changes contributing to infertility. Studies show that treatment with decitabine improves sperm motility and gene expression [87, 88], while a combination of decitabine and hormone therapy improves sperm quality and pregnancy rates [89]. Clinical trials in humans have shown the efficacy of HDACis in treating male reproductive aging, with studies showing improved sperm quality and pregnancy rates in older men with idiopathic infertility and restored spermatogenesis in men with azoospermia [81, 82]. However, the use of epigenetic drugs in male fertility is still in its early stages, and further research is needed to determine their effectiveness and safety.
MiRNAs: MiRNAs are essential in spermatogenesis, sperm function, and fertility, regulating aspects of male reproduction like germ cell differentiation, meiosis, and sperm maturation [90]. Dysregulation of miRNA expression can lead to impaired spermatogenesis, reduced sperm quality, and infertility [91]. MiRNAs also regulate epigenetic modifications, such as DNA methylation and histone modifications, crucial for genomic stability and gene expression [92]. MiRs, like miR-29b, target DNMT3A and DNMT3B, promoting spermatogonia differentiation and miR-101, increasing histone acetylation. MiRNAs play a crucial role in regulating sperm function, including motility, capacitation, and acrosome reaction [93]. They target genes like ADAM3 and SPEF, which are involved in sperm-egg fusion. MiRNAs also regulate epigenetic modifications in early embryos, impacting embryonic development and offspring health [94]. Given their role in male reproductive aging, miRNA-based therapeutic interventions hold great promise for treating age-related male infertility. MiRNA expression can be modulated using strategies like miRNA mimics, antagomiRs, and miRNA sponges, targeting epigenetic changes in male reproductive aging.
NAD + precursors: NAD + is a coenzyme involved in cellular processes like energy metabolism, DNA repair, and gene expression. Its decline with age is linked to age-related diseases [95]. NAD + precursors, which can be converted into NAD + , have therapeutic benefits like improved energy metabolism, reduced inflammation, and neurodegeneration protection [96]. NAD + also plays a role in epigenetic modifications, as it is a substrate for sirtuins, enzymes that deacetylate histones, which can indirectly affect gene expression [97]. NAD + also affects DNA methyltransferases and histone methyltransferases, which add methyl groups to DNA and histones [98]. NAD + can indirectly affect gene expression by regulating epigenetic modifiers. Therapeutic potential of NAD + precursors has been demonstrated in animal models of age-related diseases, improving energy metabolism, reducing inflammation, and protecting against neurodegeneration [99]. In aged mice, NAD + precursors improved sperm quality and fertility by increasing NAD + levels and decreasing epigenetic modifications associated with gene repression [100]. These findings suggest NAD + precursors may be a potential therapeutic intervention for male reproductive aging.
Sirtuin-activating compounds: Sirtuins play a crucial role in epigenetic modifications, such as DNA methylation, histone modifications, and non-coding RNA regulation [101]. They are involved in male reproductive aging, regulating germ cell development, spermatogenesis, and testicular function [102]. SIRT1 regulates gene expression in spermatogenesis by deacetylating histones and promoting histone H3K9me3 formation [103]. SIRT6 regulates telomere maintenance and DNA repair, protecting against age-related testicular dysfunction [104]. Aging leads to a decline in sirtuin activity, contributing to epigenetic modifications dysregulation and age-related diseases. Age-related decline in sirtuin activity is influenced by factors like reduced NAD + levels, increased oxidative stress, and mitochondrial dysfunction [104]. NAD + levels decrease due to increased consumption by NAD + -dependent enzymes and sirtuins [104]. Oxidative stress, a hallmark of aging, negatively impacts sirtuin activity by modifying sirtuins and causing DNA damage [105]. STACs, which target epigenetic changes in male reproductive aging, have emerged as promising therapeutic interventions. Resveratrol, a natural polyphenol found in red wine, is a well-studied STAC that enhances sirtuin activity by increasing their affinity for NAD + [106]. It has been shown to improve sperm quality, promote spermatogenesis, and protect against testicular dysfunction in animal models [107]. Synthetic STACs like SRT1720 and SRT2104, also enhance sirtuin activity, extending the lifespan of model organisms and improving age-related health outcomes [108].
Metformin: Studies have demonstrated that Metformin, a medication used to treat diabetes, can effectively slow down the ageing process and prevent age-related illnesses. Additionally, it has the potential to exert therapeutic benefits on the process of male reproductive ageing by specifically addressing alterations in epigenetic patterns [109]. Metformin alters DNA methylation patterns by suppressing DNA methyltransferases, resulting in overall reduction of DNA methylation [110]. As a consequence, there is an augmentation in gene expression and a postponement in the ageing process. Metformin’s therapeutic benefits on male reproductive ageing may be attributed to its regulation of enzymes, which in turn impacts histone modifications, resulting in alterations in chromatin structure and gene expression [110]. Metformin, a hormone, has been discovered to affect the expression of non-coding RNAs, specifically microRNAs and lncRNAs, which in turn control gene expression and cellular functions [111]. These alterations may contribute to its impact on the ageing process of male reproductive organs. Metformin has demonstrated efficacy in enhancing sperm quality, encompassing parameters such as count, motility, and morphology, in both animal models and human investigations [112]. In addition, it provides protection against testicular damage caused by variables such as oxidative stress, inflammation, and environmental pollutants [109]. Metformin has demonstrated efficacy in enhancing fertility, particularly in those afflicted with metabolic illnesses like as obesity and type 2 diabetes [113].
mTOR inhibitors: mTOR is a pivotal controller of cellular metabolism and has a vital involvement in male reproductive processes, such as spermatogenesis, testosterone synthesis, and preservation of sperm quality [114,115,116,117]. Sertoli cells and Leydig cells play a crucial role in this process. The process of ageing results in a reduction in mTOR signalling, which has an impact on the generation of sperm, testosterone, and the quality of sperm. This deterioration can also lead to epigenetic alterations, which are worsened by the process of ageing. Rapamycin, a mTOR inhibitor, has the potential to treat age-related disorders by modifying epigenetic alterations, as demonstrated by Chrienova et al. [118]. Rapamycin, a pharmaceutical compound that influences DNA methylation, histone changes, and the expression of non-coding RNA, has been discovered to enhance the process of spermatogenesis, increase testosterone production, and improve the quality of sperm in males experiencing reproductive ageing [114, 115]. This is due to the modulation of epigenetic modifications. mTOR inhibitors, which target epigenetic changes, may have potential applications in treating male reproductive aging, such as improving spermatogenesis, testosterone production, and sperm quality in aged mice [119]. Additionally, mTOR inhibitors may offer a new therapeutic approach for age-related male infertility, as infertility affects a significant proportion of the male population and its prevalence is expected to increase with age [120].
Antioxidant supplements: It has been suggested that antioxidant supplements could be used as therapeutic interventions to target epigenetic changes associated with male reproductive aging [26]. These supplements include taurine, coenzyme Q-10, L-arginine, vitamins, quercetin, N-acetyl-l-cysteine, and omega-3 fatty acids. These substances may influence epigenetic modifications and have antioxidant qualities, which could enhance male reproductive health.
N-acetyl-l-cysteine (NAC): It has been demonstrated that NAC possesses antioxidant and anti-inflammatory qualities [121]. It is a precursor to the antioxidant glutathione. It has been documented that NAC modifies histone modifications and DNA methylation, which may have an impact on gene expression [122]. NAC supplementation has been demonstrated to increase sperm quality and fertility in elderly mice in animal models, pointing to a possible function in regulating epigenetic changes in male reproductive aging [123].
Dasatinib and Quercetin: The two compounds, Dasatinib and Quercetin, have been shown to have potential therapeutic effects on aging and age-related diseases. Dasatinib, a tyrosine kinase inhibitor, has been used to treat chronic myeloid leukemia [124]. Quercetin, a flavonoid found in fruits and vegetables, has antioxidant and anti-inflammatory properties [117]. Both compounds have been shown to target epigenetic changes in various cell types and tissues. Dasatinib inhibits the activity of lysine-specific demethylase 1, leading to increased histone methylation and altered gene expression patterns [125]. Quercetin modulates DNA methylation by inhibiting DNA methyltransferases and promoting ten-eleven translocation enzymes [126]. A study by Zhang et al. [127] found that the combination of Dasatinib and Quercetin improved sperm quality and fertility in aged mice.
Vitamins: It has been demonstrated that vitamins, including C, E, and D, have antioxidant qualities and may influence epigenetic modifications. While vitamin E has been demonstrated to modulate DNA methylation and non-coding RNA expression, vitamin C has been found to modulate histone modifications and DNA methylation [128]. It has been documented that vitamin D influences histone modifications and DNA methylation, which may have an impact on gene expression [129]. Vitamin supplementation has been demonstrated in animal models to enhance sperm quality and fertility in elderly mice, indicating a possible function in regulating epigenetic modifications in male reproductive aging.
Taurine: An amino acid known for its antioxidant qualities [130, 131], taurine has also been demonstrated to control histone modifications and DNA methylation, which may have an impact on gene expression. Taurine supplementation has been demonstrated in animal models to enhance sperm quality and fertility in elderly mice, indicating a possible function in regulating epigenetic modifications in male reproductive aging [135].
Coenzyme Q-10 (CoQ10): An antioxidant called CoQ10 is essential to mitochondrial health [133]. CoQ10 may have an impact on gene expression because it has been shown to regulate histone modifications and DNA methylation. CoQ10 supplementation has been demonstrated in animal models to enhance sperm quality and fertility in elderly mice, pointing to a possible involvement in regulating epigenetic modifications in the aging male reproductive system [134].
L-arginine: An amino acid known as L-arginine is a precursor to the antioxidant molecule nitric oxide. It has been documented that L-arginine regulates histone modifications and DNA methylation, which may have an impact on gene expression. L-arginine supplementation has been demonstrated in animal models to enhance sperm quality and fertility in elderly mice, indicating a possible involvement in regulating epigenetic modifications in male reproductive aging [135].
Omega-3 fatty acids: It has been demonstrated that omega-3 fatty acids, such as docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), have antioxidant qualities and may influence epigenetic modifications. It has been documented that omega-3 fatty acids influence histone modifications and DNA methylation, which may have an impact on gene expression. Omega-3 fatty acid supplementation has been demonstrated in animal models to enhance sperm quality and fertility in elderly mice, indicating a possible involvement in regulating epigenetic modifications in male reproductive aging [136]. Research has shown that omega-3 fatty acids, specifically docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), has antioxidant properties and can potentially affect epigenetic changes. Research has shown that omega-3 fatty acids have an effect on histone modifications and DNA methylation, potentially influencing gene expression. The study conducted by Li et al. [136] showed that supplementing omega-3 fatty acids in aged mice improved the quality of sperm and increased fertility. This suggests that omega-3 fatty acids may play a role in controlling epigenetic alterations associated with male reproductive ageing.
Lifestyle interventions: Diet, exercise, and stress reduction are examples of lifestyle choices that can affect epigenetic changes and have been demonstrated to increase male fertility and sperm quality. Research has demonstrated that dietary changes can impact epigenetic alterations, which in turn can impact longevity and overall health [137]. Examples include SGLT2-inhibitors, spermidine, and other mimics of caloric restriction. In numerous animal models, calorie restriction—a dietary strategy that lowers caloric intake without leading to malnutrition—has been demonstrated to increase longevity and enhance overall health. Changes in epigenetic markers are thought to mediate this effect [138].
Spermidine, a polyamine, has various biological functions, including epigenetic regulation. It inhibits DNA methyltransferase activity, leading to DNA hypomethylation. It interacts with histones, promoting histone acetylation and reducing methylation. Spermidine treatment has shown potential in improving reproductive function in aged male rat [139]. In animal models, it reverses age-related DNA methylation and histone modification patterns, improves sperm quality, and enhances reproductive capacity [140]. Clinical trials have also shown that spermidine treatment improves sperm parameters; increases pregnancy rates [141] and reduces age-related epigenetic changes in sperm [139, 140].
Sodium-glucose cotransporter 2 (SGLT2) inhibitors are drugs used to treat type 2 diabetes by inhibiting glucose reabsorption in the kidneys, leading to increased glucose excretion and reduced blood glucose levels. They may also have benefits beyond glycemic control, such as cardiovascular health, kidney function, and weight loss. Studies have shown that SGLT2 inhibitors can modulate various epigenetic mechanisms, including DNA methylation, histone modifications, and non-coding RNA expression [142].
The consequences of aging on male reproductive health may be lessened by adopting healthy lifestyle measures, such as eating a well-balanced diet high in antioxidants, vitamins, and minerals [143]. The potential benefits of some diets and antioxidants in fostering positive sperm epigenetics have been studied [144]. In the context of male reproductive aging, the precise mechanisms by which dietary components and therapies regulate epigenetic alterations remain incompletely understood. Nonetheless, a number of plausible mechanisms have been suggested, such as changes in gene expression, adjustments to the enzymes involved in epigenetic modifications, and antioxidant and anti-inflammatory effects.
Exercise is a promising therapeutic intervention for male reproductive aging, as it induces beneficial epigenetic modifications that improve sperm quality and fertility [140]. Studies on young men and older men show that resistance training increases global DNA methylation and alters gene expression in sperm motility [145]. Sodium-glucose cotransporter 2 (SGLT2) inhibitors are medications used for the treatment of type 2 diabetes. They work by blocking the reabsorption of glucose in the kidneys, which results in higher amounts of glucose being excreted and lower levels of glucose in the bloodstream. In addition to glycemic control, they may also provide advantages in terms of cardiovascular health, kidney function, and weight loss. Research has demonstrated that SGLT2 inhibitors have the ability to regulate many epigenetic processes, such as DNA methylation, histone modifications, and non-coding RNA expression [142].
Adopting healthy lifestyle measures, such as consuming a well-balanced diet rich in antioxidants, vitamins, and minerals, may mitigate the negative effects of ageing on male reproductive health [143]. A study conducted by Pascoal et al. [142] has examined the possible advantages of certain diets and antioxidants in promoting favourable sperm epigenetics. The mechanisms through which dietary components and medications affect epigenetic modifications in male reproductive ageing are not fully known. However, several possible explanations have been proposed, including alterations in gene expression, modifications to the enzymes responsible for epigenetic changes, and the presence of antioxidant and anti-inflammatory properties.
Physical activity is a possible treatment for male reproductive ageing, since it triggers advantageous changes in the epigenome that enhance the quality of sperm and fertility [140]. Research conducted on both young and older men has demonstrated that engaging in resistance exercise leads to an increase in overall DNA methylation and brings about changes in gene expression that affect sperm motility [145]. Aerobic exercise improves sperm count and motility, thereby targeting epigenetic changes and restoring optimal gene expression patterns, leading to improved sperm quality and fertility [146].
Stress, a common factor in modern life, can impact epigenetic changes and contribute to reproductive aging. Chronic stress activates the hypothalamic–pituitary–adrenal axis, releasing stress hormones like cortisol, which affect gene expression and function [147]. Chronic stress triggers the activation of the hypothalamic–pituitary–adrenal axis, leading to the release of stress hormones such as cortisol. These chemicals have an impact on gene expression and overall functioning of the body [147]. Stress-induced epigenetic changes in males can lead to reduced sperm quality, impaired spermatogenesis, and altered testicular function [9]. Stress management approaches, including lifestyle modifications like exercise, a balanced diet, and adequate sleep, can be considered therapeutic interventions. Studies have shown that stress management approaches can improve sperm quality and function, such as in infertile men and healthy men [148].
Gene therapy: Gene therapy involves transferring genetic material into cells to correct gene expression, potentially correcting epigenetic modifications associated with male reproductive aging [149]. One approach is targeting specific genes involved in male fertility, as shown in studies on male mice with sperm production disorders and men with non-obstructive azoospermia [150, 151]. Another potential approach is targeting epigenetic regulators like DNA methyltransferases and histone deacetylases using gene-editing tools like CRISPR-Cas9 [152]. However, challenges remain, such as efficient and safe delivery of genetic material to target cells and the long-term effects on male fertility and overall health.
Stem cell-based therapy
Stem cell therapy is a method that involves the use of stem cell to correct or exchanges distorted body cells [153]. It has potential in male fertility to restore testes function and improve sperm production. One approach is using pluripotent stem cells, which can differentiate into any cell type, to generate sperm cells in the lab [154]. However, challenges remain, such as ensuring safety and effectiveness. Another approach is using adult stem cells, such as spermatogonial stem cells, found in testes, to extract and correct epigenetic changes in infertile men, and is currently being explored in clinical trials [154].
Conclusion
This review has delved into the intricate interplay between epigenetics and male reproductive aging, revealing a complex tapestry of molecular mechanisms that govern reproductive decline. By exploring the dynamic interplay of genetic and environmental factors, we have shed light on the critical role of epigenetic modifications in shaping male reproductive health across the lifespan. The findings highlight the importance of understanding these epigenetic modifications, particularly in the context of age-related decline in sperm quality, testicular function, and fertility. We have discussed the specific epigenetic alterations that contribute to these changes, including DNA methylation, histone modifications, and non-coding RNA regulation. These alterations, driven by factors such as oxidative stress, hormonal fluctuations, and lifestyle choices, can profoundly impact gene expression and ultimately lead to impaired reproductive capacity. Furthermore, the review has explored the potential therapeutic implications of targeting these epigenetic mechanisms. By restoring epigenetic balance and reversing age-related alterations, we may be able to mitigate the impact of male reproductive aging and potentially enhance fertility. We have discussed promising strategies, including dietary interventions, lifestyle modifications, and pharmacological approaches that aim to modulate epigenetic marks and restore reproductive function. Ultimately, this review underscores the growing recognition of epigenetics as a critical player in male reproductive aging. By unraveling the mysteries of this complex field, we are paving the way for novel therapeutic interventions that could hold the key to addressing the challenges of age-related decline in male reproductive health.
Data availability
No datasets were generated or analysed during the current study.
References
Martins da Silva S, Anderson RA (2022) Reproductive axis ageing and fertility in men. Rev Endocr Metabol Disord 23(6):1109–1121
Aitken RJ (2023) Male reproductive ageing: a radical road to ruin. Hum Reprod 38(10):1861–1871
De Jonge CJ, Barratt CL, Aitken RJ, Anderson RA, Baker P, Chan DY, Connolly MP, Eisenberg ML, Garrido N, Jørgensen N, Kimmins S (2024) Current global status of male reproductive health. Human Reprod Open 2024(2):hoae017
Catford SR, Halliday J, Lewis S, O’Bryan MK, Handelsman DJ, Hart RJ, McBain J, Rombauts L, Amor DJ, Saffery R, McLachlan RI (2022) Reproductive function in men conceived with in vitro fertilization and intracytoplasmic sperm injection. Fertil Steril 117(4):727–737
Kaltsas A, Moustakli E, Zikopoulos A, Georgiou I, Dimitriadis F, Symeonidis EN, Markou E, Michaelidis TM, Tien DM, Giannakis I, Ioannidou EM (2023) Impact of advanced paternal age on fertility and risks of genetic disorders in offspring. Genes 14(2):486
Kaltsas A, Zikopoulos A, Vrachnis D, Skentou C, Symeonidis EN, Dimitriadis F, Stavros S, Chrisofos M, Sofikitis N, Vrachnis N, Zachariou A (2024) Advanced Paternal Age in Focus: Unraveling Its Influence on Assisted Reproductive Technology Outcomes. J Clin Med 13(10):2731
Colwell ML, Townsel C, Petroff RL, Goodrich JM, Dolinoy DC (2023) Epigenetics and the exposome: DNA methylation as a proxy for health impacts of prenatal environmental exposures. Exposome. 3(1):osad001
Prandi FR, Lecis D, Illuminato F, Milite M, Celotto R, Lerakis S, Romeo F, Barillà F (2022) Epigenetic modifications and non-coding rna in diabetes-mellitus-induced coronary artery disease: pathophysiological link and new therapeutic frontiers. Int J Mol Sci 23(9):4589
Sudhakaran G, Kesavan D, Kandaswamy K, Guru A, Arockiaraj J (2024) Unravelling the epigenetic impact: oxidative stress and its role in male infertility-associated sperm dysfunction. Reprod Toxicol 2:108531
Nasrullah HA, Ahmed S, Rasool M, Shah AJ (2022) DNA methylation across the tree of life from micro to macro-organism. Bioengineered 13(1):1666–1685
Wu X, Zhang H, Zhang B, Zhang Y, Wang Q, Shen W, Wu X, Li L, Xia W, Nakamura R, Liu B (2021) Methylome inheritance and enhancer dememorization reset an epigenetic gate safeguarding embryonic programs. Sci Adv 7(52):eabl3858
Ross SE, Angeloni A, Geng FS, de Mendoza A, Bogdanovic O (2020) Developmental remodelling of non-CG methylation at satellite DNA repeats. Nucleic Acids Res 48(22):12675–12688
Ichiyanagi T, Ichiyanagi K, Miyake M, Sasaki H (2013) Accumulation and loss of asymmetric non-CpG methylation during male germ-cell development. Nucleic Acids Res 41(2):738–745
Cincotta S (2022) Glucocorticoid Receptor Signaling in the Mammalian Germline. University of California, San Francisco
Zheng H, Zhou X, Li DK, Yang F, Pan H, Li T, Miao M, Li R, Yuan W (2017) Genome-wide alteration in DNA hydroxymethylation in the sperm from bisphenol A-exposed men. PLoS ONE 12(6):e0178535
Marcho C, Oluwayiose OA, Pilsner JR (2020) The preconception environment and sperm epigenetics. Andrology 8(4):924–942
Martisova A, Holcakova J, Izadi N, Sebuyoya R, Hrstka R, Bartosik M (2021) DNA methylation in solid tumors: functions and methods of detection. Int J Mol Sci 22(8):4247
Sharma M, Verma RK, Kumar S, Kumar V (2022) Computational challenges in detection of cancer using cell-free DNA methylation. Comput Struct Biotechnol J 1(20):26–39
Cao P, Li H, Zuo Y, Nashun B (2020) Characterization of DNA methylation patterns and mining of epigenetic markers during genomic reprogramming in SCNT embryos. Front Cell Dev Biol 2(8):570107
Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, Bonneil E, Thibault P, Verreault A, Festenstein RJ (2011) H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet 7(3):e1001354
Petryk N, Reveron-Gomez N, Gonzalez-Aguilera C, Dalby M, Andersson R, Groth A (2021) Genome-wide and sister chromatid-resolved profiling of protein occupancy in replicated chromatin with ChOR-seq and SCAR-seq. Nat Protoc 16(9):4446–4493
Miller D, Brinkworth M, Iles D (2010) Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139(2):287–301
Khawar MB, Mehmood R, Roohi N (2019) MicroRNAs: recent insights towards their role in male infertility and reproductive cancers. Bosn J Basic Med Sci 19(1):31
Oluwayiose OA, Wu H, Saddiki H, Whitcomb BW, Balzer LB, Brandon N, Suvorov A, Tayyab R, Sites CK, Hill L, Marcho C (2021) Sperm DNA methylation mediates the association of male age on reproductive outcomes among couples undergoing infertility treatment. Sci Rep 11(1):3216
Huang XC, Jiang YN, Bao HJ, Wang JL, Lin RJ, Yuan J, Xian JY, Zhao Y, Chen S (2025) Role and mechanism of epigenetic regulation in the aging of germ cells: prospects for targeted interventions. Aging Dis 16(1):2
Wang JJ, Wang SX, Feng Y, Zhang RF, Li XY, Sun Q, Ding J (2022) Age-related decline of male fertility: mitochondrial dysfunction and the antioxidant interventions. Pharmaceuticals 15(5):519
Chakrabarti SK, Chattopadhyay D (2024) Expanding role of epigenetics in human health and disease. Explor Res Hypothes Med. https://doi.org/10.14218/ERHM.2023.00086
El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J (2007) Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet 122:505–514
Ashapkin V, Suvorov A, Pilsner JR, Krawetz SA, Sergeyev O (2023) Age-associated epigenetic changes in mammalian sperm: implications for offspring health and development. Hum Reprod Update 29(1):24–44
Donkin I, Barrès R (2018) Sperm epigenetics and influence of environmental factors. Mol Metabol 1(14):1–1
Santana VP, Miranda-Furtado CL, Pedroso DC, Eiras MC, Vasconcelos MA, Ramos ES, Calado RT, Ferriani RA, Esteves SC, Dos Reis RM (2019) The relationship among sperm global DNA methylation, telomere length, and DNA fragmentation in varicocele: a cross-sectional study of 20 cases. System Biol Reprod Med 65(2):95–104
Cholewa-Waclaw J, Bird A, von Schimmelmann M, Schaefer A, Yu H, Song H, Madabhushi R, Tsai LH (2016) The role of epigenetic mechanisms in the regulation of gene expression in the nervous system. J Neurosci 36(45):11427–11434
Yusipov I, Bacalini MG, Kalyakulina A, Krivonosov M, Pirazzini C, Gensous N, Ravaioli F, Milazzo M, Giuliani C, Vedunova M, Fiorito G (2020) Age-related DNA methylation changes are sex-specific: a comprehensive assessment. Aging (Albany NY) 12(23):24057
Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A (2019) The role of DNA methylation in epigenetics of aging. Pharmacol Ther 1(195):172–185
Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT (2011) Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 26(9):2558–2569
Sharma R, Agarwal A, Rohra VK, Assidi M, Abu-Elmagd M, Turki RF (2015) Effects of increased paternal age on sperm quality, reproductive outcome and associated epigenetic risks to offspring. Reprod Biol Endocrinol 13:1–20
Tatehana M, Kimura R, Mochizuki K, Inada H, Osumi N (2020) Comprehensive histochemical profiles of histone modification in male germline cells during meiosis and spermiogenesis: comparison of young and aged testes in mice. PLoS ONE 15(4):e0230930
Caponnetto A, Battaglia R, Ferrara C, Vento ME, Borzì P, Paradiso M, Scollo P, Purrello M, Longobardi S, D’Hooghe T, Valerio D (2022) Down-regulation of long non-coding RNAs in reproductive aging and analysis of the lncRNA-miRNA-mRNA networks in human cumulus cells. J Assist Reprod Genet 39(4):919–931
Dong S, Chen C, Zhang J, Gao Y, Zeng X, Zhang X (2022) Testicular aging, male fertility and beyond. Front Endocrinol 13(13):1012119
Holt JE, Stanger SJ, Nixon B, McLaughlin EA (2016) Non-coding RNA in spermatogenesis and epididymal maturation. Non-Cod RNA Reprod Sys. https://doi.org/10.1007/978-94-017-7417-8_6
Zhou F, Chen W, Jiang Y, He Z (2019) Regulation of long non-coding RNAs and circular RNAs in spermatogonial stem cells. Reproduction 158(1):R15-25
Wu D, Khan FA, Huo L, Sun F, Huang C (2022) Alternative splicing and MicroRNA: epigenetic mystique in male reproduction. RNA Biol 19(1):162–175
Bure IV, Nemtsova MV, Kuznetsova EB (2022) Histone modifications and non-coding RNAs: mutual epigenetic regulation and role in pathogenesis. Int J Mol Sci 23(10):5801
Zhou G, Zhang M, Zhang J, Feng Y, Xie Z, Liu S, Zhu D, Luo Y (2022) The gene regulatory role of non-coding RNAs in non-obstructive azoospermia. Front Endocrinol 18(13):959487
Robles V, Valcarce DG, Riesco MF (2019) Non-coding RNA regulation in reproduction: their potential use as biomarkers. Non-Coding RNA Res 4(2):54–62
Cheng Y, Vechtova P, Fussy Z, Sterba J, Linhartová Z, Rodina M, Tučková V, Gela D, Samarin AM, Lebeda I, Xin M (2021) Changes in phenotypes and DNA methylation of in vitro aging sperm in common carp Cyprinus carpio. Int J Mol Sci 22(11):5925
Santiago J, Patrício D, Silva JV. (2020). Testicular signaling: team work in sperm production. Tissue-Specific Cell Signaling. 225–55.
Zafar MI, Chen X (2024) Effects of Calorie Restriction on Preserving Male Fertility Particularly in a State of Obesity. Curr Obes Rep 15:1–9
Cescon M, Chianese R, Tavares RS (2020) Environmental impact on male (in) fertility via epigenetic route. J Clin Med 9(8):2520
Chioccarelli T, Pierantoni R, Manfrevola F, Porreca V, Fasano S, Chianese R, Cobellis G (2020) Histone post-translational modifications and CircRNAs in mouse and human spermatozoa: potential epigenetic marks to assess human sperm quality. J Clin Med 9(3):640
Bala R, Singh V, Rajender S, Singh K (2021) Environment, lifestyle, and female infertility. Reprod Sci 28:617–638
Sciorio R, Tramontano L, Adel M, Fleming S (2024) Decrease in sperm parameters in the 21st century: obesity, lifestyle, or environmental factors? an updated narrative review. J Personalized Med 14(2):198
Thompson RP, Nilsson E, Skinner MK (2020) Environmental epigenetics and epigenetic inheritance in domestic farm animals. Anim Reprod Sci 1(220):106316
Hudlikar R, Wang L, Wu R, Li S, Peter R, Shannar A, Chou PJ, Liu X, Liu Z, Kuo HC, Kong AN (2021) Epigenetics/epigenomics and prevention of early stages of cancer by isothiocyanates. Cancer Prev Res 14(2):151–164
Stuppia L, Franzago M, Ballerini P, Gatta V, Antonucci I (2015) Epigenetics and male reproduction: the consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clin Epigenetic 7:1–5
Dashwood RH, Ho E (2007) Dietary histone deacetylase inhibitors: from cells to mice to man. Inseminar Cancer Biol 17(5):363–436
Bonde JP, Tøttenborg SS, Hougaard KS (2019) Paternal environmental exposure and offspring health. Curr Opin Endocr Metabol Res 1(7):14–20
Han X, Huang Q (2021) Environmental pollutants exposure and male reproductive toxicity: The role of epigenetic modifications. Toxicology 30(456):152780
Anway MD, Rekow SS, Skinner MK (2008) Comparative anti-androgenic actions of vinclozolin and flutamide on transgenerational adult onset disease and spermatogenesis. Reprod Toxicol 26(2):100–6
Soubry A, Hoyo C, Jirtle RL, Murphy SK (2014) A paternal environmental legacy: evidence for epigenetic inheritance through the male germ line. BioEssays 36(4):359–371
Leung CT, Yang Y, Yu KN, Tam N, Chan TF, Lin X, Kong RY, Chiu JM, Wong AS, Lui WY, Yuen KW (2021) Low-dose radiation can cause epigenetic alterations associated with impairments in both male and female reproductive cells. Front Genet 2(12):710143
Nilsson E, King SE, McBirney M, Kubsad D, Pappalardo M, Beck D, Sadler-Riggleman I, Skinner MK (2018) Vinclozolin induced epigenetic transgenerational inheritance of pathologies and sperm epimutation biomarkers for specific diseases. PLoS ONE 13(8):e0202662
Nelson NG, Wu L, Maier MT, Lam D, Cheang R, Alba D, Huang A, Neumann DA, Hill T, Vagena E, Barsh GS (2022) A gene–diet interaction controlling relative intake of dietary carbohydrates and fats. Mol Metabol 1(58):101442
Kilama J, Dahlen CR, Reynolds LP, Amat S (2024) Contribution of the seminal microbiome to paternal programming. Biol Reprod. https://doi.org/10.1093/biolre/ioae068
Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, Peters AH (2014) In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 345(6198):1255903
Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ (2010) Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467(7318):963–966
Breton CV, Landon R, Kahn LG, Enlow MB, Peterson AK, Bastain T, Braun J, Comstock SS, Duarte CS, Hipwell A, Ji H (2021) Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Commun Biol 4(1):769
Bodden C, Hannan AJ, Reichelt AC (2020) Diet-induced modification of the sperm epigenome programs metabolism and behavior. Trends Endocrinol Metab 31(2):131–49
Mileva-Seitz VR, Bakermans-Kranenburg MJ, van IJzendoorn MH (2016) Genetic mechanisms of parenting. Horm Behav. https://doi.org/10.1016/j.yhbeh.2015.06.003
Bygren LO, Tinghög P, Carstensen J, Edvinsson S, Kaati G, Pembrey ME, Sjöström M (2014) Change in paternal grandmothers early food supply influenced cardiovascular mortality of the female grandchildren. BMC Genet 15:1–6
Guo T, Luo F, Lin Q (2020) You are affected by what your parents eat: diet, epigenetics, transgeneration and intergeneration. Trends Food Sci Technol 1(100):248–261
Li Z, Zhang Z, Ren Y, Wang Y, Fang J, Yue H, Ma S, Guan F (2021) Aging and age-related diseases: from mechanisms to therapeutic strategies. Biogerontology 22(2):165–187
Jin Y, Liu T, Luo H, Liu Y, Liu D (2022) Targeting epigenetic regulatory enzymes for cancer therapeutics: novel small-molecule epidrug development. Front Oncol 28(12):848221
Gonzalez-Fierro A, Dueñas-González A (2019) Emerging DNA methylation inhibitors for cancer therapy: challenges and prospects. Expert Rev Prec Med Drug Dev 4(1):27–35
la Torre A, Lo Vecchio F, Greco A (2023) Epigenetic mechanisms of aging and aging-associated diseases. Cells 12(8):1163
Wu Z, Zhang W, Qu J, Liu GH (2024) Emerging epigenetic insights into aging mechanisms and interventions. Trend Pharmacol Sci. https://doi.org/10.1016/j.tips.2023.12.002
Gallimore F, Fandy TE (2023) Therapeutic applications of azanucleoside analogs as dna demethylating agents. Epigenomes 7(3):12
Tiwari P, Yadav A, Kaushik M, Dada R (2024) Cancer risk and male Infertility: Unravelling predictive biomarkers and prognostic indicators. Clin Chim Acta 16:119670
Zavras PD, Shastri A, Goldfinger M, Verma AK, Saunthararajah Y (2021) Clinical trials assessing hypomethylating agents combined with other therapies: causes for failure and potential solutions. Clin Cancer Res 27(24):6653–6661
Goffin J, Eisenhauer E (2002) DNA methyltransferase inhibitors—state of the art. Ann Oncol 13(11):1699–1716
Milazzo G, Mercatelli D, Di Muzio G, Triboli L, De Rosa P, Perini G, Giorgi FM (2020) Histone deacetylases (HDACs): evolution, specificity, role in transcriptional complexes, and pharmacological actionability. Genes 11(5):556
Yin H, Kang Z, Zhang Y, Gong Y, Liu M, Xue Y, He W, Wang Y, Zhang S, Xu Q, Fu K (2021) HDAC3 controls male fertility through enzyme-independent transcriptional regulation at the meiotic exit of spermatogenesis. Nucleic Acid Res 49(9):5106–5123
Dai Y, Wei T, Shen Z, Bei Y, Lin H, Dai H (2021) Classical HDACs in the regulation of neuroinflammation. Neurochem Int 1(150):105182
Mishra A, Mishra PS, Bandopadhyay R, Khurana N, Angelopoulou E, Paudel YN, Piperi C (2021) Neuroprotective potential of chrysin: mechanistic insights and therapeutic potential for neurological disorders. Molecules 26(21):6456
Maes K, Mondino A, Lasarte JJ, Agirre X, Vanderkerken K, Prosper F, Breckpot K (2021) Epigenetic modifiers: anti-neoplastic drugs with immunomodulating potential. Front Immunol 30(12):652160
Koijam AS, Singh KD, Nameirakpam BS, Haobam R, Rajashekar Y (2024) Drug addiction and treatment: an epigenetic perspective. Biomed Pharmacother 1(170):115951
Kläver R, Sánchez V, Damm OS, Redmann K, Lahrmann E, Sandhowe-Klaverkamp R, Rohde C, Wistuba J, Ehmcke J, Schlatt S, Gromoll J (2015) Direct but no transgenerational effects of decitabine and vorinostat on male fertility. PLoS ONE 10(2):e0117839
Ye C, Jiang N, Zheng J, Zhang S, Zhang J, Zhou J (2023) Epigenetic therapy: Research progress of decitabine in the treatment of solid tumors. Biochim Biophys Acta Rev Cancer. https://doi.org/10.1016/j.bbcan.2023.189066
Katz TA, Huang Y, Davidson NE, Jankowitz RC (2014) Epigenetic reprogramming in breast cancer: from new targets to new therapies. Ann Med 46(6):397–408
Harchegani AB, Shafaghatian H, Tahmasbpour E, Shahriary A (2018) Regulatory functions of MicroRNAs in male reproductive health: a new approach to understanding male infertility. Reprod Sci 1:1933719118765972
Shi Z, Yu M, Guo T, Sui Y, Tian Z, Ni X, Chen X, Jiang M, Jiang J, Lu Y, Lin M (2024) MicroRNAs in spermatogenesis dysfunction and male infertility: clinical phenotypes, mechanisms and potential diagnostic biomarkers. Front Endocrinol 16(15):1293368
Yao Q, Chen Y, Zhou X (2019) The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol 1(51):11–17
Alves MB, Celeghini EC, Belleannée C (2020) From sperm motility to sperm-borne microRNA signatures: new approaches to predict male fertility potential. Front in Cell Dev Biol 21(8):791
Cai S, Quan S, Yang G, Chen M, Ye Q, Wang G, Yu H, Wang Y, Qiao S, Zeng X (2021) Nutritional status impacts epigenetic regulation in early embryo development: a scoping review. Adv Nutr 12(5):1877–1892
Chu X, Raju RP (2022) Regulation of NAD+ metabolism in aging and disease. Metabolism 1(126):154923
Amjad S, Nisar S, Bhat AA, Frenneaux MP, Fakhro K, Haris M, Reddy R, Patay Z, Baur J, Bagga P (2021) Role of NAD+ in regulating cellular and metabolic signaling pathways. Mol Metabol 1(49):101195
Schiedel M, Robaa D, Rumpf T, Sippl W, Jung M (2018) The current state of NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med Res Rev 38(1):147–200
Roberti A, Fernández AF, Fraga MF (2021) Nicotinamide N-methyltransferase: At the crossroads between cellular metabolism and epigenetic regulation. Molecular Metabolism 1(45):101165
Braidy N, Liu Y (2020) NAD+ therapy in age-related degenerative disorders: A benefit/risk analysis. Exp Gerontol 1(132):110831
Feng YQ, Liu X, Zuo N, Yu MB, Bian WM, Han BQ, Sun ZY, De Felici M, Shen W, Li L (2024) NAD+ precursors promote the restoration of spermatogenesis in busulfan-treated mice through inhibiting Sirt2-regulated ferroptosis. Theranostics 14(6):2622
Kosciuk T, Wang M, Hong JY, Lin H (2019) Updates on the epigenetic roles of sirtuins. Curr Opin Chem Biol 1(51):18–29
Loganathan C, Kannan A, Panneerselvam A, Mariajoseph-Antony LF, Kumar SA, Anbarasu K, Prahalathan C (2021) The possible role of sirtuins in male reproduction. Mol Cell Biochem 476:2857–2867
Khawar MB, Sohail AM, Li W (2022) SIRT1: A key player in male reproduction. Life 12(2):318
Zhao Y, Bai X, Jia X, Lu Y, Cheng W, Shu M, Zhu Y, Zhu L, Wang L, Shu Y, Song Y (2021) Age-related changes of human serum Sirtuin6 in adults. BMC Geriatr 21:1
Wątroba M, Szukiewicz D (2016) The role of sirtuins in aging and age-related diseases. Adv Med Sci 61(1):52–62
Abdullah A, Mohd Murshid N, Makpol S (2020) Antioxidant modulation of mTOR and sirtuin pathways in age-related neurodegenerative diseases. Mol Neurobiol 57(12):5193–5207
Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Annu Rev Biochem 73(1):417–435
Massudi H, Wu LE, Sinclair DA. Sirtuin activation by small molecules. Sirtuins. 2016:243–66.
Li R, Luo X, Li L, Peng Q, Yang Y, Zhao L, Ma M, Hou Z (2016) The protective effects of melatonin against oxidative stress and inflammation induced by acute cadmium exposure in mice testis. Biol Trace Elem Res 170:152–164
Cuyàs E, Fernández-Arroyo S, Verdura S, García RÁ, Stursa J, Werner L, Blanco-González E, Montes-Bayón M, Joven J, Viollet B, Neuzil J (2018) Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism. Oncogene 37(7):963–970
Gholami M, Klashami ZN, Ebrahimi P, Mahboobipour AA, Farid AS, Vahidi A, Zoughi M, Asadi M, Amoli MM (2023) Metformin and long non-coding RNAs in breast cancer. J Transl Med 21(1):155
Pourheydar B, Azarm F, Farjah G, Karimipour M, Pourheydar M (2021) Effect of silymarin and metformin on the sperm parameters and histopathological changes of testes in diabetic rats: an experimental study. Int J Reprod BioMed 19(12):1091
Shpakov AO (2021) Improvement effect of metformin on female and male reproduction in endocrine pathologies and its mechanisms. Pharmaceuticals 14(1):42
Oyovwi MO, Edesiri TP, Victor E, Kingsley NE, Rume RA, Faith FY, Simon OI, Oghenetega BO, Agbonifo-Chijiokwu E (2022) D-ribose-L-cysteine abrogates testicular maladaptive responses induced by polychlorinated bisphenol intoxication in rats via activation of the mTOR signaling pathway mediating inhibition of apoptosis, inflammation, and oxidonitrergic flux. J Biochem Mol Toxicol 36(10):e23161
Oyovwi MO, Simon OI, Benneth BA, Edesiri TP, Victor E, Kingsley NE (2022) D-ribose-L-cysteine activated autophagic related protein pathway: a possible therapeutic mechanism for attenuating testicular maladaptive responses. Biochem Anal Biochem 11:441
Oyovwi MO, Oghenetega OB, Faith FY, Victor E, Rume RA, Uchechukwu JG, Abioye OA (2023) Protein kinase inhibitors affect spermatogenic functions and blood testis barrier remodelling: a scoping review. Asian Pac J Reprod 12(3):97–108
Oyovwi MO, Oghenetega OB, Victor E, Faith FY, Uchechukwu JG (2023) Quercetin protects against levetiracetam induced gonadotoxicity in rats. Toxicology 1(491):153518
Chrienova Z, Nepovimova E, Kuca K (2021) The role of mTOR in age-related diseases. J Enzyme Inhib Med Chem 36(1):1678–1692
Moreira BP, Oliveira PF, Alves MG (2019) Molecular mechanisms controlled by mTOR in male reproductive system. Int J Mol Sci 20(7):1633
Jesus TT, Oliveira PF, Sousa M, Cheng CY, Alves MG (2017) Mammalian target of rapamycin (mTOR): a central regulator of male fertility? Crit Rev Biochem Mol Biol 52(3):235–253
Emojevwe V, Nwangwa EK, Naiho AO, Oyovwi MO, Ben-Azu B (2022) Toxicological outcome of phthalate exposure on male fertility: Ameliorative impacts of the co-administration of N-acetylcysteine and zinc sulfate in rats. Middle East Fertility Soc J 27(1):5
Yuan C, Wang L, Zhu L, Ran B, Xue X, Wang Z (2019) N-acetylcysteine alleviated bisphenol A-induced testicular DNA hypermethylation of rare minnow (Gobiocypris rarus) by increasing cysteine contents. Ecotoxicol Environ Saf 30(173):243–250
Jannatifar R, Parivar K, Roodbari NH, Nasr-Esfahani MH (2019) Effects of N-acetyl-cysteine supplementation on sperm quality, chromatin integrity and level of oxidative stress in infertile men. Reprod Biol Endocrinol 17:1–9
Aguilera DG, Tsimberidou AM (2009) Dasatinib in chronic myeloid leukemia: a review. Ther Clin Risk Manag 4:281–289
White JC, Pucci P, Crea F (2019) The role of histone lysine demethylases in cancer cells’ resistance to tyrosine kinase inhibitors. Cancer Drug Res 2(2):326
Wang Q, Ma C, Wang N, Mao H. Effects of quercetin on the DNA methylation pattern in tumor therapy: an updated review. Food & Function. 2024.
Garcia DN, Hense JD, Zanini BM, Isola JV, Pradiee J, Prosczek JB, Alvarado-Rincón JA, Mondadori RG, Mason JB, Brieño-Enríquez MA, Barros CC (2023) Dasatinib and quercetin increase testosterone and sperm concentration in mice. Physiology International 110(2):121–134
Brabson JP, Leesang T, Mohammad S, Cimmino L (2021) Epigenetic regulation of genomic stability by vitamin C. Front Genet 4(12):675780
Wimalawansa SJ (2019) Vitamin D deficiency: effects on oxidative stress, epigenetics, gene regulation, and aging. Biology 8(2):30
Ben-Azu B, Adebayo OG, Jarikre TA, Oyovwi MO, Edje KE, Omogbiya IA, Eduviere AT, Moke EG, Chijioke BS, Odili OS, Omondiabge OP (2022) Taurine, an essential β-amino acid insulates against ketamine-induced experimental psychosis by enhancement of cholinergic neurotransmission, inhibition of oxidative/nitrergic imbalances, and suppression of COX-2/iNOS immunoreactions in mice. Metab Brain Dis 37(8):2807–2826
Oyovwi MO, Nwangwa EK, Ben-Azu B, Rotue RA, Edesiri TP, Emojevwe V, Igweh JC, Uruaka CI (2021) Prevention and reversal of chlorpromazine induced testicular dysfunction in rats by synergistic testicle-active flavonoids, taurine and coenzyme-10. Reprod Toxicol 1(101):50–62
Freitas IN, dos Reis AT, Vettorazzi JF, Magalhães EA, Carneiro EM, Bonfleur ML, Ribeiro RA (2019) Taurine supplementation in high-fat diet fed male mice attenuates endocrine pancreatic dysfunction in their male offspring. Amino Acids 1(51):727–738
Oyovwi MO, Ben-Azu B, Agbonifo-Chijiokwu E, Moke EG, Ajayi AM, Wilson JI, Omenogor P, Nwangwa EK, Igweh JC (2022) Possible mechanisms involved in the prevention and reversal of chlorpromazine-induced testicular damage by taurine and coenzyme-Q10 in rats. Nutrir 47(2):31
Vaamonde D, Hackney AC, Algar-Santacruz C, Garcia-Moreno MJ, García-Manso JM. Coenzyme Q 10 in Fertility and Reproduction. Coenzyme Q in Aging. 2020: 283–308
Akhigbe RE, Afolabi OA, Ajayi AF (2022) L-Arginine reverses maternal and pre-pubertal codeine exposure-induced sexual dysfunction via upregulation of androgen receptor gene and NO/cGMP signaling. PLoS ONE 17(9):e0274411
Li M, Shi Q, Jiang X, Liu X, Han W, Fan X, Li P, Qi K (2022) Paternal preconceptional diet enriched with n-3 polyunsaturated fatty acids affects offspring brain function in mice. Front Nutr 28(9):969848
McKay JA, Mathers JC (2011) Diet induced epigenetic changes and their implications for health. Acta Physiol 202(2):103–118
Green CL, Lamming DW, Fontana L (2022) Molecular mechanisms of dietary restriction promoting health and longevity. Nat Rev Mol Cell Biol 23(1):56–73
Filfan M, Olaru A, Udristoiu I, Margaritescu C, Petcu E, Hermann DM, Popa-Wagner A (2020) Long-term treatment with spermidine increases health span of middle-aged Sprague–Dawley male rats. Geroscience 42:937–949
Wang JY, Ma D, Luo M, Tan YP, Zhong O, Tian G, Lv YT, Li MX, Chen X, Tang ZH, Hu LL (2022) Effect of spermidine on ameliorating spermatogenic disorders in diabetic mice via regulating glycolysis pathway. Reprod Biol Endocrinol 20(1):45
Zhang Y, Bai J, Cui Z, Li Y, Gao Q, Miao Y, Xiong B (2023) Polyamine metabolite spermidine rejuvenates oocyte quality by enhancing mitophagy during female reproductive aging. Nature Aging 3(11):1372–1386
Scisciola L, Taktaz F, Fontanella RA, Pesapane A, Surina CV, Ghosh P, Franzese M, Puocci A, Paolisso P, Rafaniello C (2023) Targeting high glucose-induced epigenetic modifications at cardiac level: the role of SGLT2 and SGLT2 inhibitors. Cardiovascular Diabetology. 22(1):24
D’Aquila P, Montesanto A, De Rango F, Guarasci F, Passarino G, Bellizzi D (2019) Epigenetic signature: Implications for mitochondrial quality control in human aging. Aging (Albany NY) 11(4):1240
Pascoal GD, Geraldi MV, Maróstica MR Jr, Ong TP (2022) Effect of paternal diet on spermatogenesis and offspring health: focus on epigenetics and interventions with food bioactive compounds. Nutrients 14(10):2150
Ingerslev LR, Donkin I, Fabre O, Versteyhe S, Mechta M, Pattamaprapanont P, Mortensen B, Krarup NT, Barrès R (2018) Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clin Epigenetic 10:1–1
Vaamonde D, Garcia-Manso JM, Hackney AC. Effect of exercise and lifestyles on male reproductive potential. InFertility, Pregnancy, and Wellness 2022 Jan 1 (pp. 131–147). Elsevier.
Milligan Armstrong A, Porter T, Quek H, White A, Haynes J, Jackaman C, Villemagne V, Munyard K, Laws SM, Verdile G, Groth D (2021) Chronic stress and A lzheimer’s disease: the interplay between the hypothalamic–pituitary–adrenal axis, genetics and microglia. Biol Rev 96(5):2209–2228
Ilacqua A, Izzo G, Emerenziani GP, Baldari C, Aversa A (2018) Lifestyle and fertility: the influence of stress and quality of life on male fertility. Reprod Biol Endocrinol 16:1–1
Verma G, Mishra MK (2016) A review on gene therapy: history, vectors, technologies and application. World J Pharm Pharm Sci 5(10):1334–1355
Kojima Y, Kurokawa S, Mizuno K, Umemoto Y, Sasaki S, Hayashi Y, Kohri K (2008) Gene transfer to sperm and testis: future prospects of gene therapy for male infertility. Curr Gene Ther 8(2):121–134
Wang YH, Yan M, Zhang X, Liu XY, Ding YF, Lai CP, Tong MH, Li JS (2021) Rescue of male infertility through correcting a genetic mutation causing meiotic arrest in spermatogonial stem cells. Asian J Androl 23(6):590–599
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X (2019) Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 4(1):62
Agrawal M, Alexander A, Khan J, Giri TK, Siddique S, Dubey SK, Patel RJ, Gupta U, Saraf S, Saraf S (2019) Recent biomedical applications on stem cell therapy: a brief overview. Curr Stem Cell Res Ther 14(2):127–136
Easley CA, Phillips BT, McGuire MM, Barringer JM, Valli H, Hermann BP, Simerly CR, Rajkovic A, Miki T, Orwig KE, Schatten GP (2012) Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep 2(3):440–446
Acknowledgements
Not applicable
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Oyovwi Mega Obukohwo & Ayodeji Folorunsho Ajayi participated in sorting and conceptualizing the manuscript and wrote the manuscript. Oyovwi Mega Obukohwo & Goodness Olatinwo organized the literature and presented ideas. Oyovwi Mega Obukohwo & Ayodeji Folorunsho Ajayi read and approved the submitted version. Oyovwi Mega Obukohwo & Akano Oyedayo Phillipsis responsible for the contribution. The author contributed to the revision of the manuscript, read and approved the submitted version.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ajayi, A.F., Oyovwi, M.O., Olatinwo, G. et al. Unfolding the complexity of epigenetics in male reproductive aging: a review of therapeutic implications. Mol Biol Rep 51, 881 (2024). https://doi.org/10.1007/s11033-024-09823-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09823-9