Abstract
Background
Preclinical and clinical studies over the past several decades have indicated the potential value of metformin, a widely utilized treatment for Type 2 diabetes, in prostate cancer therapy. Notably, these studies demonstrated metformin’s pleiotropic effects on several molecular and metabolic pathways, such as androgen signaling, cell cycle, and cellular bioenergetics. In this study we investigated the role of metformin in regulating intracellular redox status and cell survival in LNCaP prostate cancer cells.
Methods and results
The cytotoxic effects of metformin with or without the presence of SBI0206965 (AMPK inhibitor) on LNCaP cells were determined using MTT and trypan blue exclusion assays. Seahorse XP extracellular analysis, Liquid Chromatography/ Mass Spectrophotometry (LC/MS), and 2,7- and Dichlorofluoresin diacetate (DCFDA) assay were used to assess the effects of metformin on cellular bioenergetics, redox status, and redox-related metabolites. mRNA expression and protein concentration of redox-related enzymes were measured using Real Time-qPCR and ELISA assay, respectively. Independently of AMP-activated protein kinase, metformin exhibited a dose- and time-dependent inhibition of LNCaP cell survival, a response mitigated by glutathione or N-acetylcysteine (ROS scavengers) treatment. Notably, these findings were concomitant with a decline in ATP levels and the inhibition of oxidative phosphorylation. The results further indicated metformin’s induction of reactive oxygen species, which significantly decreased glutathione levels and the ratio of reduced to oxidized glutathione, as well as the transsulfuration metabolite, cystathionine. Consistent with an induction of oxidative stress condition, metformin increased mRNA levels of the master redox transcription factor Nrf-2 (nuclear factor erythroid-derived 2-like), as well as transsulfuration enzymes cystathionine beta-synthase and cystathionase and GSH synthesis enzymes γ-glutamylcysteine synthetase and glutathione synthetase.
Conclusion
Our findings highlight multiple mechanisms by which metformin-induced formation of reactive oxygen species may contribute to its efficacy in prostate cancer treatment, including promotion of oxidative stress, Nrf2 activation, and modulation of redox-related pathways, leading to its anti-survival action.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer is a malignant growth of cells of the prostate gland that predominantly affects men, with an average age at diagnosis of 66 years [1, 2]. It is the second leading cause of cancer-related death among males in the United States and remains one of the leading causes of cancer mortality in the Caribbean [1, 3, 4]. Although multiple treatment options are available, chemotherapy side effects and chemoresistance are major concerns that negatively impact patient quality of life and survival. These challenges have prompted interest in exploring new therapeutic approaches that target metabolic pathways, including mitochondrial metabolic function, that generally promote prostate cancer development and progression [5, 6] and are associated with drug resistance and poor therapeutic outcomes [5, 7]. Overall, evidence suggests that metabolic reprogramming may play an important role in the ability of prostate cancer cells to circumvent chemotherapy-induced cytotoxicity, resulting in metastatic castration-resistant prostate cancer.
Recent preclinical and clinical studies have reported the anti-cancer effects of metformin, an oral hypoglycemic drug used as a first-line treatment for type 2 diabetes mellitus [8]. Its primary mechanism of action for that use involves AMP-activated protein kinase (AMPK) activation, which enhances insulin-responsiveness and reduces hepatic gluconeogenesis [9]. Several studies have demonstrated metformin’s antitumor effects to involve direct or indirect actions on various intracellular signaling pathways [10,11,12,13,14]. Indirect anticancer effects are insulin-dependent which involves AMPK activation, resulting in the inhibition of gluconeogenesis and a subsequent decrease in blood glucose and circulating insulin [12, 13]. Direct anticancer effects are insulin-independent, involving inhibition of complex 1 of the mitochondrial electron transport chain (ETC) and the subsequent activation of AMPK, ultimately resulting in cell cycle arrest and apoptosis [13].
Reactive oxygen species (ROS) generated during mitochondrial activity are known to play a role in the activation of mitochondrial stress-induced signaling pathways and the alteration of redox homeostasis[15]. ROS-induced oxidative stress has been associated with the development and progression of prostate cancer [16]. Oxidative stress is characterized by an imbalance between ROS generation and antioxidant capacity in favor of ROS, which causes oxidative damage and adaptive metabolic responses [16]. Interestingly, recent evidence suggests that metformin exerts its anticancer effects via ROS generation [17, 18]. Several studies have shown that metformin induces apoptosis in cancer cells by altering mitochondrial activity and promoting the generation of ROS [19, 20]. These studies also demonstrated that metformin-induced ROS levels provide an anticancer effect via elevated oxidative stress. Emerging evidence also suggests that higher levels of oxidative stress increase cancer cell susceptibility to the deleterious effects of ROS [21, 22]. To our knowledge, we are the first to show that metformin regulates LNCaP’s prostate cancer cell redox status, thereby exerting its anti-proliferative effect.
Materials and methods
Cell Culture
LNCaP prostate cancer cells were given as a gift from Dr. Appu Rathinavelu at Nova Southeastern University and grown in RPMI-1640 with L-glutamine and HEPES (ATCC, Manassas, VA, USA), supplemented with 10% Fetal bovine Serum (VWR, Radnor, PA, USA) and 1%. Penicillin–Streptomycin and Amphotericin (Sigma-Aldrich, St. Louis, MO, USA). Metformin, rotenone (ROT), glutathione, N-acetylcysteine (NAC), and SBI-0206965 (SBI) were purchased from Sigma-Aldrich (St. Louis, MO, USA) and were dissolved in deionized water and RPMI media, respectively.
Cell Bioenergetic Assay
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using an Agilent XFp Extracellular Flux Analyzer (Santa Clara, CA, USA). Cells were plated at 3 × 104 cells/well and incubated overnight at 37°C. Cells were then exposed to 1 mM metformin, 1 µM SBI, and their combination for 24 h, and the medium was replaced by bicarbonate-free XF assay medium. A Cell Mito Stress Assay was performed according to the manufacturer’s protocol.
Thiol Extraction and Quantification
Cells were seeded in a 6-well plate at 2.5 × 105 cells/well and exposed to 24 h of treatment, followed by washing with ice-cold Hank’s Balanced Salt Solution (HBSS). 200ul of cell suspension was transferred to 50 µl of 0.4N perchloric acid on ice. Samples were vortexed and degassed with nitrogen. Tubes were then centrifuged at 4°C at 14, 000 RPM for 1 h. The supernatant was transferred to HPLC vials and blown with nitrogen gas for 10 s. Glutathione (GSH), cystathionine (CYST), oxidized glutathione (GSSG), cysteine, and homocysteine (HCY) ions and fragments were detected using Triple Quad LC/MS (Agilent Technologies), with an electrospray ionization (ESI) source operated in positive ion detection mode. The protein concentration in each sample was determined using a Micro BCA™ Protein Assay Kit purchased from ThermoFisher Scientific (Waltham, MA, USA, No. 23235). Thiols were normalized to sample protein concentrations.
Gene Expression Studies
Cells were seeded in a 6-well plate at 2.5 × 105 cells/well. Total RNA was extracted using the Qiagen RNeasy Mini Kit (Germantown, MD, USA), and cells were treated for 24 h with 1 mM metformin, 0.1 mM, and 2 mM NAC. cDNA synthesis was then performed using a Verso cDNA Synthesis kit (Fisher Scientific, Waltham, MA, USA) and a PTC-100™ Programmable Thermal Controller. RT-qPCR primers and TaqMan ™ Gene Expression Master Mix were purchased from ThermoFisher Scientific (Waltham, MA, USA). Amplification was carried out in a StepOnePlus™ Real-Time PCR System (ThermoFisher Scientific, Waltham, MA, USA). Fold change in expression was calculated using the 2−∆∆CT technique [23]. Data was normalized to β-actin.
MTT Assay
Cell viability was assessed with a 3-(4, 5-dimethyl-2-tetrazolyl)-2,5-diphenyl-2H tetrazolium bromide (MTT) assay. Cells were seeded in a 96-well plate at 1 × 103 cells/well. After 24 h, cells were exposed to various treatment conditions. Samples were then supplemented with 10 µl MTT (No. 475989, Sigma-Aldrich, St. Louis, MO, USA) and incubated for 4 h, after which 100 µl DMSO was added. Absorbance was determined using the BioTek Synergy H1 Hybrid plate reader (Agilent, Winooski, Vermont, USA).
Trypan Blue Exclusion Assay
Cells were seeded in a 6-well plate at 1 × 105 cells/well and were treated with 1 mM metformin or 1 µM SBI and their combination for 24, 48, and 72 h. Next, the cells were stained with trypan blue dye (Fisher Scientific, Waltham, MA, USA, No. 17-942E). Live and dead cell count was determined using an automated cell counter.
ROS Assay
ROS were quantified with 2,7-dichlorofluorecin diacetate (DCFDA) obtained from Sigma-Aldrich (St. Louis, MO, USA, No. D6883). Cells were seeded 20,000 cells per well in 96-well plate and were treated with 1 mM metformin or 0.5 µM ROT for 24 h. Next, cells were washed with PBS once and then stained with 25 µM of DCFDA for 45 min. DCFDA was removed prior to reconstituting cells with PBS. DCF fluorescence in cells was measured using the BioTek Synergy H1 Hybrid plate reader (Agilent, Winooski, Vermont, USA) with excitation/emission at 485/529 nm.
ELISA assays (CBS, CSE)
Cell extracts were prepared following 24 h of treatment with 1 mM metformin, 1 µM SBI and their combination. Cystathionine beta-synthase (CBS) and cystathionase (CSE) protein concentrations were determined using ELISA-based assays obtained from Aviva Systems Biology (San Diego, CA, USA, No. OKEH01392) and ABclonal Science, (Woburn, MA, USA, No. RK01192), respectively.
Statistical Analysis
Statistical Analysis was performed using Student’s t-test and analysis of variance with Tukey’s post hoc testing, using Prism 8.0 software (Graph-Pad Software, San Diego, CA). All experiments were replicated at least twice and expressed as mean ± standard error mean (SEM). Differences were considered significant at p < 0.05.
Results
Metformin reduces LNCaP’s cell survival in dose – and time-dependent manner independent of AMPK activation
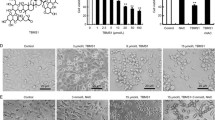
To evaluate the effect of metformin on LNCaP prostate cancer cell survival, the viability of cells exposed to metformin doses (1-10 mM) were assessed using an MTT assay. We observed that metformin significantly inhibited LNCaP cell viability in a dose-dependent manner with the 10 mM metformin reducing cell survival by 65% when compared to control (Fig. 1a). Notably, several studies demonstrated that metformin activates AMPK protein kinase which alters cellular metabolism. To determine whether AMPK activation is involved in the anti-survival effects of metformin, we also utilized the trypan blue exclusion assay to examine the effects of metformin with or without SBI, a potent AMPK inhibitor at 24, 48, and 72 h. We found that metformin significantly inhibited LNCaP cell survival in a time-dependent manner. Metformin inhibited cell survival by 31% and 26% at 48 and 72 h, respectively, when compared to baseline (Fig. 1b). Intriguingly, SBI co-treatment failed to counteract metformin’s cytotoxic effects in LNCaP cells, suggesting that metformin inhibits cell survival independent of the AMPK pathway. Importantly, our findings also showed that SBI monotherapy inhibited cell survival effects at 72 h.
Metformin decreases LNCaP prostate cancer cell survival. Cells were exposed for 24 h, a different concentration of metformin and cell viability was measured using MTT assay. b Cell survival of LNCaP cells exposed to 1 mM metformin or 1 µM SBI or their combination for 24, 48 and 72 h was measured using trypan blue exclusion assay. Data represent the mean percentage in the number of live cells from 24 h (baseline) ± SEM, N = 6. (*p < 0.05, **p < 0.001, ****p < 0.0001) indicate significant differences from the control group
Metformin inhibits mitochondrial oxidative respiration in LNCaP cells
Metformin is thought to be a mitochondrial toxin, impairing mitochondrial functions in cells. To assess the effects of metformin on mitochondrial respiration and aerobic glycolysis in LNCaP cells, OCR and ECAR were measured using an extracellular flux analyzer. LNCaP cells were treated with 1 mM metformin, 1 µM SBI, and their combination. As shown in (Fig. 2a, b), OCR was significantly reduced in the presence of metformin with or without SBI, when compared to control. To assess the basal respiration, and the oxygen consumption required to meet cellular ATP demand, the first three time points of the assay were analyzed. Our data showed that metformin significantly decreased basal respiration by 80%, indicating metformin inhibited oxidative phosphorylation (OXPHOS). In addition, proton leak and ATP production were both significantly reduced by 75% and 82%, respectively in the presence of metformin when treated with oligomycin, an ATP synthase inhibitor (Fig. 2c). In addition, our data showed that metformin significantly impaired the maximum respiration, which resulted in lower spare respiratory capacity in metformin-treated cells exposed to carbonyl cyanide p-trifluoro-methoxyphenyl hydrazone (FCCP), an uncoupler of the proton gradient that disrupts ATP synthesis (Fig. 2d). On the other hand, metformin did not produce a compensatory increase in glycolysis since there was no statistical significance in basal ECAR. However, we observed that SBI and metformin cotreatment significantly increased basal ECAR by 19%, despite a marginal decrease (p = 0.0756), compared to SBI alone. Taken together, our findings support previous studies indicating that metformin inhibits OXPHOS, and these effects are mediated locally at the mitochondrial electron transport chain. Importantly, we observed that SBI cotreatment did not reverse metformin effects on LNCaP cell’s bioenergetic status which confirms that AMPK is activated downstream in response to altered mitochondrial actions.
Metformin alters mitochondrial function in LNCaP prostate cancer cells. a OCR and b ECAR; of cells treated either with 1 mM metformin, 1 µM SBI, or their combination. c, d Average values of key mitochondrial parameters in LNCaP cells. Data represent the mean ± SEM of N = 4 in three different experiments, expressed as pmol/min. Asterisks indicate significant differences from the control group (*** p < 0.001). # (p < 0.001) indicates a significant difference between SBI vs. SBI and metformin combination
Metformin alters LNCaP cell redox status
Inhibition of oxidative phosphorylation may alter cellular redox status via increased ROS formation; hence, we analyzed thiol and thioether metabolites in the glutathione synthesis and transsulfuration pathways, redox-related pathways. Next, we treated LNCaP cells with 1 mM metformin, 1 µM SBI, or their combination for 24 h. We found that metformin significantly decreased intracellular GSH levels with or without SBI (Fig. 3b) and decreased the GSH/GSSG ratio (Fig. 3c), with no observable effect on GSSG or cysteine levels (data not shown). Despite SBI alone significantly increased GSH levels, SBI cotreatment failed to counteract metformin action on GSH levels. Since metformin significantly reduced LNCaP cells’ antioxidant capacity; Next, we evaluated the effect of metformin on the transsulfuration pathway, a supportive intermediate pathway linking the methionine cycle and glutathione synthesis (Fig. 3a). Related metabolites were measured and as shown in (Fig. 3d, e), metformin with or without SBI significantly decreased CYST level, and increased HCY levels when compared to control. Together these findings suggest that metformin regulates the redox status of LNCaP cells by altering sulfur metabolism and increasing oxidative stress; such action was mediated independent of AMPK activation.
Metformin-induced oxidative stress alters LNCaP antioxidant capacity. a Cellular redox status is regulated by GSH levels. Methionine cycle regulates cellular methylation potential by generating s-adenosylhomocysteine (SAM) via methionine adenosyltransferase (MAT2A). Methionine metabolism also plays a vital role in generating HCY which is condensed into CYST via CBS, leading to the generation of cysteine via CSE. Cysteine is further processed into GSH by glutathione synthetase (GSS) and γ-glutamylcysteine synthetase (GCL). Cysteine can also be extracellularly imported through the cystine/ glutamate antiporter (xCT) in the form of cystine. Metformin affects b GSH, c GSH/GSSG, d cystathionine, and e HCY. Data represent the mean ± SEM nmoles/mg protein (N = 3). (*p < 0.05), (***p < 0.001), and (****p < 0.0001) indicates significant differences from the control group. (# p < 0.001) indicates a significant difference between SBI vs. SBI in combination with metformin
Metformin upregulates glutathione and transsulfuration gene expression in LNCaP prostate cancer cells
Based on the effect of metformin on GSH, CYST and HCY levels, we then asked whether metformin regulates the expression of glutathione and transsulfuration-related enzymes. Thus, we measured GSS and GCL gene expression in metformin-treated cells with or without SBI treatment. As shown in Fig. 4a, b, GCL gene expression increased by 3-fold and GSS mRNA expression levels increased 2- fold in metformin-treated cells, compared to control. We also found that GCL and GSS mRNA levels were significantly increased with metformin and SBI combination by 3-fold and 2- fold, respectively. In addition, we measured CSE and CBS mRNA expression and we found that CSE (Fig. 4c) and CBS (Fig. 4d) mRNA expression were significantly increased by 3-fold and 1.4–fold, respectively. Interestingly, metformin and SBI cotreatment also caused a marked increase in CSE and CBS mRNA expression. Next, we wanted to identify if the mRNA changes also translated into protein expression changes. Hence, we assessed CSE and CBS protein concentrations using ELISA assay. Unlike RT-qPCR results, there were no observable changes in the protein expression of CSE and CBS with metformin monotherapy (Fig. 4e, f). However, metformin and SBI cotreatment significantly increased CBS protein expression at 24 h, in parallel to the mRNA gene expression levels. Overall, these findings confirm that metformin alters prostate cancer redox status with underlying changes in gene expression of glutathione and transsulfuration pathway enzymes that support the transsulfuration-glutathione axis.
Metformin alters gene expression of redox-related enzymes in LNCaP prostate cancer cells. Relative mRNA level of glutathione and transsulfuration-related genes was quantified and is represented as a GSS, b GCL, c CSE, and d CBS. e and f represent protein concentration (ELISA) of CSE and CBS, respectively. Data represent mean ± SEM and normalized to beta-actin N = 6. (*P < 0.05), (***p < 0.001), and (****p < 0.0001) represent significant differences from the control group. (# p < 0.001) indicates a significant difference between SBI vs. SBI in combination with metformin
Metformin-induced ROS inhibits LNCaP cell survival downstream of Complex 1 inhibition
To evaluate whether metformin promoted oxidative stress via ROS generation, we measured the intracellular ROS in metformin-treated cells using the DCFDA assay. Of note, several studies have proposed that complex 1 inhibition may play a role in metformin-induced ROS production [20, 24, 25]. To evaluate this hypothesis, cells were also treated with ROT, a potent complex 1 inhibitor. Our results demonstrated that both metformin and ROT significantly increased ROS levels by 98% and 74%, respectively, compared to control (Fig. 5a). In addition, we wanted to determine whether metformin-induced ROS inhibits cell survival in LNCaP cells. To this end, we measured cell viability in the presence of GSH-treated cells exposed to metformin or ROT for 24 h. We found that both metformin and ROT decrease cell viability, respectively. Moreover, our results also showed that GSH pretreatment reversed the effects of metformin and ROT on cell viability and promoted cell survival (Fig. 5b). These findings suggest that complex 1 inhibition mediates metformin-induced inhibition of LNCaP’s cell survival.
Metformin increases reactive oxygen species (ROS) production in LNCaP prostate cancer cells and inhibits cell survival. ROS production of cells exposed for 24 h to a 1 mM metformin and 0.5 µM ROT, was determined using DCFDA assay. b Viability of LNCaP cells exposed to 1 mM metformin, 0.5 µM ROT or 2 mM GSH or their combination was measured using MTT assay. Data represent the mean ± SEM N = 6. (****p < 0.001) indicate significant differences from the control group. (#p < 0.0001) indicates a significant difference between GSH vs GSH and Rotenone/Metformin combination
N-acetylcysteine decreases metformin-induced oxidative stress and protected against metformin inhibitory effects on LNCaP cell survival
To further validate the effect of metformin-induced oxidative stress on LNCaP cell survival, we incorporated the use of NAC, an indirect antioxidant that is known to promote GSH, hydrogen sulfide, and sulfane sulfur generation. Hence, we wanted to determine if NAC could also reverse metformin-induced oxidative stress and prevent its cytotoxic effects in LNCaP cells. To this end, we measured cell viability and NFE2L2 (Nrf2) gene expression in the presence of NAC using MTT and RT-qPCR, respectively. As shown in Fig. 6a, 2mM NAC significantly blocked the effects of metformin and promoted cell viability whereas 0.1 mM failed to prevent metformin cytotoxicity. We also found that 0.1 mM NAC alone had no significant effect on NFE2L2 gene expression, consistent with its lack of effect on cell viability (Fig. 6b). Moreover, we observed that NAC/metformin co-treated cells showed increased NFE2L2 expression, indicating that NAC may have potentiated the upregulation of NFE2L2 expression.
NAC prevents metformin inhibition of cell proliferation and augments the metformin-induced increase in NFE2L2 gene expression in LNCaP prostate cancer. Cells were treated with 1 mM metformin, 0.1 mM NAC or their combination. a Cellular viability was measured using MTT assay and b the mRNA level of NFE2L2 was quantified. Data represent the mean ± SEM, and mRNA expression was normalized to beta-actin. N = 6, (*** p < 0.001) indicate significant differences from control group. (# p < 0.001) indicate significant differences between metformin vs. 2 mM NAC and metformin, respectively
Discussion
Several studies have demonstrated that metformin inhibits cell proliferation and migration, as well as induces apoptosis in a variety of cancer cells [26,27,28]. Moreover, a substantial body of evidence suggest that metformin’s anticancer activities may be mediated through the activation of AMPK, a cellular energy sensor [29,30,31]. AMPK is activated in response to a change in the AMP/ATP ratio, due to declining ATP levels, which leads to adenosine monophosphate (AMP) binding to AMPK γ-subunit, causing its phosphorylation due to the conformational changes in the α-subunit at the threonine residue [13]. Besides its energy-sensing function, AMPK signaling is also associated with antioxidant production via Nrf2/ARE signaling pathway[32]. Specifically, Joo et al., reported that AMPK phosphorylates Nrf-2 on Ser550 residue, which promotes its nuclear accumulation for antioxidant response element (ARE)-associated gene transcription [33], suggesting AMPK also regulates cellular redox homeostasis. Since metformin is known to activate AMPK, in this present study we investigated the effect of metformin on the redox-related metabolic pathways as well as evaluated whether these underlying molecular pathways contribute to its anticancer actions in LNCaP prostate cancer cells. Particularly, we also wanted to explore whether these effects involved the AMPK signaling; therefore, we incorporated the use of SBI, a highly selective AMPK inhibitor compared to the commonly used AMPK inhibitor, Compound C [34]. Similar to previous studies, our results also demonstrated that metformin exerted cytotoxic effects in a dose- and time-dependent manner (Fig. 1a, b). Interestingly, these effects were unaffected by the presence of SBI, suggesting that metformin inhibits LNCaP’s cell survival independent of AMPK activation.
It is well established that metformin inhibits complex 1 of the Electron Transport Chain (ETC), suppressing mitochondrial ATP production, which leads to the activation of AMPK [13]. Moreover, previous studies showed metformin inhibited OXPHOS and yielded a compensatory increase in glycolysis, resulting in a decline in ATP production [35, 36]. Unlike these previous studies, we observed that 1 mM metformin significantly decreased OXPHOS with no significant change in glycolytic activity, as reflected in OCR and ECAR, respectively (Fig. 2a, b). Moreover, these findings were consistent with lower ATP production, proton leak, and spare respiratory capacity in metformin-treated cells, which supports metformin mitochondrial targeted actions and toxicity. As expected, we found that SBI co-treatment did not block the effects of metformin on OCR, which confirms that AMPK activation occurs downstream of complex 1 inhibition. Activated AMPK plays a crucial role in metabolic regulation by downregulating ATP-consuming pathways such as protein synthesis and upregulating ATP-generating pathways such as glycolysis enabling metabolic adaptation [37]. Of note, studies have reported that prostate cancer cells are metabolically flexible, supporting a mixed oxidative-glycolytic phenotype [38, 39]. Here, our results were consistent with earlier findings indicating that LNCaP prostate cancer cells exhibit a more oxidative phenotype than a glycolytic one, resulting in a higher sensitivity to mitochondrial modulators [40, 41] and metformin. On the other hand, a recent in vitro study showed that metformin increased glucose uptake and glycolytic activity in cisplatin-resistant liver cancer, by increasing glycolytic enzymes, such as hexokinase 2 [42]. S. Andrzejewski et.al [43] demonstrated that complex 1 inhibition stimulated glycolysis as a compensatory response to restore cellular ATP levels in metformin-treated breast cancer cells. Similar findings were observed in metformin-treated mouse intestinal organoids and 2D monolayers, which showed that metformin down-regulated mitochondrial activities and increased intestinal glycolysis [44]. Contrary to these findings, we found that ECAR levels were unaffected despite metformin inhibiting OXPHOS in metformin-treated cells. SBI and metformin cotreatment significantly increased glycolytic activity in uncharacterized matter, despite the lower basal ECAR levels observed when SBI was alone.
In order to determine whether mitochondrial dysfunction alters prostate cancer redox status, we assessed redox-related metabolites. Mitochondria are a major source of ROS production in mammalian cells and is viewed as a double-edged sword for cancer progression [21]. Emerging evidence depicts a complex role of ROS signaling in cancer, which may promote survival or in contrast can also induce apoptosis via protein-thiol modifications [45, 46], supporting a potential role of redox signaling in cancer progression as a potential target for therapy. We found that 1 mM metformin significantly decreased CYST (Fig. 3d) and GSH (Fig. 3b) levels, in association with a decrease in cellular redox capacity, as reflected in the GSH/GSSG ratio (Fig. 3c). This action was consistent with underlying changes in the gene expression of GCL and GSS (Fig. 4a, b). Notably, these findings are aligned with previous reports indicating that metformin alters cellular redox metabolomics [47] as well as increase ROS levels, supporting its action in reducing GSH levels, a major antioxidant [48, 49]. Moreover, a previous study reported that rotenone, a complex 1 inhibitor, stimulates mitochondrial ROS production and induces apoptosis in leukemia cells [25], suggesting complex 1 inhibition may be involved in metformin-induced ROS generation. It is noteworthy to mention that the effect of metformin on complex 1 is reversible [50]. Although the mechanism involved in complex 1 inhibition leading to ROS production is unclear, it has been proposed that mitochondrial iron-sulfur clusters can mediate electron transfer to oxygen molecules, thereby producing ROS. Another mechanism is thought to involve the formation of coenzyme Q semiquinone radicals that readily interact with molecular oxygen in the presence of protons [24]. Such mechanisms might be involved in metformin-induced ROS production and should be explored in future studies.
Nrf2 can be activated during oxidative stress conditions as an adaptive mechanism [51]. Nrf2, encoded by the NFE2L2 gene, is a transcription factor that up-regulates cellular antioxidant production, in part by increasing intracellular cysteine which supports GSH synthesis [52]. We showed that metformin up-regulated NFE2L2 gene expression in LNCaP cells (Fig. 6b) and that this occurred in conjunction with an increase in mRNA of the Nrf2-regulated gene GCL; which encodes for GCL, as shown in Fig. 4a. These findings, coupled with lower GSH levels, suggest that metformin-induced oxidative stress may upregulate NFE2L2 expression as a compensatory response to ROS produced in LNCaP cells. Besides Nrf2 regulation, GSH production relies on cysteine availability, highlighting the importance of cysteine/cystine transporters and the transsulfuration pathway[52]. The transsulfuration pathway involves the transfer of a sulfur atom, originally derived from methionine to yield cysteine via two sequential reactions. The first is the condensation of HCY and serine by CBS to generate CYST. In the second reaction, CSE cleaves CYST into cysteine and alpha-ketobutyrate, and the cysteine derived from this pathway can be utilized for GSH production. A. Prudova et al. [53] demonstrated that testosterone-treated prostate cancer cells exhibit lower levels of CBS, which consequently resulted in lower cysteine production, highlighting two important aspects: (i) The transsulfuration pathway is highly functional in prostate cancer cells, and (ii) Activity of the transsulfuration pathway can be affected by testosterone levels. Our current results showed that metformin significantly increased CSE (Fig. 4c) and CBS (Fig. 4d) mRNA expression, respectively. Up-regulation of CSE mRNA expression with metformin exposure was consistent with decreased CYST levels. Unlike our mRNA expression data, we did not observe changes in CSE and CBS protein levels at 24 h, suggesting that this difference might be because of the differential time-point between mRNA and protein expression (Fig. 4e, f) which should be assessed in future studies at longer treatment exposure. Notably, studies have reported that metformin increases HCY and decreases cellular antioxidant capacity, which is consistent with our study [47, 54]. However, research by Sun and colleagues showed that metformin significantly increased expression levels of glutathione and transsulfuration-related enzymes which upregulated GSH and cysteine levels in liver tissue of rats [55]. Particularly, the authors proposed that metformin exhibited protective effect against oxidative insults on hepatic tissue caused by bisphenol A, an environmental toxin. It is possible that differences in physiological conditions and pathological state could explain these controversial effects of metformin. Nonetheless, the rewiring of the transsulfuration pathway we observed, in conjunction with decreased GSH and GSH/GSSG, implies that metformin impairs redox status in LNCaP cells. Overall, SBI treatment did not reverse the effects of metformin in this current study, suggesting that AMPK signaling may not be involved in inhibiting cell survival in our cell model.
Next, we assessed cell viability in GSH-treated cells in the presence of metformin or ROT to determine whether metformin exerts its cytotoxic effects through the inhibition of complex 1. We found that GSH pretreatment blocked the cytotoxic effects of metformin and ROT which supported our hypothesis that metformin-induced oxidative stress inhibits LNCaP cell viability via the inhibition of complex 1 (Fig. 5a, b). This finding was also validated by incorporating the NAC treatment as well as measuring the gene expression of the transcription factor Nrf2, which up-regulates antioxidant production in response to oxidative stress. Our results suggested that metformin upregulated NFE2L2 gene expression as a compensatory response to increased oxidative conditions due to redox imbalance. Contrary to previous studies showing NFE2L2 gene expression downregulated in the presence of NAC and oxidative stress inducers, we found that NFE2L2 gene expression was upregulated with NAC and metformin co-treatment (Fig. 5b) [56, 57]. A possible reason for this unexpected observation is that 0.1 mM NAC concentration may have been inadequate to counteract metformin-induced oxidative stress (Fig. 6a). This notion is supported by the reversibility of metformin’s cytotoxic effects observed with a 20-fold higher NAC treatment.
There is a vast amount of literature supporting the anti-aging and cytoprotective effects of metformin on benign cells [58], and numerous studies document the utility of metformin as an adjunctive agent for a variety of malignancies[8]. Our study did not focus on the beneficial effects of metformin on benign cells but highlighted its cytotoxic effects on LNCaP cancer cells, which favors its use in conjunction with more cytotoxic therapies, like chemotherapy, many of which promote oxidative stress. The current study provides evidence that metformin regulates LNCaP prostate cancer redox status by modulating mitochondrial activities and GSH-related parameters downstream of ETC inhibition (Fig. 7). Our findings also provide preliminary evidence that metformin-induced oxidative stress appears to stimulate a compensatory response to maintain LNCaP intracellular antioxidant capacity, albeit, at lower levels that are not conducive for cellular proliferation.
Metformin-induced oxidative stress downregulates LNCaP antioxidant capacity and inhibits cell survival. Metformin is intracellularly uptake via organic cation transporters (OCT) and inhibits complex I (CI) of the electron transport chain in the mitochondrion, which subsequently downregulates OXPHOS and mitochondrial membrane potential. OXPHOS downregulation favors the sequential reduction of oxygen (O2) in the mitochondrion, leading to ROS formation. ROS produced is dependent upon the overall rate of electron flow in the ETC. Elevated ROS levels along with downregulated antioxidant capacity increase oxidative stress and decreased cell survival; such metabolic changes might further support its anti-proliferative actions
Conclusion
In summary, our findings suggest that metformin-induced oxidative stress impairs LNCaP prostate cancer cell redox status, viability, and growth, all of which are most likely to occur downstream of its mitochondrial effects. Our current study provides additional mechanistic understanding to support further exploration of metformin in prostate cancer therapy based upon its ability to induce oxidative stress and decrease LNCaP’s cell survival.
Data availability
The data are available from the corresponding author on reasonable request.
Abbreviations
- AMPK:
-
AMP-activated protein kinase
- ARE:
-
Antioxidant response element
- CBS:
-
Cystathionine beta-synthase
- CSE:
-
Cystathionase
- CYST:
-
Cystathionine
- DCFDA:
-
Dichlorofluoresin diacetate
- ECAR:
-
Extracellular acidification rate
- ESI:
-
Electrospray ionization
- ETC:
-
Electron Transport Chain
- FCCP:
-
Carbonyl cyanide p-trifluoro-methoxyphenyl hydrazone
- GSH:
-
Glutathione
- GSSG:
-
Oxidized glutathione
- HCY:
-
Homocysteine
- MAT2A:
-
Methionine adenosyltransferase
- MTT:
-
3-(4, 5-Dimethyl-2-tetrazolyl)-2,5-diphenyl-2H tetrazolium bromide assay
- NAC:
-
N-acetylcysteine
- Nrf-2:
-
Nuclear factor erythroid-derived 2-like
- OCR:
-
Oxygen consumption rate
- OXPHOS:
-
Oxidative phosphorylation
- ROS:
-
Reactive oxygen species
- SAM:
-
S-adenosylhomocytseine
References
Rawla P (2019) Epidemiology of prostate cancer, world. J Oncol 10:63–89. https://doi.org/10.14740/wjon1191
Syrigos KN, Karapanagiotou E, Harrington KJ (2005) Prostate cancer in the elderly. Anticancer Res 25:4527–4533
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics. CA Cancer J Clin 71(2021):7–33. https://doi.org/10.3322/CAAC.21654
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global Cancer STATISTICS 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/CAAC.21660
Ahmad F, Cherukuri MK, Choyke PL (2021) Metabolic reprogramming in prostate cancer. Br J Cancer 125(9):1185–1196. https://doi.org/10.1038/s41416-021-01435-5
Farhadi P, Yarani R, Dokaneheifard S, Mansouri K (2020) The emerging role of targeting cancer metabolism for cancer therapy. Tumor Biology 42:1–18. https://doi.org/10.1177/1010428320965284
Park JH, Pyun WY, Park HW (2020) Cancer metabolism: phenotype, signaling and therapeutic targets. Cells 9:2308. https://doi.org/10.3390/CELLS9102308
Hua Y, Zheng Y, Yao Y, Jia R, Ge S, Zhuang A (2023) Metformin and cancer hallmarks: shedding new lights on therapeutic repurposing. J Transl Med 21:403. https://doi.org/10.1186/s12967-023-04263-8
Rena G, Hardie DG, Pearson ER (2017) The mechanisms of action of metformin. Diabetologia 60:1577–1585. https://doi.org/10.1007/s00125-017-4342-z
Demir U, Koehler A, Schneider R, Schweiger S, Klocker H (2014) Metformin anti-tumor effect via disruption of the MID1 translational regulator complex and AR downregulation in prostate cancer cells. BMC Cancer 14:1–9. https://doi.org/10.1186/1471-2407-14-52
Hirsch A, Hahn D, Kempná P, Hofer G, Nuoffer JM, Mullis PE, Flück CE (2012) Metformin inhibits human androgen production by regulating steroidogenic enzymes HSD3B2 and CYP17A1 and complex I activity of the respiratory chain. Endocrinology 153:4354–4366. https://doi.org/10.1210/en.2012-1145
Whitburn J, Edwards CM, Sooriakumaran P (2017) Metformin and prostate cancer: a new role for an old drug. Curr Urol Rep 18:46. https://doi.org/10.1007/s11934-017-0693-8
Zaidi S, Gandhi J, Joshi G, Smith NL, Khan SA (2019) The anticancer potential of metformin on prostate cancer. Prostate Cancer Prostatic Dis 22:351–361. https://doi.org/10.1038/s41391-018-0085-2
Zingales V, Distefano A, Raffaele M, Zanghi A, Barbagallo I, Vanella L (2017) Metformin: a bridge between diabetes and prostate cancer. Front Oncol 7:1–7. https://doi.org/10.3389/fonc.2017.00243
Zhang B, Pan C, Feng C, Yan C, Yu Y, Chen Z, Guo C, Wang X (2022) Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep 27:45–52. https://doi.org/10.1080/13510002.2022.2046423
Shiota M (2021) Oxidative stress and prostate cancer. Cancer. Elsevier, pp 15–26. https://doi.org/10.1016/B978-0-12-819547-5.00002-X
Li B, Zhou P, Xu K, Chen T, Jiao J, Wei H, Yang X, Xu W, Wan W, Xiao J (2020) Metformin induces cell cycle arrest, apoptosis and autophagy through ROS/JNK signaling pathway in human osteosarcoma. Int J Biol Sci 16:74. https://doi.org/10.7150/IJBS.33787
Warkad MS, Kim CH, Kang BG, Park SH, Jung JS, Feng JH, Inci G, Kim SC, Suh HW, Lim SS, Lee JY (2021) Metformin-induced ROS upregulation as amplified by apigenin causes profound anticancer activity while sparing normal cells. Sci Rep 11(1):1–13. https://doi.org/10.1038/s41598-021-93270-0
Gao ZY, Liu Z, Bi MH, Zhang JJ, Han ZQ, Han X, Wang HY, Sun GP, Liu H (2016) Metformin induces apoptosis via a mitochondria-mediated pathway in human breast cancer cells in vitro. Exp Ther Med 11:1700–1706. https://doi.org/10.3892/etm.2016.3143
Warkad MS, Kim CH, Kang BG, Park SH, Jung JS, Feng JH, Inci G, Kim SC, Suh HW, Lim SS, Lee JY (2021) Metformin-induced ROS upregulation as amplified by apigenin causes profound anticancer activity while sparing normal cells. Sci Rep 11:14002. https://doi.org/10.1038/S41598-021-93270-0
Perillo B, Di Donato M, Pezone A, Di Zazzo E, Giovannelli P, Galasso G, Castoria G, Migliaccio A (2020) ROS in cancer therapy: the bright side of the moon. Exp Mol Med 52:192. https://doi.org/10.1038/S12276-020-0384-2
Van Loenhout J, Peeters M, Bogaerts A, Smits E, Deben C (2020) Oxidative stress-inducing anticancer therapies: taking a closer look at their immunomodulating effects. Antioxidants 9:1–19. https://doi.org/10.3390/antiox9121188
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/METH.2001.1262
Fato R, Bergamini C, Bortolus M, Maniero AL, Leoni S, Ohnishi T, Lenaz G (2009) Differential effects of mitochondrial complex I inhibitors on production of reactive oxygen species. Biochim Biophys Acta 1787:384–392. https://doi.org/10.1016/J.BBABIO.2008.11.003
Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP (2003) Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem 278:8516–8525. https://doi.org/10.1074/jbc.M210432200
Lee J, Hong EM, Kim JH, Jung JH, Park SW, Koh DH, Choi MH, Jang HJ, Kae SH (2019) Metformin induces apoptosis and inhibits proliferation through the AMP-activated protein kinase and insulin-like growth factor 1 receptor pathways in the bile duct cancer cells. J Cancer 10:1734–1744. https://doi.org/10.7150/jca.26380
Zhou X, Kuang Y, Liang S, Wang L (2019) Metformin inhibits cell proliferation in SKM-1 cells via AMPK-mediated cell cycle arrest. J Pharmacol Sci 141:146–152. https://doi.org/10.1016/j.jphs.2019.10.003
Chen Y-H, Yang S-F, Yang C-K, Tsai H-D, Chen T-H, Chou M, Hsiao Y-H (2021) Metformin induces apoptosis and inhibits migration by activating the AMPK/p53 axis and suppressing PI3K/AKT signaling in human cervical cancer cells. Mol Med Rep 23(1):88. https://doi.org/10.3892/mmr.2020.11725
Ikhlas S, Ahmad M (2017) Metformin: insights into its anticancer potential with special reference to AMPK dependent and independent pathways. Life Sci 185:53–62. https://doi.org/10.1016/j.lfs.2017.07.029
Rocha GZ, Dias MM, Ropelle ER, Osório-Costa F, Rossato FA, Vercesi AE, Saad MJA, Carvalheira JBC (2011) Metformin amplifies chemotherapy-induced AMPK activation and antitumoral growth. Clin Cancer Res 17:3993–4005. https://doi.org/10.1158/1078-0432.CCR-10-2243
Chen C, Wang H, Geng X, Zhang D, Zhu Z, Zhang G, Hou J (2021) Metformin exerts anti-AR-negative prostate cancer activity via AMPK/autophagy signaling pathway. Cancer Cell Int 21:404. https://doi.org/10.1186/s12935-021-02043-2
Petsouki E, Cabrera SNS, Heiss EH (2022) AMPK and NRF2: Interactive players in the same team for cellular homeostasis? Free Radic Biol Med 190:75–93. https://doi.org/10.1016/j.freeradbiomed.2022.07.014
Joo MS, Kim WD, Lee KY, Kim JH, Koo JH, Kim SG (2016) AMPK facilitates nuclear accumulation of Nrf2 by phosphorylating at serine 550. Mol Cell Biol 36:1931–1942. https://doi.org/10.1128/mcb.00118-16
Dite TA, Langendorf CG, Hoque A, Galic S, Rebello RJ, Ovens AJ, Lindqvist LM, Ngoei KRW, Ling NXY, Furic L, Kemp BE, Scott JW, Oakhill JS, Peter S, MacKillop M (2018) AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965 the and the The atomic coordinates and structure factors (code 6BX6) have been deposited in the Protein Data Bank. J Biol Chem 293(23):8874–8885. https://doi.org/10.1074/jbc.RA118.003547
De Souza Silva FM, Rocha Alves Da Silva MH, Bracht A, Eller GJ, Constantin RP, Yamamoto NS (2010) Effects of metformin on glucose metabolism of perfused rat livers. Mol Cell Biochem 340:283–289. https://doi.org/10.1007/s11010-010-0429-2
Owen MR, Doran E, Halestrap AP (2000) Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochemical Journal 348:607–614. https://doi.org/10.1042/bj3480607
Goel S, Singh R, Singh V, Singh H, Kumari P, Chopra H, Sharma R, Nepovimova E, Valis M, Kuca K, Bin Emran T (2022) Metformin: activation of 5′ AMP-activated protein kinase and its emerging potential beyond anti-hyperglycemic action. Front Genet 13:1022739. https://doi.org/10.3389/fgene.2022.1022739
Ashton TM, Gillies McKenna W, Kunz-Schughart LA, Higgins GS (2018) Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res 24:2482–2490. https://doi.org/10.1158/1078-0432.CCR-17-3070
Peitzsch C, Gorodetska I, Klusa D, Shi Q, Alves TC, Pantel K, Dubrovska A (2020) Metabolic regulation of prostate cancer heterogeneity and plasticity. Semin Cancer Biol. https://doi.org/10.1016/j.semcancer.2020.12.002
Higgins LH, Withers HG, Garbens A, Love HD, Magnoni L, Hayward SW, Moyes CD (2009) Hypoxia and the metabolic phenotype of prostate cancer cells. Biochim Biophys Acta Bioenerg 1787:1433–1443. https://doi.org/10.1016/j.bbabio.2009.06.003
Tao Y, Liu S, Lu J, Fu S, Li L, Zhang J, Wang Z, Hong M (2022) FOXO3a-ROS pathway is involved in androgen-induced proliferation of prostate cancer cell. BMC Urol 22:70. https://doi.org/10.1186/s12894-022-01020-9
Cai L, Jin X, Zhang J, Li L, Zhao J (2020) Metformin suppresses Nrf2-mediated chemoresistance in hepatocellular carcinoma cells by increasing glycolysis. Aging 12:17582–17600. https://doi.org/10.18632/AGING.103777
Andrzejewski S, Gravel S-P, Pollak M, St-Pierre J (2014) Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab 2:12. https://doi.org/10.1186/2049-3002-2-12
Yang M, Darwish T, Larraufie P, Rimmington D, Cimino I, Goldspink DA, Jenkins B, Koulman A, Brighton CA, Ma M, Lam BYH, Coll AP, O’Rahilly S, Reimann F, Gribble FM (2021) Inhibition of mitochondrial function by metformin increases glucose uptake, glycolysis and GDF-15 release from intestinal cells. Sci Rep 11:1–20. https://doi.org/10.1038/s41598-021-81349-7
Shah MA, Rogoff HA (2021) Implications of reactive oxygen species on cancer formation and its treatment. Semin Oncol 48:238–245. https://doi.org/10.1053/j.seminoncol.2021.05.002
Coffey RNT, Watson RWG, Hegarty NJ, O’Neill A, Gibbons N, Brady HR, Fitzpatrick JM (2000) Thiol-mediated apoptosis in prostate carcinoma cells. Cancer 88:2092–2104. https://doi.org/10.1002/(SICI)1097-0142(20000501)88:9%3c2092::AID-CNCR15%3e3.0.CO;2-9
Liu X, Romero IL, Litchfield LM, Lengyel E, Locasale JW (2016) Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab 24:728–739. https://doi.org/10.1016/j.cmet.2016.09.005
Pecinova A, Drahota Z, Kovalcikova J, Kovarova N, Pecina P, Alan L, Zima M, Houstek J, Mracek T (2017) Pleiotropic effects of biguanides on mitochondrial reactive oxygen species production. Oxid Med Cell Longev 2017:7038603. https://doi.org/10.1155/2017/7038603
Yang J, Zhou Y, Xie S, Wang J, Li Z, Chen L, Mao M, Chen C, Huang A, Chen Y, Zhang X, Khan NUH, Wang L, Zhou J (2021) Metformin induces ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J Exp Clin Cancer Res 40:206. https://doi.org/10.1186/S13046-021-02012-7
Vial G, Detaille D, Guigas B (2019) Role of mitochondria in the mechanism(s) of action of metformin. Front Endocrinol (Lausanne) 10:1–8. https://doi.org/10.3389/fendo.2019.00294
Bae T, Hallis SP, Kwak MK (2024) Hypoxia, oxidative stress, and the interplay of HIFs and NRF2 signaling in cancer. Exp Mol Med. https://doi.org/10.1038/s12276-024-01180-8
McBean G (2017) Cysteine, glutathione, and thiol redox balance in astrocytes. Antioxidants 6:62. https://doi.org/10.3390/antiox6030062
Prudova A, Matthias A, Zachary B, Lin A, Vitvitsky V, Baneriee R (2007) Testosterone regulation of homocysteine metabolism modulates redox status in human prostate cancer cells. Antioxid Redox Signal 11:1875–1881. https://doi.org/10.1089/ars.2007.1712
Janati S, Behmanesh MA, Najafzadehvarzi H, Kassani A, Athari N, Poormoosavi SM (2022) Changes of serum level of homocysteine and oxidative stress markers by metformin and inositol in infertile women with polycystic ovary syndrome: a double blind randomized clinical trial study. Int J Fertil Steril 16:102–107. https://doi.org/10.22074/IJFS.2021.530040.1125
Sun Y, Wang X, Zhou Y, Zhang J, Cui W, Wang E, Du J, Wei B, Xu X (2021) Protective effect of metformin on BPA-induced liver toxicity in rats through upregulation of cystathionine β synthase and cystathionine γ lyase expression. Sci Total Environ 750:141685. https://doi.org/10.1016/j.scitotenv.2020.141685
Sun P, Nie X, Chen X, Yin L, Luo J, Sun L, Wan C, Jiang S (2018) Nrf2 signaling elicits a neuroprotective role against PFOS-mediated oxidative damage and apoptosis. Neurochem Res 43:2446–2459. https://doi.org/10.1007/s11064-018-2672-y
Ren J, Su D, Li L, Cai H, Zhang M, Zhai J, Li M, Wu X, Hu K (2020) Anti-inflammatory effects of Aureusidin in LPS-stimulated RAW264.7 macrophages via suppressing NF-κB and activating ROS- and MAPKs-dependent Nrf2/HO-1 signaling pathways. Toxicol Appl Pharmacol 387:114846. https://doi.org/10.1016/j.taap.2019.114846
Mohammed I, Hollenberg MD, Ding H, Triggle CR (2021) A critical review of the evidence that metformin is a putative anti-aging drug that enhances healthspan and extends lifespan. Front Endocrinol (Lausanne) 12:933. https://doi.org/10.3389/FENDO.2021.718942/BIBTEX
Acknowledgements
The authors gratefully acknowledge Dr. Appu Rathinavelu for his generous gift of LNCaP cells. The authors would also like to thank Ms. Kimberly Whyte for her assistance with Fig. 7 and Ms. Natasha Rose for her technical assistance.
Funding
The authors declare that no funds, grants, or other support were received during the publication of this manuscript.
Author information
Authors and Affiliations
Contributions
S.D and M.T wrote the main text of the manuscript. All authors reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to Publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dixon, S., Tran, A., Schrier, M.S. et al. Metformin-induced oxidative stress inhibits LNCaP prostate cancer cell survival. Mol Biol Rep 51, 729 (2024). https://doi.org/10.1007/s11033-024-09662-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09662-8