Abstract
Background
Coccidiosis is the most common and pathogenic intestinal disease caused by different species of Eimeria is chicken. In this study, we describe the prevalence, molecular diagnosis and evolutionary insight of Eimeria tenella in chicken of Meghalaya’s sub-tropical mountainous area.
Methods and results
Faecal samples (337 no.) and dead chicks (298 no.) were collected every month from January to July’ 2023 from poultry farms (4nos.) in and around Umiam, Ri-Bhoi, Meghalaya. The chicks were categorized into different age groups viz. < 3, 3–6 and > 6 weeks. Samples were examined by flotation techniques and post-mortem. The oocysts were sporulated in 2.5% potassium dichromate solution. Eimeria tenella’s 18 S rRNA gene genomic DNA was extracted, amplified, and sequenced. Fecal sample and postmortem examinations revealed 24.04% and 33.22% infections of Eimeria sp., respectively. Oocyst per gram (OPG) was recorded highest and lowest in July (26,500) and February (9800), respectively. Amplification of the 18 S rRNA small subunit gene (SSU) by Polymerase Chain Reaction (PCR) revealed a 1790 bp band size. The amplicon was sequenced and deposited in the NCBI database. BLAST analyses of the SSU rRNA gene of E. tenella, Umiam, Meghalaya isolate (OR458392.1) revealed sequence similarities of more than 99% with SSU rRNA gene sequences available in the NCBI database. Pair wise alignment exhibited nucleotide homology ranging from 71.59 to 100.0% with the maximum sequence homology (100.0%) shared with the E. tenella isolate from Turkey (HQ680474.1) and the lowest homology of 95.6% with UK (HG994972.1). Umiam isolate were found to have 97.08% and 100.0% nucleotide similarities with E. tenella from both the UK (AF026388.1) and the USA (U40264.1), respectively. However, nucleotide similarities of 98.24%, 85.33%, 84.75% and 81.35% were observed with E. tenella strain Bangalore (JX312808.1), E. tenella isolate Kerala-1 (JX093898.1), E. tenella isolate Kerala-3 (JX093900.1) and E. tenella isolate Kerala-2 (JX093899.1), respectively. Phylogenetic analysis of SSU rRNA sequences of E. tenella Umiam, Meghalaya isolate with cognate sequences throughout the world revealed these sequences are distinct but at the same time share a close phylogenetic relationship with Indian isolates from Bangalore and Andhra Pradesh. In addition, the distant phylogenetic relationship was observed with cognate gene sequences of United States of America, Canada, China.
Conclusion
Phylogenetic analysis of SSU rRNA sequences of E. tenella Umiam, Meghalaya isolate with cognate sequences throughout the world revealed these sequences are distinct but at the same time share a close phylogenetic relationship with Indian isolates from Bangalore and Andhra Pradesh. Distant phylogenetic relationship was observed with cognate gene sequences of United States of America, Canada, China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coccidiosis is a common intracellular intestinal parasitic disease in chicken caused by different species of Eimeria. In poultry birds, nine Eimeria species have been identified, with E. brunetti, E. maxima, E. necatrix and E. tenella being the most pathogenic; E. acervulina, E. mitis and E. mivati being less pathogenic, and E. praecox and E. hagani being the least pathogenic [1, 2]. Eimeria sp. spreads via the fecal–oral pathway. Infection begins with the ingestion of sporulated (infectious) oocysts, and after asexual and sexual replications, un-sporulated oocysts are expelled with feces [3]. Commercial poultry producers tend to be concerned about coccidiosis due to the costs associated with effective chemoprophylaxis and immunoprophylaxis as well as the losses endured as a result of acute infection, digestion difficulty, decrease in egg production, morbidity and mortality as high as 80% [4, 5]. Furthermore, Mesa-Pineda et al. [6] state that the chicken enterprise is one of the primary source of protein, but it faces various obstacles, including coccidiosis, one of the diseases with the greatest impact on production performance. According to Blake et al. [7], the global financial cost of coccidiosis in chickens is estimated to be £10.4 billion.
Caecal coccidiosis is caused by E. tenella and is characterized by diarrhoea and severe caecal haemorrhages. It is a substantial threat to birds that are between 15 and 50 days old and is associated with morbidity of 50–70% [8, 9]. The afflicted birds’ growth and feed consumption are severely hampered, which reduces output and inflicting enormous financial losses to the farm enterprise [10]. It damages the intestinal epithelium due to deep penetration of the cecum tissues, causing cecal subepithelial hemorrhagic lesions [11] and anaemia [12, 13]. Secondary Clostridium perfringens infection may ensue, predisposing infected birds to additional gut infections such as necrotic enteritis. Its prevalence in poultry is increasing as a result of higher stocking densities and intense husbandry practices [14].
ITS-1 and ITS-2 genomic area sequences, as well as 18 S rDNA, the small subunit rRNA, are commonly utilized for parasite identification, ecological genetic research, and phylogenetic and evolutionary analysis at the taxonomic level, including Eimeria [10, 15, 16]. Though there have been reports on the incidence and phylogenetic analysis of E. tenella in chickens from various parts of the world viz. Bangladesh [17], Vietnam [18], Pakistan [19, 20], China [10], North India [21], Haryana [22], South India [23] and Tamil Nadu [24]. Phylogenetic analysis helps us to estimate the relationship of the isolate from the region with other isolates of the world. It will also provide useful information for prevention and control of caecal coccidiosis in the region. However, no report on the phylogenetic analysis of E. tenella in chickens of Meghalaya and North East region of India is available. Thus, the present investigation was attempted for molecular diagnosis and phylogenetic evaluation of E. tenella in chickens of the Meghalaya’s subtropical mountainous terrain.
Materials and methods
Study area and period
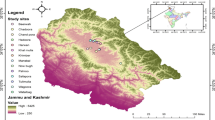
The current investigation was carried out in and around Umiam, Meghalaya’s Ri-Bhoi district, which lies between 25°6768′ North latitude and 91°9270′ East longitude. The study was conducted from April’2022 to October’2022.
Sample collection
Fresh fecal droppings and dead chicks were gathered every month from chicken farms (4nos.) within and around Umiam, Ri-Bhoi, Meghalaya in the clearly labelled plastic pouches/vials during January-July’2023. All of the chicks were divided into three groups based on their age: 3 weeks, 3–6 weeks, and > 6 weeks. In total, 337nos. fecal samples from chicken chicks of different ages have been collected viz. < 3 weeks (106nos.), 3–6 weeks (119nos.) and > 6 weeks (112nos.). 298nos. of dead chick samples from various age groups have been collected viz. < 3 weeks (83nos.), 3–6 weeks (123nos.) and > 6 weeks (92nos.).
Sample examination
To identify Eimeria oocysts in the specimens of chick feces, the samples have been examined using the direct flotation technique with saturated salt and sucrose solution [25]. Samples that were not tested on the same day were refrigerated (4 °C) for analysis the following day. Sporulation of the oocyst was accomplished by combining positive samples containing Eimeria oocysts in a 1:5 volume ratio with 2.5% potassium dichromate solution [26, 27]. The oocysts were morphologically characterized using an Olympus BX51 microscope at 100x and 400x magnifications [25]. Post-mortem examination of dead chicks was done. Wet smears were made for microscopic evaluation of Coccidia oocysts with deep scrapings from the duodenum, jejunum, ileum, and caecum as per the method described by Flek and Moody [28].
Genomic DNA extraction
Before DNA extraction, repeated centrifugation and resuspension of sporulated oocysts in distilled water was performed to remove potassium dichromate. Oocysts are cleaned 3–4 times by centrifugation at 13,000 rpm for 5 min with distilled water. The pellet having 5000 oocysts was subsequently processed by glass bead DNA extraction protocol [29] with slight modification i.e. repeated thawing (57 °C) and freezing (− 80 °C) till oocyst wall gets ruptured. In the oocyst pellet, 200 µL of STES buffer (0.1% SDS (w/v), 0.2 M Tris-HCl, EDTA 0.01 M, pH 7.6) was added followed by freezing (− 20 °C) and thawing (57 °C) 3–4 times. Then 100 µL phenol-chloroform-isoamyl alcohol and a few glass beads (0.5 mm) were added to the suspension, and vortexed for 5 min. The solution was then centrifuged for 5 min at 13,000 rpm, the supernatant was extracted and passed to a fresh sterile tube, where it was precipitated with cold 2 V (volume) absolute ethanol and 0.1 V 5 M NaCl. After a period of 30 min, the solution was spun in a centrifuge for 10 min at 12,000 rpm. The pellet had been washed two times with 70% ethanol before being air dried. The genomic DNA eluted in 100 µL of 1X TE buffer. Thermo-Fisher Scientific’s NanoDrop 1000 UV–Vis Spectrophotometer was used to quantify genomic DNA concentration. Further confirmation was done by using gel electrophoresis on an ethidium bromide-stained agarose gel (0.8%).
Amplification and sequencing of 18 S rRNA of Eimeria tenella
The extracted DNA’s small subunit (SSU) of the 18 S rRNA was amplified utilising species-specific primers (EtF: 5ʹ-ACCTGGTTGATCCTGCCAG-3ʹ and reverse EtR: 5ʹ-CTTCCGCAGGTTCACCTACGG-3ʹ) as already stated by Schwarz et al. [30]. The amplification has been carried out in a 25 µL reaction mixture comprising 12.5 µL of master mix (DreamTaq Green PCR Master Mix (2X), 2 µL of genomic DNA template, 1 µL of each primer, along with 8.5 µL of nuclease free water. A thermal cycler was used for amplification with cyclic settings: 5 min of initial denaturation at 95 °C, 60 s of denaturation at 95 °C, annealing at 62° for 60 s, 120 s extension at 72 °C and final extension at 72 °C for 5 min. The PCR result was separated by electrophoresis on a 1% agarose gel, stained by ethidium bromide, and visualized using a UV transilluminator. The size of the PCR product was identified by using a 1Kb DNA ladder. The sequencing of PCR amplified fragments was performed by the Sanger sequencing method.
Sequence alignment and phylogenetic analysis
The nucleotide sequence of E. tenella obtained after sequencing was established by using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/) and then multi-aligned utilizing the CLUSTAL W algorithm [31] from the suite of Molecular Evolutionary Genetic Analysis (MEGA X) program package. The evolutionary history was deduced using the Maximum Parsimony and Neighbor-Joining Method [32], respectively with a bootstrap value of 1000 replicates [33] to depict the evolutionary history of the taxa analyzed.
Results and discussion
Fecal sample and postmortem examinations revealed 24.04% and 33.22% Eimeria tenella infections in chicken, respectively of the sub-tropical hilly region of Meghalaya. Fecal sample examination revealed 10.38% (< 3), 43.70% (3–6) and 16.07% (> 6) infections in chicks, statistically significant (P < 0.05). However, post-mortem examination revealed 6.02%, 61.79% and19.57% infections in < 3, 3–6 and > 6 weeks old chicks, respectively which was significant statistically (P < 0.05) (Table 1). Oocyst per gram (OPG) was recorded highest and lowest in July (26,500) and February (9800), respectively. Earlier Kalita et al. [34], Thenmozhi et al. [23], Mares et al. [35] and Hassan et al. [36] reported 62.5%, 46.6%, 10.84% and 5.88% E. tenella infections in chicken, respectively. The discrepancy in percent prevalence observed in the current study could be attributed to differences in climate, environment and geographical region. E. tenella infects and destroys the epithelial cells of Lieberkhun’s caecal crypts, causing various degrees of hemorrhage depending on the amount of the parasite’s infective dosage and host characteristics such as age, genotype, and pre-exposure [37]. This parasite may trigger moderate to extreme morbidity, reduction in weight or gain, dehydration, diarrhoea, blood loss, and, in severe cases, death [38, 39]. According to Choi et al. [40], E. tenella infection reduces cecal volatile fatty acid (VFA) production, which harms feed efficiency and small intestine health. VFA is linked to feed efficiency by supplying more energy to the host or affecting chicken metabolism.
The use of species-specific primers to amplify the small subunit (SSU) of the 18 S rRNA gene from isolated genomic DNA confirmed Eimeria tenella infection in chicken. An expected size of 1790 bp PCR product was visualized on agarose gel electrophoresis (Fig. 1).
The amplicon of SSU rRNA gene of E. tenella isolate from Umiam, Meghalaya was sequenced and deposited in the GenBank database of the National Centre for Biotechnology Information (NCBI). The sequence has been verified to be from the SSU rRNA gene of E. tenella, Umiam, Meghalaya isolate (Accession No. OR458392.1) using the Basic Local Alignment Search Tool (BLAST). Analyses revealed that these sequences shared similarities of more than 99% with SSU rRNA gene sequences of E. tenella available in the NCBI database with pair wise similarities ranging from 71.59 to 100.0%. The highest sequence homology of 100.0% was discovered between the Umiam isolate of E. tenella from Meghalaya and the E. tenella isolate from Turkey (HQ680474.1), and the lowest homology of 95.6% was found in the E. tenella genome assembly, chromosome: 12, UK (HG994972.1). The SSU rRNA gene sequences of the Umiam isolate were found to have 97.08% and 100.0% nucleotide similarities with E. tenella from both the UK (AF026388.1) and the USA (U40264.1), respectively (Fig. 2).
However, nucleotide similarities of 98.24%, 85.33%, 84.75% and 81.35% were observed with E. tenella strain Bangalore (JX312808.1), E. tenella isolate Kerala-1 (JX093898.1), E. tenella isolate Kerala-3 (JX093900.1) and E. tenella isolate Kerala-2 (JX093899.1), respectively. A similar range of nucleotide homology was also reported by Thenmozhi et al. [24], from ITS-1 sequences of E. tenella, isolated from feces from farms in Tamil Nadu, India with the American and European strains with nucleotide homology ranging from 95 to 100% respectively. A comparable sequence homology of SSU rRNA as observed in the present study was also reported amongst different E. tenella isolates from Kerala [23].
Phylogenetic analysis of SSU rRNA sequences of E. tenella Umiam, Meghalaya isolate with cognate sequences throughout the world revealed these sequences are distinct but at the same time share a close phylogenetic relationship with Indian isolates from Bangalore and Andhra Pradesh (Fig. 3).
In addition, the distant phylogenetic relationship was observed with cognate gene sequences of United States of America, Canada, China. Similar observations were also made by other researchers for ITS-l sequences of E. tenella Indian isolates, whereby Chinese E. tenella strains were clustered differently from the Indian isolates [24]. Interestingly, a rather distant relationship was observed between the E. tenella Umiam, Meghalaya isolates (OR458392.1) and three Kerala isolates. Furthermore, phylogenetic analysis of SSU rRNA sequences of E. tenella Umiam, Meghalaya isolate, with other Indian isolates showed three different clades (Fig. 3). Except for the Umiam, Meghalaya isolate (OR458392.1), being an out-group, three Kerela isolates (JX093898.1), (JX093900.1) and (JX093899.1) were clustered together in one group. Similarly, a monophyletic clade was observed between E. tenella strain Bangalore (JX312808.1) and E. tenella Andhra Pradesh isolate (JX312810.1) indicating a close relationship between these two isolates. Furthermore, the cladogram showed that the E. tenella Umiam, Meghalaya isolate (OR458392.1), is genetically distinct but shows some genetic relatedness with the E. tenella strain Bangalore (JX312808.1) and E. tenella Andhra Pradesh isolate (JX312810.1), but was observed to be genetically distant from the three Kerala isolates (JX093898.1, JX093900.1 and JX093899.1). A distant relationship between the E. tenella Umiam, Meghalaya isolate (OR458392), and the E. tenella Kerala isolates (JX093898.1, JX093900.1, JX093899.1) was observed with more than 2% of divergence based on the distance matrix.
Conclusions
The current investigation concluded that Eimeria tenella infection is common in different age groups of chickens of Meghalaya. E. tenella 18 S rRNA gene amplification and sequencing revealed that SSU rRNA gene of E. tenella, Umiam, Meghalaya isolate (OR458392.1) have sequence similarities of more than 99% with sequences available in the NCBI database. Pair wise alignment exhibited nucleotide homology ranging from 71.59 to 100.0% with the maximum sequence homology (100.0%) shared with the E. tenella isolate from Turkey (HQ680474.1). Phylogenetic analysis with cognate sequences throughout the world revealed that SSU rRNA sequences of E. tenella Umiam, Meghalaya isolate are distinct but also share a close phylogenetic relationship with Indian isolates from Bangalore and Andhra Pradesh. This report on phylogenetic analysis of E. tenella may be considered as the first report from Meghalaya as well as North East region of India.
Data availability
All data obtained during this study are included in the manuscript.
References
Jadhav BN, Nikam SV, Bhamre SN, Jaid EL (2011) Study of Eimeria necatrix in broiler chicken from Aurangabad District of Maharashtra state India. Int Multidiscip Res J 1(11):11–12
Nematollahi A, Moghaddam GH, Niyazpour F (2008) Prevalence of Eimeria spp. among broiler chicks in Tabriz (Northwest of Iran). Res J Poult Sci 2:72–74
Gaboriaud P, Sadrin G, Guitton E, Fort G, Niepceron A, Lallier N, Rossignol C, Larcher T, Sausset A, Guabiraba R (2020) The absence of gut microbiota alters the development of the apicomplexan parasite Eimeria tenella. Front Cell Infect Microbiol 10:926. https://doi.org/10.3389/fcimb.2020.632556
Fayer R (1980) Epidemiology of protozoan infections- Coccidia. Vet Parasitol 6:75–103
Quiroz-Castaneda RE (ed) (2018) Chap 7. Avian coccidiosis, new strategies of treatment. Farm animals diseases, recent omic trends and new strategies of treatment; books on demand. Norderstedt. Germany. 119.
Mesa-Pineda C, Navarro-Ruíz JL, López-Osorio S, Chaparro-Gutiérrez JJ, Gómez-Osorio LM (2021) Chicken coccidiosis: from the parasite lifecycle to control of the disease. Front Vet Sci 8:787653. https://doi.org/10.3389/fvets.2021.787653
Blake DP, Knox J, Dehaeck B, Huntington B, Rathinam T, Ravipati V, Ayoade S, Gilbert W, Adebambo AO, Jatau ID, Raman M, Parker D, Rushton J, Tomley FM (2020) Re-calculating the cost of coccidiosis in chickens. Vet Res 51(1):115. https://doi.org/10.1186/s13567-020-00837-2
Clark EL, Tomley FM, Blake DP (2017) Are Eimeria genetically diverse, and does it matter? Trends Parasitol 33(3):231–241
Fornace KM, Clark EL, Macdonald SE, Namangala B, Karimuribo E, Awuni JA, Thieme O, Blake DP, Rushton J (2013) Occurrence of Eimeria species parasites on small-scale commercial chicken farms in Africa and indication of economic profitability. PLoS One 8(12):e84254
Tan L, Li Y, Yang X, Ke Q, Lei W, Mughal MN, Fang R, Zhou Y, Zhao BSJ (2017) Genetic diversity and drug sensitivity studies on Eimeria tenella field isolates from Hubei Province of China. Parasit Vectors 10:137
Vrba V, Poplstein M, Pakandl M (2011) The discovery of the two types of small subunit ribosomal RNA gene in Eimeria mitis contests the existence of E. mivati as an Independent species. Vet Parasitol 183:47–53
Lillehoj HS, Trout JM (1993) Coccidia: a review of recent advances on immunity and vaccine development. Avian Pathol 22:3–21. https://doi.org/10.1080/03079459308418897
Whitmarsh S (1997) Protozoan poultry diseases. Poultry science homepage, College of Agricultural and Life Sciences, Mississippi State University. http://www.misstate.edu/dept/poultry/disproto.htm
Nnadi PA, George SO (2010) A cross-sectional survey on parasites of chickens in selected villages in the sub humid zones of South-Eastern Nigeria. J Parasitol Res 141:1–6
Khodakaram-Tafti A, Hashemnia M, Razavi SM, Sharifiyazdi H, Nazifi S (2013) Genetic characterization and phylogenetic analysis of Eimeria arloingi in Iranian native kids. Parasitol Res 112:3187–3192
Ogedengbe ME, El-Sherry S, Ogedengbe JD, Chapman HD, Barta JR (2018) Phylogenies based on combined mitochondrial and nuclear sequences conflict with morphologically defined genera in the eimeriid coccidia (Apicomplexa). Intern J Parasitol 48(1):59–69
Alam MZ, Dey AR, Aqter S, Shahnaz R, Akter PS (2022) Phylogenetic analysis of Eimeria tenella isolated from the litter of different chicken farms in Mymensingh. Bangladesh Vet Med Sci 8:1563–1569
Nguyen HH, Le TV, Lu TA, Bao TNH (2021) Morphological and molecular characterization of coccidiosis in local chickens of Mekong Delta in Vietnam. J World’s Poult Res 11(4):506–512
Shahid SRA, Shah MA, Riaz A, Malik AM, Hasan MU, Xiangrui L, Babar W, Shahid S (2020) Identification and molecular characterization of Eimeria tenella based on EtMic5 gene in Pakistan. Pak Vet J 40(4):443–448
Sultan R, Aslam A, Tipu MY, Rehman H, Usman S, Anjum A, Imran MS, Usman M, Iqbal MZ (2021) Pathology and molecular characterization of Eimeria tenella isolated from clinically infected broiler chickens in District Lahore, Pakistan. Pak J Zool. https://doi.org/10.17582/journal.pjz/20200622030642
Kumar S, Garg R, Banerjee PS, Ram H, Kundu K, Kumar S, Mandal M (2015) Genetic diversity within ITS-1 region of Eimeria species infecting chickens of North India. Infect Genet Evol 36:262–267
Kundu K, Banerjee PS, Garg R, Kumar S, Mandal M, Maurya PS, Tomley F, Blake D (2015) Cloning and sequencing of beta-tubulin and internal transcribed spacer-2 (ITS-2) of Eimeria tenella isolate from India. J Parasit Dis 39(3):539–544
Thenmozhi V, Veerakumari L, Raman M (2014) Preliminary genetic diversity study on different isolates of Eimeria tenella from South India. Intern J Adv Vet Sci Tech 3(1):114–118
Thenmozhi V, Sureshkumar M, Raman M, Gomathinayagam S, Veerakumari L (2013) Molecular characterization of Coimbatore isolates of Eimeria tenella by ITS-1 based nested PCR. Indian Vet J 90(11):22–23
Soulsby EJ (1982) Helminths, arthropods and protozoan’s of domesticated animals, 7th edn. Bailliere Tindall, London
MAFF (1986) Ministry of agriculture, fisheries and food. manual of veterinary parasitological techniques. Her Majesty’s Stationery Office, London
Sloss MW, Kemp RL, Zajac AM (1994) Veterinary clinical parasitology, 6th edn. Iowa State University Press, Ames
Fleck SL, Moody AH (1993) Diagnostic technique in medical parasitology. Cambridge University Press, Cambridge
Reginato CZ, Braunig P, Portella LP, Mortari APG, Minuzzi CE, Sangioni LA, Vogel, F S F (2020) DNA extraction methods for molecular detection of Eimeria spp. in cattle and sheep. Pesqui Vet Brasil 40(7):514–518. https://doi.org/10.1590/1678-5150-PVB-6625
Schwarz RS, Jenkins MC, Klopp S, Miska KB (2009) Genomic analysis of Eimeria spp. populations in relation to performance levels of broiler chicken farms in Arkansas and North Carolina. J Parasitol 95:871–880
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstruction of phylogenetic trees. Mol Biol Evol 4:406–425
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Kalita A, Kakati P, Sarmah PC (2021) Isolation and molecular identification of Eimeria species circulating in broiler chicken flocks of Assam (India). Haryana Vet 60(2):251–254
Mares MM, Al-Quraishy S, Abdel-Gaber R, Murshed M (2023) Morphological and molecular characterization of Eimeria spp. infecting domestic poultry Gallus gallus in Riyadh City, Saudi Arabia. Microorganisms 11:795
Hassan TA, El-Gawady HM, El-Gayer AK, Sallam NH (2023) Morphological and molecular studies of ecto and endoparasites infested chicken in Ismailia Province, Egypt. J Adv Vet Res 13(3):352–359
Macdonald SE, Nolan MJ, Harman K, Boulton K, Hume DA, Tomley FM, Stabler RA, Blake DP (2017) Effects of Eimeria tenella infection on chicken caecal microbiome diversity, exploring variation associated with severity of pathology. PLoS One 12(9):e018489
Allen PC, Fetterer R (2002) Recent advances in biology and immune biology of Eimeria species and in diagnosis and control of infection with these coccidian parasites of poultry. Clin Microbiol Rev 15(1):58–65
Conway DP, McKenzie ME (2007) Poultry coccidiosis: diagnostic and testing procedures. Wiley, Hoboken. https://doi.org/10.1002/9780470344620
Choi J, Ko H, Tompkins YH, Teng PY, Lourenco JM, Callaway TR, Kim WK (2021) Effects of Eimeria tenella infection on key parameters for feed efficiency in broiler chickens. Animals 11:3428. https://doi.org/10.3390/ani11123428
Acknowledgements
The authors are inclined to express their gratitude to the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India, New Delhi for providing financial support and the Director, ICAR Research Complex for NEH Region, Umiam, Meghalaya for executing the research work under the project (OXX5395).
Funding
This work was supported by Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India, New Delhi (No. BT/PR45350/SPD/24/861/2022).
Author information
Authors and Affiliations
Contributions
MD: conceptualization, funding acquisition, drafted manuscript. NM: methodology, data analysis. MMM: sample collection, data curation.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethical approval
This study was done as per the guidelines of the Institute Ethics (OXX5395) and consent to publish manuscript approved by Institute. Owners of the poultry farm gave consent to us for collections of samples from chicks used in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Das, M., Masharing, N. & Makri, M.M. Phylogenetic analysis of Eimeria tenella isolates from chicken of sub-tropical mountains of Meghalaya, India. Mol Biol Rep 51, 110 (2024). https://doi.org/10.1007/s11033-023-09181-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-09181-y