Abstract
Background
Detection of high-risk human papillomaviruses (hrHPV) is widely used at the first line of cervical cancer screening, requiring rigorous validation of the clinical performance of commercial kits designed for this indication.
Methods
Performance of the AmpFire HPV Screening 16/18/HR test (AF, Atila Biosystems) and the Hybrid Capture 2 test (HC2, Qiagen) for detecting hrHPV was cross-compared in 200 cervical samples in our institution.
Results
The global percentage of agreement between the 2 techniques was 95.0% (95%CI 92–98%) with a Cohen’s kappa coefficient of 0.85 (95%CI 0.75–0.94). Ten samples showed discordant results between the 2 techniques in both directions (5 HC2+/AF- and 5 HC2-/AF+). Among possible explanations for these discrepancies was the detection of HPV66 and HPV53 genotypes in two samples, since these genotypes are targeted by the Ampfire test but not by the HC2 test, as well as intrinsic differences in analytical performance to target specific genotypes.
Conclusions
A high level of agreement was observed between the two techniques, which encourages further testing in order to definitively validate the use of the Ampfire kit for primary cervical cancer screening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human papillomaviruses (HPV) are a leading cause of anogenital and oropharyngeal cancers worldwide and represent a major burden in low-income countries [1]. Among these HPV-induced cancers, the higher incidence is observed for cervical cancers, which exceeded 600,000 cases in 2020, responsible for more than 340,000 deaths, mainly in low-income countries (Globocan, 2020). Cervical cancers typically appear several decades after sexual contamination by HPV, in case of non-effective immune response and viral persistence inside the cervical mucosae [2].
More than 400 HPV types (or “genotypes”) are currently described [3] but only 13 of them, all belonging to the genus Alphapapillomavirus, are considered high-risk (hrHPV) oncogenic types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68) and some are considered possibly high-risk (such as HPV53 and 66). All other types are considered low-risk types but may be involved in benign lesions such as warts and condylomas. Among hrHPV, HPV16 is the most prevalent and the most potent carcinogenic type, followed by HPV18 in the case of cervical cancers [4].
Since the detection of hrHPV is now widely used at the first line of primary cervical cancer screening, numerous HPV assays have been developed. However, only a small part of these tests fulfilled the clinical criteria of sensitivity and specificity required for cervical cancer screening [5]. These criteria were primarily based on the clinical performance of Hybrid Capture 2 HPV DNA test (HC2, Qiagen) or GP5+/6 + PCR- Enzyme Immunoassay [6]. Therefore, the relative clinical accuracy of any new HPV commercial kit has to be evaluated in comparison with one of these 2 reference tests. Commercial HPV kits also differ in their technology and practicability. This later criteria can have a significant impact on facilities with poor molecular biology equipment.
AmpFire HPV Screening 16/18/HR test (Atila Biosystems, USA) is an isothermal nucleic acid amplification assay allowing the DNA detection of 15 high risk HPV (16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66 and 68) with a specific genotyping of HPV16 and HPV18. Here, we cross-compared the performance of the AmpFire HPV Screening 16/18/HR test with the HC2 test for detecting hrHPV in a collection of cervical samples.
Materials and methods
Samples
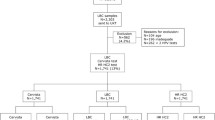
Two hundred cervical samples, obtained from the Department of Gynaecology and Obstetrics, Besançon University Hospital, France, were used.
Samples were obtained by scraping the cervix with a cytobrush, secondarily discharged into 1 mL of a specific transport medium (Digene Specimen Transport Medium, Qiagen, Germany). Samples were transported to the laboratory within 24 h, and stored at 4 °C until processing for hrHPV testing by HC2 in the context of routine biological diagnosis in our institution. The samples were then stored at -20 °C.
Hybrid capture 2 test and HPV16/18 viral loads
The HC2 test is designed to detect in bulk the complete genome of 13 hrHPV genotypes (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68). The test was used according to the manufacturer’s instructions and its principle has been described elsewhere [7]. Results were expressed in Relative Light Units (RLU) and standardized with respect to the response for 1 pg/mL HPV16 DNA (Positive Control, PC). Samples with a RLU/PC ≥ 1 were considered positive.
In case of positive result by the HC2 screening test, the sample was tested by an in-house real-time PCR assay, targeting E6 HPV16 and HPV18 genes (Jacquin et al., 2013). PCR results were expressed in log10 HPV copies per mL (log10 cp/mL).
AmpFire HPV screening 16/18/HR protocol
HPV DNA was extracted from 100 µL of cervical samples using the QIAamp DNA Mini Kit (Qiagen) with a final elution in 80 µL. AmpFire HPV Screening 16/18/HR test was used from this extracted DNA according manufacturer’s instructions (of note, other protocols are proposed by the manufacturer without this extraction step). Briefly, 9.5 µL of eluted DNA was diluted by half in water and 1 µL of 20X lysis buffer was added. This preparation (20 µL) was incubated 20 min at room temperature and 2 µL of it were added to 23 µL of the supplied master mix.
DNA amplification was performed in a CFX96 Real-Time System (Bio-Rad) with an isothermal reaction setting for 60 min at 60 °C. Fluorescence was read in the FAM (other HPV) / HEX (cellular control) / CY5 (HPV16) / ROX (HPV18) channels and amplification curves were analyzed using the BioRad CFX Manager Software 1.6.
INNO-LiPA® HPV genotyping extra II test
Samples with discordant results between HC2 and Ampfire tests were analyzed by the INNO-LiPA HPV Genotyping Extra II kit (Fujirebio), according to the manufacturer’s instructions. This technique allows the identification of 32 different HPV genotypes (including high-risk, possibly high-risk, and some low-risk genotypes) by reverse hybridization after a step of HPV genome amplification with SPF10 primers.
Ethics
All used samples were stored into a biobank for which a declaration of preparation and storage of human samples for research use has been sent to the « Ministère de l’Enseignement Supérieur et de la Recherche » (n°DC-2014-2086).
Statistical analysis
The number of samples to be analyzed was estimated before the study. Comparison between the 2 techniques was performed by determining the global percentage of agreement, the percentage of agreement in positive HC2 samples, negative HC2 sample, and finally using Cohen’s kappa coefficient [8]. Confidence intervals for proportions and Cohen’s kappa coefficient were determined as described previously [9]. All analyses were performed with R version 4.1 for Windows (R Core Team, 2021; R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. www.R-project.org/).
Results and discussion
Two hundred cervical samples, first analyzed by the HC2 assay, were secondarily tested by the Ampfire technique. A positive result for hrHPV was obtained in 41 (20.5%) samples by each technique (Table 1). Despite this equivalent rate of positivity among the tested samples, some discrepancies were observed between the 2 techniques (Table 1), since 5 samples were negative by HC2 (HC2-) but positive by Ampfire (AF+), and 5 samples had the opposite result (HC2+/AF-).
Thus, the global percentage of agreement between the 2 techniques was 95.0% (95% confidence interval, 95%CI 92–98%) and the Cohen’s kappa coefficient was 0.85 (95%CI 0.75–0.94). The positive and negative percentage of agreement were 88% (n = 41; 95%CI 78–98%) and 97% (n = 159; 95%CI 94–100%), respectively.
The 10 samples that showed discordant results were further characterized by the INNO-LiPA HPV Genotyping Extra II technique (Fujirebio) and by an in-house HPV16 and HPV18 real-time PCR. Complete results and interpretations are shown in Table 2.
Among the 5 [HC2+/AF-] samples, 2 were considered as false positive by HC2 (samples 1 and 2; since they contained the low risk HPV26, 54, and 61) and 3 were considered as false negative by AmpFire (samples 3 to 5; since the contained the high risk HPV52 or 16).
Among the 5 [HC2-/AF+] samples, 2 were considered as false positives by AmpFire (samples 6 and 7; since they contained the low risk HPV70 or no detectable HPV by the other technique). The 3 remaining samples (samples 8, 9, and 10) were difficult to interpret and could correspond (i) to false negatives by HC2 (since they contained the high risk HPV51, 52, and 68 but with unknown viral loads), (2) to false positives by AmpFire (due to an inappropriate detection of low risk HPV62 and 81 or to an inappropriate detection of hrHPV that would be below the threshold of clinical relevance), (3) to the detection of HPV66 (sample 9) and HPV53 (sample 10) that are not targeted by the HC2 test or (4) to a mix of these different situations.
This comparison between the results of the HC2 and the Ampfire techniques, performed from 200 cervical samples collected in our institution, showed a high global percentage of agreement (95.0%), with a Cohen’s kappa coefficient of 0.85, despite some discrepancies between the 2 techniques.
The HC2 test is a classical comparator to validate the performance of other kits designed to be used for cervical cancer screening. In this screening context, aiming to detect high-grade cervical lesions and cervical cancers without inducing unnecessary or excessive follow-up procedures for HPV-infected women, candidate HPV tests should reach an optimal balance between clinical sensitivity and specificity [6]. Therefore, guidelines have been proposed to validate these potential new tests. However, this validation requires the use of a large number of fully characterized samples to determine the clinical sensitivity and specificity of the test (at least 860 samples), and its intra/inter-laboratory reproducibility (at least 500 samples) [6]. Here, we performed a smaller-scale comparison to obtain initial performance data for the Ampfire test.
The INNO-LiPA HPV genotyping Extra II kit and a specific in-house PCR targeting HPV16 and HPV18 were used to help resolving discrepancies. The INNO-LiPA HPV genotyping Extra II kit is a highly sensitive technique (due to the small size of the amplicon) able to detect and identify 32 different HPV genotypes. The main drawback of this technique is an insufficient clinical specificity for cervical cancer screening [10]. Thus, this technique can detect “traces” of HPV that are considered too weak to be clinically relevant in the context of cervical cancer screening. However, as used in our study, it can be of great help to detect and identify a HPV genome in case of discordance between 2 other techniques.
Among most of the samples with discordant results (with the notable exception of samples 8 to 10), the INNO-LiPA technique allowed us to provide the most probable explanation. In two cases (samples 3 and 4), we suggested that the Ampfire test failed to detect HPV52. This should be confirmed by testing a larger number of samples harboring this genotype (ideally with different viral loads). The non-detection of HPV16 in sample 5 is surprising but the result was verified on a second experiment to rule out any technical problems during the manipulation. In 2 cases (samples 6 and 7), the Ampfire test was positive although no hrHPV could be detected by the other techniques, suggesting a false positive result with the Ampfire test. Interestingly, the detection signal of the Ampfire test was weak in both cases that may question the clinical relevance of the cut-of used to interpret a positive result.
The results for samples 8 to 10 were more difficult to interpret. It is possible that the Ampfire test have succeeded to detect the 3 hrHPV (HPV51, 52, 68) harbored by the samples, thanks to a better sensitivity than the HC2 test for these genotypes. However, this better sensitivity could be at the cost of a lower specificity in case of using the kit for cervical cancer screening. This is probably not the case as a recent study conducted in a screening population reported that the Ampfire test had clinical sensitivity and specificity similar to the validated Cobas® 4800 HPV test [11]. For samples 9 and 10, another simple explanation could be the presence of HPV66 and HPV53 (both classified as a “possibly high-risk” types), respectively, since the Ampfire technique is able to detect these genotypes while the HC2 technique do not target them. Alternatively, a false negative reaction by the HC2 test or a false positive reaction by the Ampfire test (against another HPV genotype) for these 3 samples cannot be completely ruled out.
In conclusion, the results of the Ampfire and HC2 tests showed very high agreement. However, further evaluations of the Ampfire test are needed, especially an intra- and inter-laboratory evaluation of the reproducibility to fulfil all Meijer’s criteria [6] for its use in cervical cancer screening.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
de Martel C, Plummer M, Vignat J, Franceschi S (2017) Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer 141:664–670. https://doi.org/10.1002/ijc.30716
Schiffman M, Doorbar J, Wentzensen N et al (2016) Carcinogenic human papillomavirus Infection. Nat Rev Dis Primer 2:16086. https://doi.org/10.1038/nrdp.2016.86
Bzhalava D, Eklund C, Dillner J (2015) International standardization and classification of human papillomavirus types. Virology 476:341–344. https://doi.org/10.1016/j.virol.2014.12.028
de Sanjosé S, Serrano B, Tous S et al (2018) Burden of human papillomavirus (HPV)-Related cancers attributable to HPVs 6. JNCI Cancer Spectr 2 and 58:pky045. 11/16/18/31/33/45/52
Arbyn M, Simon M, Peeters E et al (2021) 2020 list of human papillomavirus assays suitable for primary Cervical cancer screening. Clin Microbiol Infect off Publ Eur Soc Clin Microbiol Infect Dis 27:1083–1095. https://doi.org/10.1016/j.cmi.2021.04.031
Meijer CJLM, Berkhof J, Castle PE et al (2009) Guidelines for human papillomavirus DNA test requirements for primary Cervical cancer screening in women 30 years and older. Int J Cancer 124:516–520. https://doi.org/10.1002/ijc.24010
Debernardi A, Jarassier W, Soret C et al (2021) Repurposing the Hybrid capture 2 (HC2) screening test for whole-genome sequencing of human papillomaviruses. Arch Virol 166:3421–3425. https://doi.org/10.1007/s00705-021-05259-9
Cohen J (1960) A coefficient of Agreement for Nominal scales. Educ Psychol Meas 20:37–46. https://doi.org/10.1177/001316446002000104
Fleiss JL, Cohen J, Everitt BS (1969) Large sample standard errors of kappa and weighted kappa. Psychol Bull 72:323
Xu L, Padalko E, Oštrbenk A et al (2018) Clinical evaluation of INNO-LiPA HPV genotyping EXTRA II assay using the VALGENT Framework. Int J Mol Sci 19:2704. https://doi.org/10.3390/ijms19092704
Zhang W, Du H, Huang X et al (2020) Evaluation of an isothermal amplification HPV detection assay for primary Cervical cancer screening. Infect Agent Cancer 15:65. https://doi.org/10.1186/s13027-020-00328-1
Acknowledgements
The authors would like to thank the Fujirebio Company for providing the kits used for this comparison.
Funding
Inserm CIC1431 received funding from Fujirebio for statistical analysis. The funder had no role in the design and interpretation of the data.
Author information
Authors and Affiliations
Contributions
AK and AB: Investigation MD: conceptualization, statistical analysis, data curation KD: statistical analysis, data curation LP: writing review and editing QL: statistical analysis, data curation, writing original draft JLP: conceptualization, Resources, writing review and editing, project administration.
Corresponding author
Ethics declarations
Ethical approval
This study was conducted in accordance with the amended Declaration of Helsinki. All used samples were stored into a biobank for which a declaration of preparation and storage of human samples for research use has been sent to the « Ministère de l’Enseignement Supérieur et de la Recherche » (n°DC-2014-2086).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Koussouri, A., Baraquin, A., Desmarets, M. et al. Comparison of high-risk HPV detection by the AmpFire® HPV Screening 16/18/HR technique (Atila Biosystems) and the hybrid capture 2 test (Qiagen). Mol Biol Rep 51, 52 (2024). https://doi.org/10.1007/s11033-023-08939-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-08939-8