Abstract
SARS-CoV-2, a novel coronavirus within the Coronaviridae family, is the causative agent behind the respiratory ailment referred to as COVID-19. Operating on a global scale, COVID-19 has led to a substantial number of fatalities, exerting profound effects on both public health and the global economy. The most frequently reported symptoms encompass fever, cough, muscle or body aches, loss of taste or smell, headaches, and fatigue. Furthermore, a subset of individuals may manifest more severe symptoms, including those consistent with viral pneumonitis, which can be so profound as to result in fatalities. Consequently, this situation has spurred the rapid advancement of disease diagnostic technologies worldwide. Predominantly employed in diagnosing COVID-19, the real-time quantitative reverse transcription PCR has been the foremost diagnostic method, effectively detecting SARS-CoV-2 viral RNA. As the pandemic has evolved, antigen and serological tests have emerged as valuable diagnostic tools. Antigen tests pinpoint specific viral proteins of SARS-CoV-2, offering swift results, while serological tests identify the presence of antibodies in blood samples. Additionally, there have been notable strides in sample collection methods, notably with the introduction of saliva-based tests, presenting a non-invasive substitute to nasopharyngeal swabs. Given the ongoing mutations in SARS-CoV-2, there has been a continuous need for genomic surveillance, encompassing full genome sequencing and the identification of new variants through Illumina technology and, more recently, nanopore metagenomic sequencing (SMTN). Consequently, while diagnostic testing methods for COVID-19 have experienced remarkable progress, no test is flawless, and there exist limitations with each technique, including sensitivity, specificity, sample collection, and the minimum viral load necessary for accurate detection. These aspects are comprehensively addressed within this current review.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
December 12th, 2019 marked the initial report of an infection caused by a highly contagious respiratory virus in Wuhan, located in Hubei Province, China. This virus was subsequently identified as a member of the Coronaviridae family [1,2,3,4,5]. Rapidly, the virus spread to neighboring countries, swiftly evolving into one of the most substantial pandemics within a mere six-month span. This led to successive waves of infection across the globe, ultimately culminating in the largest pandemic witnessed in the past five decades [6,7,8,9]. It’s worth mentioning that the International Committee on Taxonomy of Viruses designated the virus as SARS-CoV-2, signifying “severe acute respiratory syndrome coronavirus 2.“ Concurrently, as per the World Health Organization (WHO), the official nomenclature for the associated illness was “coronavirus disease 2019” (COVID-19) [10,11,12], after the publication of its genome [13]. The rapid transmission of the infection occurs through person-to-person contact, and SARS-CoV-2 is responsible for inducing a range of severe symptoms, including fever, intense bodily discomfort, headaches, and gastrointestinal issues, among others [1, 14]. SARS-CoV-2 has the potential to significantly impact the respiratory system, leading to conditions such as desaturation and general hypoxemia [15].
As of August 2023, COVID-19 has resulted in more than 6,955,141 deaths out of 769,774,646 confirmed cases globally, and a total of 13,498,570,620 vaccine doses have been administered (source: WHO; https://www.worldometers.info/coronavirus/) [16]. It’s important to highlight that due to the elevated prevalence and mortality rates associated with COVID-19, molecular biology has faced the challenge of enhancing and developing faster diagnostic techniques for the detection of this virus. These efforts aim to effectively curb the virus’s progression within the population. Consequently, the strategies for detecting COVID-19 have evolved, transitioning from real-time PCR to rapid tests utilizing samples such as bronchoalveolar lavage fluid and saliva [1, 17]. n this review, we provide an overview of the origin and primary challenges encountered during the global progression of COVID-19. We also offer an extensive description of the key molecular methods utilized for detecting COVID-19. Furthermore, we delve into the significant pathophysiological repercussions that COVID-19 inflicts on the respiratory system.
Infection overview
t’s widely recognized that SARS-CoV-2 is neither the sole nor the initial member of the Coronaviridae virus family, specifically belonging to the subgroup B known as betacoronavirus, which has induced severe infections in humans. Instances of novel coronaviruses leading to emergent situations have been recurrent throughout human history. An illustrative case is the bat HKU2 virus, which surfaced in 2018 and is accountable for triggering acute diarrhea syndrome in pigs [18]. Several members of the Coronaviridae family, including 229E, OC43, NL63, and HKU1, are known to induce mild cold-like symptoms in humans [19]. A notable aspect to highlight is that the HKU2 virus, situated within the expansive Coronaviridae family, demonstrates a broad host range. This virus has the capability to infect a diverse array of creatures, encompassing birds and various mammals such as camelids, bats, civets, rats, mice, dogs, and cats. This wide spectrum of potential hosts facilitates the transmission and dissemination of the virus.
In 2002, a pivotal event occurred when the first fatal virus for humans originating from the Coronaviridae family emerged in China. This virus was labeled SARS-CoV-1, derived from “severe acute respiratory syndrome.“ Notably, SARS-CoV-1 managed to infect approximately 8,000 individuals, resulting in a mortality rate of 10%. In 2003, despite its initial disappearance, this virus resurfaced in Guangzhou, located in Guangdong province, China. Given the significant mortality and the rapid rate of transmission associated with SARS-CoV-1, exhaustive efforts were undertaken to combat its spread. These collective endeavors ultimately led to the successful eradication of the virus by 2004 [12].
n 2012, a consequential occurrence unfolded with the emergence of another lethal virus known as MERS-CoV, denoted as “Middle East respiratory syndrome.“ This virus first manifested in Saudi Arabia. The World Health Organization (WHO) has recorded a total of 2,574 confirmed cases, resulting in 886 fatalities. Geographically, the virus has spread across 12 countries within the Middle East region. A significant aspect to highlight is the high mortality rate exhibited by MERS-CoV, reaching 34.4%. However, this rate is notably lower than that observed for other viruses like the Ebola virus (50%) and the rabies virus (95%) [12].
Origin of the SARS-CoV-2 virus
It’s widely acknowledged that all human coronaviruses trace their origins back to zoonotic sources, a pattern shared by numerous pathogenic viruses affecting humans. The prevailing body of evidence strongly indicates that the novel coronavirus SARS-CoV-2 likely originated from a primary reservoir, such as the horseshoe bat. It’s believed to have subsequently passed through an intermediate reservoir, possibly leading to an outbreak within wildlife. This transition may have occurred in settings like the Huanan market in Wuhan, China, in 2019, where the trading of animal wildlife takes place [9, 20]. Supporting this perspective, environmental samples taken from the Wuhan market exhibited positive results for SARS-CoV-2. However, when samples were sourced from live animals and the carcasses of wildlife, there were no indications of SARS-CoV-2 positivity. This observation raises the possibility that SARS-CoV-2 may be fatal to various other species, including pets such as dogs and cats, as well as wild animals like deer, lynx, and monkeys. Additionally, this could encompass different types of large felines, such as lions, tigers, and snow leopards. The fact that these animals exhibit similar pathological characteristics to humans could indicate an alternative source for the virus, suggesting that the initial detection in Wuhan might not necessarily be its point of origin [21,22,23].
Phylogenetically, SARS-CoV-2 can be categorized into two distinct lineages, namely A and B. These two lineages likely coexisted concurrently during the initial outbreak phases [24]. Lineage B was initially identified in the initial cases connected to the Huanan market, and it subsequently gained global predominance. Conversely, lineage A was observed in instances associated with different markets and subsequent occurrences in Wuhan City as well as other regions across China [12]. The emergence of SARS-CoV-2 entails one or potentially several instances of contact with infected animals or individuals. This includes the likelihood of various indirect scenarios, such as the transfer of live animals to different markets within Wuhan [25].
A noteworthy observation is that SARS viruses were identified prior to the emergence of SARS-CoV-2, exhibiting significant infection rates, seroprevalence, and genetic diversity. These SARS viruses were detected in animals from both the Dongmen market in Shenzhen and the Xinyuan market in Guangzhou, China [26, 27]. During the initial phase of the 2019 pandemic, the genetic evolution of SARS-CoV-2 remained relatively limited. However, a significant development occurred with the emergence of a globally dominant variant known as D614G. This particular variant was linked to increased transmissibility compared to earlier forms of the virus [28]. Numerous variants of SARS-CoV-2 have been characterized, and several among them are categorized as variants of concern due to their significant impact on public health. According to the epidemiological update as of October 8, 2022, five SARS-CoV-2 variants of concern (COV) have emerged since the onset of the pandemic: Alpha (B.1.1.7) from the United Kingdom, Beta (B.1.351) from South Africa, Gamma (P.1) from Brazil, Delta (B.1.617.2) from India, and Omicron (B.1.1.529). However, as of March 20, 2023, the Omicron variant is the sole strain designated as a variant of concern (VOC), while the others are categorized as variants being monitored (VBM) [29].
Through virus genome sequencing and evolutionary analysis, it has been determined that the SARS-CoV-2 genome bears a 96.2% resemblance to the CoV-RaTG13 strain found in bats, particularly Rhinolophus affinis in Yunnan. This contrasts with SARS-CoV, which demonstrates a 79.5% similarity. These findings suggest that bats could serve as the natural host for the original virus. For instance, the bat viruses RmYN02, RpYN06, and PrC31 exhibit greater genetic proximity to most parts of the SARS-CoV-2 genome, particularly the ORF1ab region. This closer resemblance points to a more recent shared ancestor with SARS-CoV-2 [9, 30, 31]. SARS-CoV-2 might have been transmitted from bats to humans through intermediary hosts that were previously undiscovered [9]. Nonetheless, recent investigations into the origin of SARS-CoV-2 have indicated that pangolins might be plausible candidates as the intermediate hosts for the virus [32, 33]. The precise route through which the virus infects humans via pangolins remains unclear. Neither of the viruses found in these mammalian species closely resemble SARS-CoV-2 enough to be considered its direct ancestor. Furthermore, the heightened vulnerability of minks and cats to SARS-CoV-2 implies that other animal species could potentially serve as reservoirs for the virus. A visual representation of the plausible origin of the SARS-CoV-2 virus has been summarized in Fig. 1.
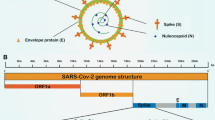
SARS-CoV-2 hypothetical origin scenarios and invasion mechanisms. (A) Zoonotic spillover is the leading hypothesis suggesting that the virus originated in animals and was transmitted to humans. Coronaviruses have been found in various animal species, including bats, which are considered a natural reservoir for many coronaviruses. It is speculated that an intermediate animal host, such as a pangolin, could have played a role in transmitting the virus from bats to humans. The specific mechanisms by which this spillover event occurred are still under investigation, but recombination has been identified as a possible factor in the emergence of SARS-CoV-2. It is important to note that the investigation into the origin of SARS-CoV-2 is ongoing. (B) Representation of the reference genome of SARS-CoV-2, where the coding regions for essential proteins for diagnostic studies are shown in detail. These consist of ORF1ab (RdRp), Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N). The entry of SARS-CoV-2 into host cells involves several steps, primarily mediated by the spike protein (S) on the virus’s surface, priming by proteases, membrane fusion, genome release, translation, replication, assembly, and release of virions. It’s important to note that the entry mechanism and viral replication process can vary in different cell types and tissues. Created with BioRender.com
SARS-CoV-2 virus genome and mechanisms entry into the cells
Coronaviruses are enveloped viruses with a genome composed of a single positive-sense, single-stranded RNA molecule. Unlike other biological systems, viruses exhibit a wider range of mutation rates [34, 35]. The replication accuracy of viral RNA molecules is consistently lower compared to DNA, resulting in RNA viruses accumulating more mutations and adapting to new hosts more rapidly than their DNA genome counterparts. The SARS-CoV-2 virus possesses an RNA genome and has been demonstrated to exhibit a mutation rate that is 1,000 times slower than that of flu or HIV [9].
SARS ARS-CoV-2 falls within the coronavirus family situated in the Nidovirales order, constituting one of the virus groups characterized by the longest RNA genomes. Specifically, its genome is composed of a single strand of positive-sense RNA (+ ssRNA) spanning approximately 30,000 base pairs [36], with a guanine and cytosine content of 38%. The genome comprises no less than fifteen open reading frames (ORFs), of which twelve are functional. The viral replicase gene is located within ORF1ab, housing the largest ORF in the genome (~ 20,000 bp). These ORFs are organized into replicase and protease (ORF1a-ORF1b) segments, along with significant protein components (S, E, M, and N), as well as accessory protein genes (HE, 3, 7a, among others) (Fig. 1). These proteins play essential roles in facilitating virus entry, fusion, and the virus’s ability to thrive within host cells [7, 37, 38]. The ensuing discussion outlines some of the key proteins of SARS-CoV-2:
Spike protein
The spike protein, denoted as protein S with a size of 180–200 kDa, has been elucidated in SARS-CoV, MERS, and SARS-CoV-2. In these viruses, protein S contains 1,104 to 1,273 amino acids and is composed of two distinct subunits: an (N)-terminal subunit termed S1 (located extracellularly) and a C-terminal subunit designated S2 (positioned intracellularly) [39,40,41]. Fundamentally, this protein is essential for facilitating the virus’s entry into the host cell.
Envelope protein
Among all structural proteins, protein E holds the distinction of being the smallest, with a size of 8–12 kDa. It plays a multifaceted role within a broad functional range [42]. While it is expressed in abundance within infected cells during the replication cycle, only a minor fraction is integrated into the virion envelope. The majority of this protein is found within intracellular pathways, including the Golgi complex, where it contributes to viral particle assembly. Its significance extends to the virus’s production and maturation processes [43].
Membrane protein
This protein stands out as the most prevalent structural protein, playing a pivotal role in shaping the virion. The monomeric form of M (25 to 30 kDa) functions as a membrane protein integrated into the envelope through three transmembrane domains. The N-terminal segment comprises a small ectodomain, while the C-terminal endodomain represents the most substantial portion of the molecule. It is situated within the virion or on the intracellular membrane’s cytoplasmic side. Glycosylation can modify the ectodomain, influencing both the organ tropism for infection and the interferon (IFN)-inducing capacity of certain coronaviruses. Moreover, this protein contributes to the attachment of the nucleocapsid to internal structures’ membranes, such as the Golgi complex. It plays a role in transmembrane nutrient transport, virion release, and the formation of the envelope field [44,45,46].
Nucleocapsid protein
The N protein, weighing 43 to 50 kDa, takes on the role of forming the helical nucleocapsid, encompassing the entirety of the viral genome (Wang et al., 2003). This protein comprises two domains, each capable of recognizing viral RNA. Additionally, the N protein has been found to bind nsp3 (nonstructural protein 3), facilitating the guiding of the genome toward the replication and transcription complex and aiding in nucleocapsid packaging. It also serves as an interferon antagonist and a suppressor of virus-encoded RNA interference. Another crucial function involves its interaction with protein M [45, 47, 48].
Among the accessory proteins, the S protein stands out as one of the most crucial viral structures in the infection process of SARS-CoV. The S1 subunit takes on the role of binding to angiotensin 2 receptors [49]. Upon interaction with the host cell, the S protein undergoes structural reconfiguration, leading to the exposure of the S2′ cleavage site within the S2 subunit. This structural change enables the virus to fuse with the host cell membrane [50]. The cleavage at the S2′ site transpires once the virus has bound to angiotensin-converting enzyme 2 (ACE2) on the target cell. Interestingly, the spike-shaped protein structures (S) are adorned with polysaccharide molecules, which could potentially function as a form of camouflage, aiding the virus in evading detection by the host’s immune system during invasion [51, 52].
The mentioned processes occur within host cells, particularly in cases where there is an insufficient expression of transmembrane protease serine 2 (TMPRSS2), or the virus ACE2 complexes fail to encounter TMPRSS2. In scenarios where the virus ACE2 complex is internalized through clathrin-mediated endocytosis into endolysosomes, the S2’ cleavage is carried out by cathepsins [50]. Subsequently, the virus releases its genetic material into the cell’s cytoplasm, leveraging the cellular machinery of the target cell for replication. Grasping the functionalities of these proteins and their interactions with the host is pivotal for the development of therapeutics, vaccines, and diagnostics aimed at combatting COVID-19.
SARS-CoV-2 virus diagnostic methods
COVID-19 does not stand alone as the first or final pandemic our human society has experienced. In response to this situation, a swift acceleration in diagnostic technology has transpired on a global scale, effectively transforming the Earth into an extensive laboratory dedicated to thwarting the progression of the pandemic.
Characteristics of the samples to be collected
or the diagnosis of COVID-19 through molecular techniques, respiratory materials collected from primarily symptomatic patients are commonly employed. The materials consist of upper respiratory samples, including nasopharyngeal swabs (NPS), oropharyngeal swabs (OPS), and nasal washes [53]. Additionally, in cases of more severe respiratory conditions, samples from the lower respiratory tract such as sputum, endotracheal aspirate, or bronchoalveolar lavage can be utilized (Fig. 2) [54]. It’s noteworthy that the virus has also been identified in samples derived from both blood and feces [9, 54].
Scheme for taking nasopharyngeal samples. Sampling method to be used in diagnostic techniques for identifying SARS-CoV-2 and the different types of most used viral transport media. Viral Transport Medium (VTM) is specifically designed to maintain the viability and stability of respiratory viruses, including SARS-CoV-2. It often contains balanced salt solutions, protein stabilizers, antibiotics, and antifungal agents. VTM-N: Viral Transport Medium with Non-inactivated Virus; eNAT: Extraction-Free Nucleic Acid Transport; PBS: Phosphate-Buffered Saline. Created with BioRender.com
An intriguing observation is that certain authors have conducted a comparison between saliva and nasopharyngeal samples to evaluate the efficacy of diagnosing the SARS-CoV-2 virus through RT-qPCR tests. Their findings indicated that saliva samples exhibit higher sensitivity and consistency in detecting the virus’s presence throughout the infection’s progression. This suggests that saliva could serve as a sample type facilitating the monitoring of changes in SARS-CoV-2 titers over time. Importantly, using saliva samples carries the advantages of greater acceptability among the population and a reduced risk of contagion for healthcare personnel during sample collection, especially in the presymptomatic phase [55,56,57,58]. Notably, viral titers in saliva were observed to be five times higher than in nasopharyngeal samples. However, while some studies propose that saliva is more effective for early presymptomatic cases, its diagnostic efficacy appears to be similar to nasopharyngeal samples during the course of infection. Even among preschool children, virus detection efficiency has been found to be notably high in saliva samples. Moreover, a comparison between saliva and nasopharyngeal samples demonstrated comparable diagnostic effectiveness using the Nucleic Acid Amplification Test (NAAT), particularly in ambulatory settings [59].
While the sample collection protocol is of utmost importance in COVID-19 detection, the standard confirmation of COVID-19 cases hinges on identifying distinct RNA sequences of the virus through nucleic acid amplification testing, such as RT-qPCR. When needed, this is further verified through nucleic acid sequencing [60]. It’s crucial to highlight that the effectiveness of diagnosis is contingent upon various factors, including the sample’s quality, the medium used for transport, the viral load present, the possibility of contamination post-sample collection, and the specific days post-infection when the sample was obtained [61].
Nucleic acid detection for PCR
RT-qPCR, a method that detects the virus’s RNA, stands as the primary diagnostic tool for identifying SARS-CoV-2 in human patients. In certain countries, like Chile, as of March 17, 2023, a total of 4,391 new COVID-19 cases were documented. Within this context, 27,695 tests were administered, encompassing both PCR and antigen tests. Notably, 65% of diagnoses were accomplished via antigen tests, 27% were initiated through active case search (BAC), and 9% of those notified were asymptomatic [62]. The sensitivity of this approach is intrinsically tied to the viral load present in the sample [63]. In summary, the method involves utilizing samples predominantly extracted from the respiratory tract, including nasopharyngeal exudate, nasal exudate, tracheobronchial aspirate, and sputum. The World Health Organization (WHO) recommends the partial amplification of specific genes, such as the RNA-dependent RNA Polymerase gene (RdRp), to confirm the virus’s presence. Furthermore, genes encoding the virus’s envelope proteins (E), nucleocapsid proteins (N), and membrane proteins (M) are targeted to verify the virus’s existence. To mitigate the risk of false positives or negatives, two or three genes are concurrently amplified for each sample during the experiment [12].
Significantly, in the latter months of 2021, the emergence of new variants triggered an upswing in false negatives across multiple diagnostic methods, including RT-qPCR. This phenomenon was attributed to mutations within the virus’s genome, primarily concentrated in the S protein. These mutations had the potential to cause the absence of bands, thereby impeding the successful amplification of fragments. This was an outcome of the original primer designs being based on the initial genome sequence identified in Wuhan in 2019. Notably, one of the early-identified variants was the British variant B.1.1.7 (20I/501Y.V1), which carried over 20 mutations, the majority clustered within the S protein. Some of these mutations included Spike ΔH69-V70, ΔY144, N501Y, A570D, P681H, T716I, S982A, and D1118H. This S protein, a primary target for neutralizing antibodies, was thus heavily affected [64,65,66]. These mutations can reduce the diagnostic sensitivity of PCR tests that target the gene encoding the S protein. Of note, ΔH69/V70 deletion mutation not only interfered with detection methods of the variant B.1.1.7, if not could promote the propagation of the virus. Currently, there have been advancements in the development of primers that can identify various emerging variables. Consequently, these mutations had the potential to reduce the diagnostic sensitivity of PCR tests that targeted the gene encoding the S protein. Particularly noteworthy was the ΔH69/V70 deletion mutation, which not only impacted the detection methods of the B.1.1.7 variant but also potentially facilitated the virus’s propagation. Notably, there have been advancements in the development of primers capable of detecting various emerging variants. Notable examples include the D614G and N501Y mutations, which appear to heighten the interaction between the S protein and the ACE2 receptor [67, 68]. Moreover, the combination and accumulation of mutations have enabled the identification of specific variants through the absence of certain bands. For instance, the combination of mutations 484E and 501 N denotes the wild strain, whereas the pairing of 484E and 501Y designates the British variant. This underscores the evolving nature of diagnostics to encompass the recognition of these emerging variants.
Likewise, the Brazilian variant is marked by the presence of both 484 K and 501Y mutations. Distinctly, the South African variant is characterized by the mutations 417 N, while the Californian variant bears the L452R mutation. Additionally, the Indian variants, including B.1.617, B.1.617.1, and B.1.617.3, exhibit a combination of the L452R mutation and E484Q [69]. The delta variant, known as B.1.617.2 and originating from India, presents an extensive array of mutations, with particular significance attributed to mutations compromising the S protein at T19R, L452R, E484Q, T478K, P681R, and D950N. Among these, E484Q and L452R are situated in the receptor binding domain (RBD) of the S protein, a notable reason behind its colloquial moniker as the “double mutant” (Fig. 1) within the Pango lineage [69, 70].
Rapid detection tests (RDTs)
In addition to RT-qPCR, the determination of antigens is employed through several biosensor systems. This method involves detecting either the SARS-CoV-2 virus protein or the antibodies that develop in a patient after being infected. These assays can be conducted using blood, plasma, or serum samples from infected individuals. This type of detection represents a rapid diagnostic technique that was initially proposed by the WHO in 1981. It’s considered accurate and crucial for immediate patient care [71]. Quick diagnostic methods encompass various approaches, including immunofluorescence (IF), immunochromatography (IC), and enzyme-linked immunosorbent assay (ELISA). All these techniques employ monoclonal antibodies directed against different viral antigens, enabling the detection of the virus within a short timeframe. Importantly, these methods can identify certain non-viable viruses present in the sample [17].
A notable advantage of utilizing rapid antigen detection tests (Ag-RDTs) is their ability to identify antigens from samples through a widely adopted immunochromatography system. These Ag-RDTs, designed for SARS-CoV-2 detection, consist of a nitrocellulose strip coated with immobilized anti-SARS-CoV-2 gold conjugate antibodies. To serve as a control, the membrane contains anti-chicken IgY monoclonal antibodies. In essence, Ag-RDTs directly identify SARS-CoV-2 antigens by recognizing the virus nucleocapsid proteins via the conjugated anti-SARS-CoV-2 gold antibody on the nitrocellulose membrane [72]. A noteworthy point is that most Ag-RDTs intended for SARS-CoV-2 detection necessitate nasopharyngeal swab samples [73].
Similarly to all diagnostic techniques, rapid diagnostic tests (RDTs) have their limitations. One of the primary challenges associated with immunological tests based on the detection of antibodies is their comparatively lower sensitivity and specificity. Notably, antibodies like IgM and IgG become detectable 7 to 49 days after viral infection, making it challenging to differentiate between an infected and a healthy individual [74]. Given the relatively lower sensitivity of Ag-RDTs in detecting positive cases of SARS-CoV-2, current research has concentrated on devising novel, cost-effective, and highly sensitive RDTs for virus detection. For instance, Huang et al. (2021) introduced an innovative nanoplasmonic biosensor integrated into a chip cartridge to swiftly detect SARS-CoV-2. This detection approach capitalizes on the surface plasmon resonance method to rapidly identify viral particles. The nanoplasmonic sensor chip is designed to recognize SARS-CoV-2 by facilitating interactions between immobilized SARS-CoV-2 monoclonal antibodies on the surface of the resonance sensor chips and the spike protein of the virus. This recognition prompts alterations in wavelengths or intensity in the resonance sensor, which can be quantified by an optical sensing system linked to a smartphone. The remarkable sensitivity and reliability of the nanoplasmonic sensor chips were established by evaluating various concentrations of pseudo-SARS-CoV-2 diluted in PBS, demonstrating successful detection of the virus at low concentrations (370 vp/ml) within a mere 15 min [75, 76]. An explanatory diagram of the rapid test procedure is presented in Fig. 3.
Immunoassay diagnostic test scheme. Rapid detection of SARS-CoV-2 with the recognition of IgG and IgM-type antibodies can be achieved using serological tests known as rapid antibody tests or rapid diagnostic tests (RDTs). RDTs include (A) blood, serum, or plasma sample collection. (B, C) Test procedure: the sample is applied to a test strip or cartridge containing specific antigens derived from SARS-CoV-2). (D) Antibody Binding: If the individual has been infected with SARS-CoV-2 and has developed an immune response, IgG and IgM antibodies will bind to the viral antigens on the test strip. (E) Results Interpretation: the test strip includes a visual indicator, such as colored lines or dots, that show the presence or absence of IgG and IgM antibodies. (F) The appearance of specific sequences or dots indicates a positive result for the corresponding antibody. Created with BioRender.com
Loop-mediated isothermal amplification (LAMP)
The Loop-Mediated Isothermal Amplification (LAMP) technique is a molecular testing method that revolves around amplifying specific genes using a DNA polymerase. This method employs primers designed to target six distinct sequences of viral DNA, and it operates under isothermal conditions [77, 78]. Presently, it’s feasible to combine the LAMP test with reverse transcription in a single reaction, forming the RT-LAMP method [3]. Notably, this approach has demonstrated its efficacy in detecting SARS-CoV-2 mRNA in samples collected from symptomatic patients. The RT-LAMP test exhibits superior sensitivity and specificity compared to RT-qPCR tests while being notably rapid and cost-effective [78]. Nonetheless, it’s crucial to evaluate its diagnostic accuracy in the context of a novel virus like SARS-CoV-2 and potential emerging variants.Researchers have conducted comparisons with the 2019-nCoV CDC USA kit (IDT, USA), which serves as the gold standard for SARS-CoV-2 detection through the RT-PCR technique [36, 79,80,81]. Results have indicated that the Isopollo kit demonstrates a 100% detection rate due to its broad specificity, although its overall sensitivity in comparison to the Center for Disease Control (CDC) protocol is 61.9%. This means that out of 168 positive samples for SARS-CoV-2, only 104 were correctly identified using the Isopollo kit [82], showcasing relatively lower effectiveness when contrasted with the traditional RT-PCR method. Nevertheless, it’s essential to acknowledge that whole genome sequencing methods stand as the benchmark for accurately identifying SARS-CoV-2 variants [71].
Metagenomic sequencing by nanopore technology (SMTN)
In 1989, the American biologist David Deamer introduced the concept of sequencing DNA molecules using nanopores. The fundamental principle behind this technique involves the passage of DNA molecules through nanopore channels, typically created using proteins and embedded in a matrix. As the DNA molecule traverses the nanopore, each nitrogenous base within it causes a disruption in the flow of ions, resulting in a reduction of electric current. The extent of this current reduction is determined by both the size of the molecule and the time it spends within the channel. Because the nitrogenous bases within DNA possess distinct chemical structures and molecular sizes, each of them triggers a unique alteration in the electric current as it moves through the nanopore. This distinct response enables the identification of the precise order or sequence in which these bases are arranged along the DNA chain. This revolutionary nanopore-based sequencing method has since gained considerable attention and is being continually refined for various applications in genomics and molecular biology [83, 84].
The Oxford Nanopore Company has developed and commercialized nanopore technology, offering various models to the biomedical market. One of their notable devices is the MinION, a portable platform designed for the execution and analysis of up to 96 genomes. These devices utilize a flow cell equipped with 512 nanopore channels, allowing for the real-time sequencing of DNA or RNA molecules. This technology enables researchers to directly observe the sequencing process as it happens, providing valuable insights into genetic information. For further details, please refer to Fig. 4 for a visual representation of the MinION device and its components.
Metagenomic sequencing by nanopore technology. Metagenomic sequencing by nanopore technology refers to applying nanopore sequencing platforms to analyze complex microbial communities in various samples. Types of equipment available with MiniOn technology, from highest to lowest sequencing capacity (MiniON, MiniON Mk1C, and GridiON), are illustrated. Created with BioRender.com
Summary of diagnostic methods for the identification of SARS-CoV-2. Most used methods for the effective diagnosis of SARS-CoV-2. The choice of diagnostic approach depends on the stage of infection, the purpose of testing, and the availability of resources. RT-PCR is primarily used for diagnosing active infection, while antibody tests are helpful for retrospective analysis, seroprevalence studies, and assessing immune response. Multiple testing approaches, including molecular and serological methods, may enhance diagnostic accuracy and provide a comprehensive understanding of SARS-CoV-2 infections
The advancement of nanopore sequencing technology, particularly through platforms like the GridION and PromethION, has enabled the identification and analysis of various respiratory viruses, including Zika, Ebola, Dengue, and SARS-CoV-2. By amplifying and sequencing specific gene fragments for each virus on the nanopore platform, researchers have been able to target virulence genes recommended by organizations like the CDC. The sequencing fragments typically range in size between 600 and 900 base pairs, allowing for accurate identification and analysis of these viruses [54, 85].
Clinical metagenomics, a field that originally used microarrays in the early 2000s, has evolved significantly due to the emergence of next-generation sequencing (NGS) technologies around 2005. However, the recent integration of third-generation sequencing, particularly nanopore technology, has taken clinical metagenomics to new heights. This approach allows for the targeted sequencing of specific gene sequences from a diverse group of etiological agents. It has found applications in clinical diagnosis, epidemiological surveillance, and establishing control measures against infectious pathogens [86,87,88,89,90,91].
Nanopore technology’s strengths lie in its ability to detect unique sequences rapidly and simultaneously from various respiratory virus gene fragments. It’s highly adaptable, user-friendly, and scalable, making it suitable for massive pathogen detection efforts. The sequencing results obtained using this technology contribute significantly to timely patient diagnosis, treatment, and epidemiological surveillance. Additionally, the ability to amplify and sequence fragments of varying sizes enables the identification of strains, variants, and the creation of epidemiological distribution maps. Furthermore, nanopore technology’s capacity to generate complete genomes of pathogens enhances its utility in various fields [54, 92]. This approach is a valuable tool in addressing public health challenges, particularly in the context of infectious disease outbreaks.
Genomic surveillance, particularly using advanced sequencing technologies like Illumina and nanopore sequencing, has become a central method for monitoring and understanding pandemics. This approach involves the systematic sequencing of the genomes of pathogens, such as viruses, to track their evolution, spread, and the emergence of new variants. It provides valuable insights into how diseases are spreading, mutating, and adapting over time.
SMTN has played a pivotal role in the prevention, control, and monitoring of new SARS-CoV-2 variants. Amid the COVID-19 outbreak, diverse researchers affiliated with hospitals and research laboratories employed MinION nanopore sequencing to procure high-quality genomes from COVID-19-positive patient samples [84, 93]. The speed and reliability of nanopore technology in capturing specific gene sequences of SARS-CoV-2 have facilitated the identification of a considerable number of SARS-CoV-2 mutations. Notably, this includes those linked to SARS-CoV-2 VOCs with elevated transmissibility and virulence. This advancement enables swift tracking and early detection of numerous SARS-CoV-2 variants circulating globally [84, 92, 94].
Molecular diagnostic test selection criteria
Several crucial criteria dictate the selection of the most suitable diagnostic test for each patient (Fig. 5). Key considerations include whether the patient exhibits symptoms and, if so, the duration since symptom onset [95, 96]. These factors play a pivotal role in guiding the test choice. Antigen testing might not yield optimal results if more than ten days have transpired since symptom initiation [97, 98]. In cases where patients necessitate urgent medical attention due to their symptoms, rapid point-of-care testing should be promptly employed to expedite patient triage.
Selecting a diagnostic test involves careful consideration of specific questions and criteria. These encompass clinical and infection control factors, such as the urgency of obtaining results and the acceptable turnaround time from test to result. The testing environment and sample collection location are also crucial aspects, along with the feasibility of individuals quarantining while awaiting results. Resource availability is another critical facet: Which diagnostic tools are at hand, and how are they prioritized? What testing capacity is feasible in each context? Is sample quality verified, or will swabbing be supervised? Lastly, the prevalence of infection must be factored in when making the test selection [99].
Developing diagnostic tests for individuals with symptoms is crucial in containing disease transmission [100]. The severity of a patient’s symptoms should guide test selection. For severely ill patients requiring hospitalization, rapid diagnostic tests like antigen tests are recommended for swift identification, facilitating timely health interventions [101]. Real-time PCR results yield cycle threshold (CT) values. Lower CT values [12,13,14,15, 17,18,19] suggest high viral loads, potentially indicating a greater likelihood of severe symptoms and future hospitalization [102].
Hospitalized patients necessitate diagnostic testing to prevent infections and transmission within healthcare facilities, enabling isolation until results are available [103]. Patients who have recovered from COVID-19 must undergo PCR diagnostic testing and remain isolated until discharge, emphasizing the importance of these measures now and in the future [99]. When selecting a test, additional considerations include sampling time, sample storage conditions, potential contamination, use of unvalidated assays, low viral load due to disease stage, and viral gene recombination or mutation [104,105,106].
Pathophysiological consequences of COVID-19 disease
Patients infected with SARS-CoV-2 typically exhibit initial flu-like symptoms such as fever, cough, nasal congestion, and fatigue [107, 108]. As the viral infection progresses, patients may encounter dyspnea and persistent symptoms of viral pneumonitis, including lowered oxygen saturation, lymphopenia, and chest imaging revealing ground-glass opacities and alveolar exudates with intralobular engagement [107, 108]. In more severe cases, patients can develop acute respiratory distress syndrome (ARDS), a severe condition of acute lung injury [109, 110].
SARS-CoV-2 is well known to infect human cells through a process called receptor recognition, wherein the virus’s spike protein binds to the ACE2 receptor present on the surface of human cells. This interaction allows the virus to enter the cell’s internal compartment, where it can replicate and propagate the infection to other cells within the human body [9, 111, 112]. As a result of this cellular invasion and infection, the body triggers an inflammatory response involving both innate and adaptive immune mechanisms [113,114,115]. According to data released by the World Health Organization (WHO), it is estimated that out of all patients diagnosed with COVID-19, around 20% will develop a severe clinical condition [12]. This heightened severity could be attributed to the fact that the SARS-CoV-2 virus predominantly infects the lower airway epithelial pulmonary cells, particularly alveolar pneumocytes of both type I and type II. These cells are characterized by a higher expression level of ACE2, which serves as the primary entry point for SARS-CoV-2 to infect the host cell [50].
It’s worth noting that reports from hospitalized COVID-19 patients have revealed a potential explanation for the progression to a severe clinical profile. SARS-CoV-2 seems to have developed a strategy to evade the host immune response by expressing various proteins that interfere with the interferons (IFNs) signaling pathway and IFN-stimulated genes [116, 117]. The IFNs signaling pathway is crucial for initiating cellular and molecular events that inhibit viral replication and activate the adaptive immune response. However, lower levels of IFNs are observed in the lungs and peripheral blood circulation, which leads to uncontrolled replication and spread of SARS-CoV-2. This reduction in IFNs contributes to an intense inflammatory response characterized by the abnormal release of proinflammatory cytokines, including interleukin-1 (IL-1), interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α), a phenomenon often referred to as a “cytokine storm.“ This pro-inflammatory environment results in the influx of monocytes, neutrophils, and the activation of T cells, which collectively contribute to damage in the alveolar tissue [117,118,119]. From a pathophysiological perspective, the uncontrolled levels of pro-inflammatory cytokines lead to vascular dysfunction. This dysfunction triggers the activation of coagulation factors and a decrease in the expression of activator plasminogen-1 and thrombomodulin. This disruption in the vasculature’s integrity contributes to disseminated intravascular coagulation, which has widespread effects and can lead to dysfunction in various organs, including the heart, lungs, liver, and brain [120,121,122,123,124].
Alongside the respiratory effects, individuals with severe COVID-19 may also exhibit gastrointestinal symptoms, including diarrhea, nausea, and vomiting [125]. Another notable manifestation is the loss of taste and smell (anosmia and ageusia) [126]. Consequently, the severe clinical presentation of COVID-19 patients can give rise to complications such as widespread inflammation and injury to the alveolar epithelium, resulting in compromised oxygen uptake and impaired elimination of carbon dioxide. Moreover, individuals with severe COVID-19 can progress to conditions like pneumonia, acute respiratory distress syndrome (ARDS), and respiratory failure.
Apart from the pathophysiological consequences of SARS-CoV-2 infection, post-COVID-19 sequelae have been observed in patients who have recovered from the disease. The potential role of various treatments, including physical training, has been investigated. However, exercise regimens for COVID-19 patients may need to be tailored, considering that some individuals exhibit muscular and respiratory issues both at baseline and during exercise due to the effects of SARS-CoV-2 infection [15]. Recent systematic reviews have highlighted the importance of early rehabilitation for COVID-19 patients recovering from severe respiratory failure [127]. Recommendations for those with mild sequelae stemming from COVID-19 include neuromotor rehabilitation, home-based rehabilitation, and telerehabilitation, along with light physical activities such as yoga and tai chi [128]. or outpatient management of mild cases, approaches like pulmonary rehabilitation, education, airway clearance techniques, physical exercises, and breathing exercises can be considered [9, 15]. Activities like yoga and tai chi, which involve coordinated postural movements with controlled breathing, have been suggested [129]. Recent findings indicate that exercise training interventions, particularly light exercise, can significantly improve the mortality rate among COVID-19 patients with hypertension (HTN). This suggests that incorporating light exercise could be a primary approach for managing COVID-19 in patients with HTN [15].
On the contrary, while many individuals experience mild to moderate symptoms and recover within a few weeks, a subset of patients continues to endure persistent symptoms for an extended duration, a condition commonly termed as “Long COVID” or “post-acute sequelae of SARS-CoV-2 infection” [130]. Long COVID encompasses an array of symptoms affecting various organs and systems, including the respiratory, cardiovascular, neurological, and musculoskeletal systems [131]. The precise pathophysiology of Long COVID remains incompletely understood but is known to be multifactorial. Direct viral damage from SARS-CoV-2, immune dysregulation, and persistent inflammation are believed to be major contributors to the development of Long COVID [132]. SARS-CoV-2’s affinity for ACE2 receptors in multiple tissues enables it to directly infect and harm cells. This can lead to conditions such as pulmonary fibrosis and compromised gas exchange in the lungs. Within the cardiovascular system, it might result in myocardial inflammation and endothelial dysfunction [133]. Neurological manifestations such as brain fog, headaches, and neuropathies could arise from the virus’s direct invasion or immune-mediated responses. Additionally, disordered immune reactions are suspected to contribute to the pathology of Long COVID. Prolonged viral presence and continuous immune activation may lead to a state of chronic inflammation, resulting in tissue damage and dysfunction [134]. Autoimmune responses triggered by the initial infection or molecular mimicry might also play a role in the persistent symptoms observed in Long COVID patients [135].
Long COVID symptoms exhibit a wide range and can endure for several months following the acute phase of infection. Common presentations encompass lingering fatigue, dyspnea, chest discomfort, cognitive impairment, and musculoskeletal issues. Continued respiratory symptoms like cough and shortness of breath can persist due to factors such as lung fibrosis, heightened airway reactivity, or damage to respiratory muscles [135,136,137]. Notably, Long COVID involves the cardiovascular system, as patients often encounter symptoms such as palpitations, chest pain, and reduced exercise tolerance [138]. Potential mechanisms underlying this cardiac involvement include myocardial inflammation, microvascular dysfunction, and disturbances in autonomic regulation [139]. Neurological manifestations, often referred to as “brain fog,“ encompass cognitive deficits, memory challenges, headaches, dizziness, and mood disorders. These symptoms may result from factors such as neuroinflammation, microvascular dysfunction, and disrupted immune responses [140]. Frequently reported musculoskeletal symptoms comprise joint pain, myalgia, and muscle weakness. These issues can be attributed to persistent systemic inflammation, muscle damage, and physical deconditioning [135].
In sum, the emergence of Long COVID can be attributed to a variety of mechanisms. These encompass viral persistence, disruptions in immune function, endothelial dysfunction, and imbalanced cytokine responses [135]. Additionally, host-related factors like pre-existing health conditions, age, gender, and genetic susceptibility might impact the probability and intensity of experiencing Long COVID. Gaining a comprehensive understanding of the pathogenesis and fundamental mechanisms of Long COVID is pivotal for enhancing patient care, devising effective management strategies, and exploring potential interventions.
Conclusions
Similar to many viral agents, SARS-CoV-2 has undergone ongoing evolutionary changes since the onset of the COVID-19 pandemic. The World Health Organization (WHO) has classified several variants of concern (VOCs) and variants of interest based on their potential to spread and replace older variants, trigger new waves of infections, and necessitate adjustments in public health strategies. As of February 2022, the Omicron variant constitutes over 98% of publicly available viral sequences. This variant serves as the genetic foundation from which novel SARS-CoV-2 variants are likely to emerge. These emerging variants could originate from previous VOCs or even entirely new mutations. In comparison to earlier variants, the Omicron variant has continued to evolve genetically and antigenically. It has given rise to various sublineages, characterized by their ability to evade pre-existing immunity within the population and a tendency to primarily infect the upper respiratory tract instead of the lower respiratory tract. The significance of developing effective diagnostic techniques cannot be understated. Such efforts directly contribute to informed decision-making and the formulation of sound public health policies.
Data Availability
No data were used to support this study.
Change history
13 October 2023
The city name in the second affiliation is corrected from “Manila” to “Iquique”.
Abbreviations
- COVID-19:
-
Coronavirus disease 2019
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- SARS-CoV-1:
-
Severe acute respiratory syndrome coronavirus 1
- MERS-CoV:
-
Middle East respiratory syndrome
- PCR:
-
Polymerase chain reaction
- DNA:
-
Deoxyribonucleic acid
- RNA:
-
Ribonucleic acid
- WHO:
-
World Health Organization
- RaTG13:
-
Bat coronavirus
- ORFs:
-
Open reading frames
- +ssRNA:
-
Single-stranded RNA of positive polarity
- nsp3:
-
Nonstructural protein 3
- SMTN:
-
Nanopore metagenomic sequencing
- NGS:
-
Next-generation sequencing
- Spike protein:
-
Protein S
- Envelope protein:
-
Protein E
- Membrane protein:
-
Protein M
- Nucleocapsid protein:
-
Protein N
- nsp3:
-
Nonstructural protein 3
- ACE2:
-
Angiotensin-converting enzyme 2
- TMPRSS2:
-
Transmembrane protease serine 2
- NPS:
-
Nasopharyngeal swabs
- OPS:
-
Oropharyngeal swabs
- RT-qPCR:
-
Quantitative reverse transcription polymerase chain reaction
- BAC:
-
Active case search
- RdRp:
-
RNA-dependent RNA Polymerase gene
- RBD:
-
Receptor binding domain
- RDTs:
-
Rapid detection tests
- IF:
-
Immunofluorescence
- IC:
-
Immunochromatography
- EIA:
-
Enzyme-linked Immunoassay
- Ag-RDTs:
-
Rapid antigen detection tests
- LAMP:
-
Loop-mediated isothermal amplification
- CDC:
-
Center for Disease Control
- Ct:
-
Cycle threshold
- ARDS:
-
Acute respiratory distress syndrome
- INFs:
-
Interferons
- IL-1:
-
Interleukin- 1
- IL-6:
-
Interleukin-6
- TNF-α:
-
Tumor factor necrosisα
- EX:
-
Exercise training
- HTN:
-
Hypertension
- VOCs:
-
Variants of concern
References
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y et al (2020) Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 382(13):1199–1207
Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K et al (2020) Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol 92(5):491–494
Huang WE, Lim B, Hsu CC, Xiong D, Wu W, Yu Y et al (2020) RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol 13(4):950–961
Pagani I, Ghezzi S, Alberti S, Poli G, Vicenzi E (2023) Origin and evolution of SARS-CoV-2. Eur Phys J Plus 138(2):157
Domingo JL (2022) An updated review of the scientific literature on the origin of SARS-CoV-2. Environ Res 215(Pt 1):114131
Dong E, Du H, Gardner L (2020) An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20(5):533–534
Wu JT, Leung K, Leung GM (2020) Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 395(10225):689–697
Xu S, Li Y (2020) Beware of the second wave of COVID-19. Lancet 395(10233):1321–1322
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273
Rai NK, Ashok A, Akondi BR (2020) Consequences of chemical impact of disinfectants: safe preventive measures against COVID-19. Crit Rev Toxicol 50(6):513–520
Ren LL, Wang YM, Wu ZQ, Xiang ZC, Guo L, Xu T et al (2020) Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) 133(9):1015–1024
WHO. Laboratory biosafety guidance related to coronavirus disease 2019 (COVID-19): interim guidance, 24 March 2022 2022 [World Health Organization. Retrieved from https://apps.who.int/iris/handle/10665/340374.]
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ et al (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res 7(1):11
Colizzi M, Bortoletto R, Silvestri M, Mondini F, Puttini E, Cainelli C et al (2020) Medically unexplained symptoms in the times of COVID-19 pandemic: a case-report. Brain Behav Immun Health 5:100073
Fernandez F, Vazquez-Muñoz M, Canals A, Arce-Álvarez A, Salazar-Ardiles C, Alvarez CR-C, Rodrigo et al (2023) Intrahospital supervised exercise training improves survival rate among hypertensive patients with COVID-19. J Appl Physiol J Appl Physiol 134(3):678–684
WHO. Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update (2023) [Available from: https://www.who.int/emergencies/disease/novel-coronavirus-2019/situation-reports/
Calvo RC, García M, C F B. P. Infecciones respiratorias virales 2020 [Available from: https://www.aeped.es/sites/default/files/documentos/irsv.pdf
Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W et al (2018) Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556(7700):255–258
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J et al (2016) Epidemiology, genetic recombination, and pathogenesis of Coronaviruses. Trends Microbiol 24(6):490–502
Cohen J (2020) Mining coronavirus genomes for clues to the outbreak’s origins. Science, New York, NY
Gerhards NM, Gonzales JL, Vreman S, Ravesloot L, van den Brand JMA, Doekes HP et al (2023) Efficient direct and limited environmental transmission of SARS-CoV-2 Lineage B.1.22 in domestic cats. Microbiol Spectr 11(3):e0255322
Health EPA, Welfare, Nielsen SS, Alvarez J, Bicout DJ, Calistri P et al (2023) SARS-CoV-2 in animals: susceptibility of animal species, risk for animal and public health, monitoring, prevention and control. EFSA J 21(2):e07822
USDA. Confirmed Cases of SARS-CoV-2 in Animals in the United States 2023 [Available from: https://www.aphis.usda.gov/aphis/dashboards/tableau/sars-dashboard
Rambaut A, Holmes EC, O’Toole A, Hill V, McCrone JT, Ruis C et al (2020) A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5(11):1403–1407
Xiao T, Lu J, Zhang J, Johnson RI, McKay LGA, Storm N et al (2021) A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat Struct Mol Biol 28(2):202–209
Yaqing HE (2004) Surveillance of SARS coronavirus among wild animal sold in Dongmen market in Shenzhen city. Disease Surveillance. 19(8):287–91
Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL et al (2003) Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302(5643):276–278
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W et al (2020) Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182(4):812–27e19
CDC. SARS-CoV-2 variant classifications and definitions 2023 [Available from: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html#anchor_1632158885160
Li LL, Wang JL, Ma XH, Sun XM, Li JS, Yang XF et al (2021) A novel SARS-CoV-2 related coronavirus with complex recombination isolated from bats in Yunnan province, China. Emerg Microbes Infect 10(1):1683–1690
Lytras S, Hughes J, Martin D, Swanepoel P, de Klerk A, Lourens R et al (2022) Exploring the Natural Origins of SARS-CoV-2 in the light of recombination. Genome Biol Evol. 14(2)
Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC et al (2020) Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285
Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ et al (2020) Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583(7815):286–289
Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R (2010) Viral mutation rates. J Virol 84(19):9733–9748
Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46(D1):D708–D17
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574
Mossel EC, Wang J, Jeffers S, Edeen KE, Wang S, Cosgrove GP et al (2008) SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology 372(1):127–135
Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A et al (2020) Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim et Biophys Acta Mol Basis Disease. 1866(10)
Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77(16):8801–8811
Knipe DM, Howley P (2013) Fields Virology (6a ed.). Wilkins LW, editor
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O et al (2020) Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. bioRxiv.
Schoeman D, Fielding BC (2019) Coronavirus envelope protein: current knowledge. Virol J 16(1):69
Schoeman D, Fielding BC (2020) Is there a Link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front Microbiol 11:2089
de Wit E, Neeltje. vD, Falzarano D, Munster VJ (2016) SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 18(8)
Cui J, Wang L-F (2015) Genomic mining reveals deep evolutionary relationships between bornaviruses and bats. Viruses 7(11):5792–5800
Oliva J (2020) The receptor binding domain of the spike protein of SARS-CoV-2: a critical target for vaccines and therapeutic antibodies. Front Immunol 11:2190
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods in molecular biology. (Clifton NJ) 1282:1–23
Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS (2010) An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol 84(19):10276–10288
Wang J, Ji J, Ye J, Zhao X, Wen J, Li W et al (2003) The structure analysis and antigenicity study of the N protein of SARS-CoV. Genomics Proteom Bioinf 1(2):145–154
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23(1):3–20
Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5(4):562–569
Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M (2020) Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369(6501):330–333
Piras A, Rizzo D, Longoni E, Turra N, Urru S, Saba PP et al (2020) Nasopharyngeal swab collection in the suspicion of Covid-19. Am J Otolaryngol 41(5):102551
Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z et al (2020) Nanopore targeted sequencing for the Accurate and Comprehensive Detection of SARS-CoV-2 and other respiratory viruses. Small 16(32):e2002169
Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P et al (2020) Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. bioRxiv.
Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A et al (2020) Saliva is a reliable tool to detect SARS-CoV-2. J Infect 81(1):e45–e50
Delaunay-Moisan A, Guilleminot T, Semeraro M, Briand N, Bader-Meunier B, Berthaud R et al (2022) Saliva for molecular detection of SARS-CoV-2 in pre-school and school-age children. Environ Microbiol 24(10):4725–4737
Lai J, German J, Hong F, Tai SS, McPhaul KM, Milton DK et al (2022) Comparison of saliva and midturbinate swabs for detection of SARS-CoV-2. Microbiol Spectr 10(2):e0012822
Butler-Laporte G, Lawandi A, Schiller I, Yao M, Dendukuri N, McDonald EG et al (2021) Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and Meta-analysis. JAMA Intern Med 181(3):353–360
WHO. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 19 March 2020: World Health Organization (2020); [Available from: https://apps.who.int/iris/handle/10665/331329
Jayamohan H, Lambert CJ, Sant HJ, Jafek A, Patel D, Feng H et al (2021) SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal Bioanal Chem 413(1):49–71
Minsal Reporte Diario Covid-19 2023 [Available from: from https://www.gob.cl/coronavirus/cifrasoficiales/
Yu F, Du L, Ojcius DM, Pan C, Jiang S (2020) Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect 22(2):74–79
Walensky RP, Walke HT, Fauci AS (2021) SARS-CoV-2 variants of concern in the United States-Challenges and Opportunities. JAMA 325(11):1037–1038
MWGR. on SARS-CoV-2 variants and implications for global COVID-19 vaccination programs Multidisciplinary Working Group Report 2020 [Available from: https://www.ciencia.gob.es/site-web/Ministerio/Coronavirus/informes-cientificos.html
Rambaut A, Loman. N, Pybus O, Barclay W, Barrett J, Carabelli A et al (2020) Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.
Mascola JR, Graham BS, Fauci AS (2021) SARS-CoV-2 viral Variants-Tackling a moving target. JAMA 325(13):1261–1262
Meng B, Kemp SA, Papa G, Datir R, Ferreira I, Marelli S et al (2021) Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the alpha variant B.1.1.7. Cell Rep 35(13):109292
Matic N, Lowe CF, Ritchie G, Stefanovic A, Lawson T, Jang W et al (2021) Rapid Detection of SARS-CoV-2 variants of concern, including B.1.1.28/P.1, British Columbia, Canada. Emerg Infect Dis 27(6):1673–1676
Salimović-Bešić I, Bihorac A, Jusufović E, Marić S (2022) SARS-CoV-2 Delta variant B.1.617.2-Double mutation. Acta Informatica Medica 30(1):57–60
WHO. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays: Interim guidance World Health Organization2021 [Available from: https://apps.who.int/iris/handle/10665/334254
Vashist SK (2020) In Vitro Diagnostic assays for COVID-19: recent advances and emerging Trends. Diagnostics (Basel). 10(4)
Gestoso-Pecellín L, García-Flores Y, González-Quintana P, Marrero-Arencibia JL (2021) Recomendaciones y uso de los diferentes tipos de test para detección de infección por SARS-COV-2. Enfermeria Clin 31(1):40–48
Datta M, Singh DD, Naqvi AR (2021) Molecular Diagnostic Tools for the detection of SARS-CoV-2. Int Rev Immunol 40(1–2):143–156
Dang T, Hu W, Zhang W, Song Z, Wang Y, Chen M et al (2019) Protein binding kinetics quantification via coupled plasmonic-photonic resonance nanosensors in generic microplate reader. Biosens Bioelectron 142:111494
Huang L, Ding L, Zhou J, Chen S, Chen F, Zhao C et al (2021) One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens Bioelectron 171:112685
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N et al (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):E63
Kashir J, Yaqinuddin A (2020) Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med Hypotheses. 141(109786)
CDC. Answers to frequently asked questions». Centers for Disease Control and Prevention Centres Disease Control and Prevention2020 [Available from:. https://www.cdc.gov/coronavirus/2019-nCoV/index.html
Rhoads DD, Wrona A, Foutz A, Blevins J, Glisic K, Person M et al (2020) Diagnosis of prion diseases by RT-QuIC results in improved surveillance. Neurology 95(8):e1017–e26
Nalla AK, Casto AM, Huang MW, Perchetti GA, Sampoleo R, Shrestha L et al (2020) Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol. 58(6)
Freire-Paspuel B, Garcia-Bereguiain MA (2021) Low clinical performance of the Isopollo COVID-19 detection kit (M monitor, South Korea) for RT-LAMP SARS-CoV-2 diagnosis: a call for action against low quality products for developing countries. Int J Infect Dis 104:303–305
Deamer DW, Akeson M (2000) Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol 18(4):147–151
Chen P, Sun Z, Wang J, Liu X, Bai Y, Chen J et al (2023) Portable nanopore-sequencing technology: Trends in development and applications. Front Microbiol 14:1043967
ONT. Choose nanopore sequencing Oxford nanopores technology 2021 [Available from: https://nanoporetech.com/products
Miller MB, Tang YW (2009) Basic concepts of microarrays and potential applications in clinical microbiology. Clin Microbiol Rev 22(4):611–633
Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP et al (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300(5624):1394–1399
Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360(8):790–800
Arroyo Muhr LS, Dillner J, Ure AE, Sundstrom K, Hultin E (2021) Comparison of DNA and RNA sequencing of total nucleic acids from human cervix for metagenomics. Sci Rep 11(1):18852
Voelkerding KV, Dames SA, Durtschi JD (2009) Next-generation sequencing: from basic research to diagnostics. Clin Chem 55(4):641–658
Wooley JC, Ye Y, Metagenomics (2009) Facts and artifacts, and computational Challenges*. J Comput Sci Technol 25(1):71–81
Zheng P, Zhou C, Ding Y, Liu B, Lu L, Zhu F et al (2020) Nanopore sequencing technology and its applications. MedComm 2023;4(4):e316
Anwar MZ, Lodhi MS, Khan MT, Khan MI, Sharif S (2022) Coronavirus genomes and unique mutations in Structural and non-structural proteins in pakistani SARS-CoV-2 Delta Variants during the Fourth Wave of the pandemic. Genes (Basel). 13(3)
Gauthier NPG, Nelson C, Bonsall MB, Locher K, Charles M, MacDonald C et al (2021) Nanopore metagenomic sequencing for detection and characterization of SARS-CoV-2 in clinical samples. PLoS ONE 16(11):e0259712
CDC, Testing for COVID-19 Centers for Disease Control and Prevention2022 [Available from: https://www.cdc.gov/coronavirus/2019-ncov/testing/index.html
Arevalo-Rodriguez I, Seron P, Buitrago-Garcia D, Ciapponi A, Muriel A, Zambrano-Achig P et al (2021) Recommendations for SARS-CoV-2/COVID-19 testing: a scoping review of current guidance. BMJ Open 11(1):e043004
Mallett S, Allen AJ, Graziadio S, Taylor SA, Sakai NS, Green K et al (2020) At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med 18(1):346
Engelmann I, Alidjinou EK, Ogiez J, Pagneux Q, Miloudi S, Benhalima I et al (2021) Preanalytical issues and cycle threshold values in SARS-CoV-2 real-time RT-PCR testing: should test results include these? ACS Omega 6(10):6528–6536
Baldanti F, Ganguly NK, Wang G, Mockel M, O’Neill LA, Renz H et al (2022) Choice of SARS-CoV-2 diagnostic test: challenges and key considerations for the future. Crit Rev Clin Lab Sci 59(7):445–459
Singanayagam A, Hakki S, Dunning J, Madon KJ, Crone MA, Koycheva A et al (2022) Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis 22(2):183–195
Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S et al (2020) Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 8(8):CD013705
Shah VP, Farah WH, Hill JC, Hassett LC, Binnicker MJ, Yao JD et al (2021) Association between SARS-CoV-2 cycle threshold values and clinical outcomes in patients with COVID-19: a systematic review and meta-analysis. Open Forum Infectious Diseases 8(9):ofab453
Möckel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC et al (2021) SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 26(3):213–220
Mitchell SL, St George K, Rhoads DD, Butler-Wu SM, Dharmarha V, McNult P et al (2020) Understanding, verifying, and Implementing Emergency Use Authorization Molecular Diagnostics for the detection of SARS-CoV-2 RNA. J Clin Microbiol. 58(8)
Erbak Yilmaz H, Iscan E, Oz O, Batur T, Erdogan A, Kilic S et al (2022) Considerations for the selection of tests for SARS-CoV-2 molecular diagnostics. Mol Biol Rep 49(10):9725–9735
Lippi G, Simundic AM, Plebani M (2020) Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med 58(7):1070–1076
Velavan TP, Meyer CG (2020) Mild versus severe COVID-19: Laboratory markers. Int J Infect Dis 95:304–307
Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y et al (2005) Multiple organ infection and the pathogenesis of SARS. J Exp Med 202(3):415–424
Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A et al (2019) Acute respiratory distress syndrome. Nat Rev Dis Primers 5(1):18
Meyer NJ, Gattinoni L, Calfee CS (2021) Acute respiratory distress syndrome. Lancet 398(10300):622–637
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271–80e8
Bal A, Destras G, Gaymard A, Bouscambert-Duchamp M, Valette M, Escuret V et al (2020) Molecular characterization of SARS-CoV-2 in the first COVID-19 cluster in France reveals an amino acid deletion in nsp2 (Asp268del). Clin Microbiol Infect 26(7):960–962
Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H et al (2020) Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 130(5):2620–2629
Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y et al (2020) Comparative replication and Immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an Ex vivo study with implications for the Pathogenesis of COVID-19. Clin Infect Dis 71(6):1400–1409
Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M et al (2020) Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584(7821):463–469
Iwasaki A, Yang Y (2020) The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 20(6):339–341
Nile SH, Nile A, Qiu J, Li L, Jia X, Kai G (2020) COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev 53:66–70
Biying Z, Rong L, Ming Z, Jingyu H, Qun Y, Rui W et al (2020) The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int J Antimicrob Agents 55(5):105954
Merad M, Blish CA, Sallusto F, Iwasaki A (2022) The immunology and immunopathology of COVID-19. Science 375(6585):1122–1127
Siddiqi HK, Libby P, Ridker PM (2021) COVID-19 - a vascular disease. Trends Cardiovasc Med 31(1):1–5
Lang M, Som A, Carey D, Reid N, Mendoza DP, Flores EJ et al (2020) Pulmonary vascular manifestations of COVID-19 pneumonia. Radiol Cardiothorac Imaging 2(3):e200277
Wichmann D, Sperhake JP, Lutgehetmann M, Steurer S, Edler C, Heinemann A et al (2020) Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med 173(4):268–277
Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC (2020) Pathophysiology, transmission, diagnosis, and treatment of Coronavirus Disease 2019 (COVID-19): a review. JAMA 324(8):782–793
Yao XH, Luo T, Shi Y, He ZC, Tang R, Zhang PP et al (2021) A cohort autopsy study defines COVID-19 systemic pathogenesis. Cell Res 31(8):836–846
Pan F, Ye T, Sun P, Gui S, Liang B, Li L et al (2020) Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology 295(3):715–721
Mao L, Jin H, Wang M, Hu Y, Chen S, He Q et al (2020) Neurologic manifestations of hospitalized patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol 77(6):683–690
Negrini F, de Sire A, Andrenelli E, Lazzarini SG, Patrini M, Ceravolo MG et al (2021) Rehabilitation and COVID-19: a rapid living systematic review 2020 by Cochrane Rehabilitation Field. Update as of October 31st, 2020. Eur J Phys Rehabil Med 57(1):166–170
de Sire A, Andrenelli E, Negrini F, Lazzarini SG, Patrini M, Ceravolo MG et al (2020) Rehabilitation and COVID-19: the Cochrane Rehabilitation 2020 rapid living systematic review. Update as of August 31st, 2020. Eur J Phys Rehabil Med 56(6):839–845
Ngai SP, Jones AY, Tam WW (2016) Tai Chi for chronic obstructive pulmonary disease (COPD). Cochrane Database Syst Rev 2016(6):CD009953
Proal AD, VanElzakker MB (2021) Long COVID or post-acute sequelae of COVID-19 (PASC): an overview of biological factors that may contribute to persistent symptoms. Front Microbiol 12:698169
Raman B, Bluemke DA, Lüscher TF, Neubauer S (2022) Long COVID: post-acute sequelae of COVID-19 with a cardiovascular focus. Eur Heart J 43(11):1157–1172
Kopanska M, Barnas E, Blajda J, Kuduk B, Lagowska A, Banas-Zabczyk A (2022) Effects of SARS-CoV-2 inflammation on selected Organ Systems of the human body. Int J Mol Sci. 23(8)
Dhakal BP, Sweitzer NK, Indik JH, Acharya D, William P (2020) SARS-CoV-2 infection and Cardiovascular Disease: COVID-19 Heart. Heart Lung Circ 29(7):973–987
Peluso MJ, Lu S, Tang AF, Durstenfeld MS, Ho HE, Goldberg SA et al (2021) Markers of Immune activation and inflammation in individuals with Postacute sequelae of severe Acute Respiratory Syndrome Coronavirus 2 infection. J Infect Dis 224(11):1839–1848
Davis HE, McCorkell L, Vogel JM, Topol EJ (2023) Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol 21(3):133–146
Ceban F, Ling S, Lui LMW, Lee Y, Gill H, Teopiz KM et al (2022) Fatigue and cognitive impairment in Post-COVID-19 syndrome: a systematic review and meta-analysis. Brain Behav Immun 101:93–135
Swank Z, Senussi Y, Manickas-Hill Z, Yu XG, Li JZ, Alter G et al (2023) Persistent circulating severe acute respiratory syndrome coronavirus 2 spike is associated with post-acute coronavirus disease 2019 sequelae. Clin Infect Diseases: Official Publication Infect Dis Soc Am 76(3):e487–e90
Xie Y, Xu E, Bowe B, Al-Aly Z (2022) Long-term cardiovascular outcomes of COVID-19. Nat Med 28(3):583–590
Spudich S, Nath A (2022) Nervous system consequences of COVID-19. Science 375(6578):267–269
Holdsworth DA, Chamley R, Barker-Davies R, O’Sullivan O, Ladlow P, Mitchell JL et al (2022) Comprehensive clinical assessment identifies specific neurocognitive deficits in working-age patients with long-COVID. PLoS ONE 17(6):e0267392
Acknowledgements
This work is dedicated to my dear professor Dr. Jorge E. Araya Rojas, with whom we worked together during the pandemic on projects dedicated to the genomic sequencing of SARS-CoV-2, and from there, the idea of this review was born. Unfortunately, he passed away during the development of this work. C.T. was supported by the Agencia Nacional de Investigación y Desarrollo (ANID) through Fondeyt de Iniciación # 11220962.
Funding
This study was supported by the Agencia Nacional de Investigación y Desarrollo (ANID) through Anillo ACT210083. DCA was funded by grant Fondeyt de Iniciación #11220870 and Minera Escondida Ltda. MEL2203.
Author information
Authors and Affiliations
Contributions
C.S-A. contributed to the draft, preparation of the manuscript, and the concept of the project. L.S.R.; C.C-G; D.A.; M.V-M.; C.T.; and D.C.A. contributed to the preparation of the manuscript. D.C.A., contributed to the concept of the project. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salazar-Ardiles, C., Asserella-Rebollo, L., Cornejo, C. et al. Molecular diagnostic approaches for SARS-CoV-2 detection and pathophysiological consequences. Mol Biol Rep 50, 10367–10382 (2023). https://doi.org/10.1007/s11033-023-08844-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08844-0