Abstract
Introduction
Nosocomial infections (NIs) are a major challenge worldwide. Identification of antibiotic resistance pattern extended spectrum beta-lactamases (ESBLs) and carbapenem-resistant Enterobacteriaceae (CRE) were the objectives of this study.
Methods
In this cross-sectional study, the antimicrobial susceptibility pattern of bacterial isolates collected from patients with NIs in ICU was determined. Overall, 42 Escherichia coli and Klebsiella pneumoniae isolates from different infection sites were used to determine phenotypic tests of ESBLs, Metallo-β-lactamases (MBLs) and CRE. Detection of ESBLs, MBLs and CRE genes were performed by the polymerase chain reaction (PCR) method.
Results
From 71 patients with NIs, 103 different bacterial strains were isolated. The most frequently isolated bacteria were E. coli (n = 29; 28.16%), Acinetobacter baumannii (n = 15; 14.56%), and K. pneumoniae (n = 13; 12.26%). Also, the rate of multidrug-resistant (MDR) isolates was 58.25% (60/103). Based on phenotypic confirmation tests, 32 (76.19%) isolates of E. coli and K. pneumoniae produced ESBLs, and 6 (14.28%) isolates were identified as CRE producers. PCR showed the high prevalence of the blaCTX-M (n = 29; 90.62%) in ESBL genes. In addition, blaNDM was detected in 4 (66.66%), blaOXA-23 in 3 (50%), and blaOXA-48 gene in 1 (16.66%) isolates. The blaVIM, blaKPC, and blaIMP genes were not detected in any of the isolates.
Conclusion
The Gram-negative bacteria E. coli, A. baumannii, and K. pneumoniae with high resistance levels were the most common bacteria causing NIs in the ICU. This study for the first time identified blaOXA-11, blaOXA-23, and blaNDM-1 genes in E. coli and K. pneumoniae in Ilam city of Iran.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nosocomial infections (NIs) are one of the most significant concerns of medical centers worldwide [1,2,3]. About 80% of the NIs are in the form of nosocomial urinary tract infections (UTIs), nosocomial bloodstream infection (NBSIs), nosocomial surgical site infections (SSIs), and nosocomial pneumonia infection (NPNEU) [4]. The increasing rate of NIs causes more drugs usage, accordingly leading to economic burdens. In spite of global control efforts, NIs still remains a prevalent issue and a cause of antibiotic resistance. The rapid spread of this resistance and its dissemination and burden has become an important health problem throughout the world [5]. According to the Centers for Disease Control and Prevention (CDC) reports, more than 70% of the bacterial agents of NIs are resistant to at least one of the medications used [6].These infections occur 48–72 h after the patient’s admission to different wards of a hospital, particularly the intensive care unit (ICU) [1]. The presence of underlying diseases or dysfunction of organs, immune system disorders, and immunosuppressive drugs, as well as the need for respiratory, cardiac and renal support, makes the patients prone to such infections. Moreover, the use of long-term invasive methods, such as intravenous and arterial catheterization, urinary catheterization, tracheal intubation, and mechanical ventilation and also the use of injective antibiotics have made these infection unavoidable in the ICU [2, 7].
Antibiotic resistance has emerged as a main determinant of outcome in ICU patients, mostly due to the administration of inadequate antibiotic treatment. Gram-positive bacteria (e.g. Staphylococcus species) or mostly Gram-negative bacteria (e.g. Enterobacteriaceae, Pseudomonas, and Acinetobacter species) are the cause of 70% of NIs in the ICUs [8, 9]. Specific Gram-negative bacteria such as Enterobacteriaceae producing extended -spectrum β-lactamases (ESBLs), carbapenemases, multidrug-resistant (MDR) Pseudomonas aeruginosa, and carbapenem-resistant Acinetobacter baumannii are the main reasons for high levels of resistance to antibiotics [2]. Hence, the issue of increasing antibiotic resistance of Gram-negative bacteria in the ICU is greatly worrying and remains a threat to the public health [10, 11].
Enterobacteriaceae, the most prevalent Gram-negative bacteria in NIs, are associated with resistance to several groups of antibiotics, frequently complicating treatment options. Escherichia coli and Klebsiella pneumoniae are the most opportunistic Gram-negative opportunistic organisms and the most common pathogens in NIs [9, 12]. In the Enterobacteriaceae family, the largest producers of ESBLs are often found in E. coli and K. pneumoniae [13,14,15]. The ESBL production by these bacteria has led to resistance of multiple classes of antibiotics and limitation of treatment choices [16]. ESBLs as one of the most significant mechanisms of resistance among Gram-negative bacteria have become a major threat to human health [17].
Carbapenems are still used as a first-line treatment and a therapeutic alternative for severe infections caused by Enterobacteriaceae, especially MDR and ESBLs E. coli [18]. However, the widespread use of these antibiotic agents has led to the development of carbapenem-hydrolyzing enzyme, namely carbapenem-resistant Enterobacteriaceae (CRE). One of the main challenges for ICU physicians, in addition to the prevention of NIs, is to identify the cause of infection and to select an appropriate experimental antibiotic regimen [2, 9, 19].
The type and rate of resistance vary in different regions; therefore, identification of bacterial agents and their resistance act a key role in determining their strategy and decreased bacterial resistance (7,19). Today, NIs and bacterial resistance to antibiotics have dramatically increased, and treatment-resistant NIs are growing. By focusing on the alarming facts, mentioned above, we conducted the present study to evaluate the prevalence of NIs and their antimicrobial susceptibility pattern in the ICUs at Imam Khomeini Hospital, Ilam, Iran.
Materials and methods
Study design and setting
This descriptive cross-sectional study was conducted on patients admitted to the ICUs 1 and 2 at Imam Khomeini Hospital, Ilam, Iran, from July to January 2020. Inclusion criteria for NIs were defined according to the Centers for Disease Control and Prevention (CDC) [20]. Written informed consents were received from each patient.
Phenotypic characterization of the Isolates
Sampling was performed on patients who had been admitted to the ICU for 48–72 h. Clinical samples such as respiratory secretions, blood, middle urine, and other body fluids, as well as wounds were collected. Bacterial identification was carried out using phenotypic and biochemical examinations with standard methods [21]. In brief, clinical specimens were first transferred to the laboratory and then cultured on blood and MacConkey agars (both from Merck Co., Germany). Thereafter, differential tests were used to identify bacterial species and genus.
Antimicrobial susceptibility testing
Kirby-Bauer disk diffusion method was performed on Müller-Hinton agar (MHA) (Merck Co.) in accordance with the Institute of Clinical and Laboratory Standards guidelines (CLSI 2020) [22]. Antimicrobial disks used for Gram-negative bacteria included Amikacin, trimethoprim/sulfamethoxazole, ciprofloxacin, imipenem, meropenem, aztreonam, cefotaxime, ceftazidime, ceftriaxone, and cefpodoxime, 30 μg for each (TAV, Turkey). Linezolid, clindamycin, tigecycline, penicillin, amikacin, gentamicin, cefoxitin, and trimethoprim/sulfamethoxazole (30 μg for each) were antimicrobial disks used for Gram-positive bacteria. To evaluate the quality control of the discs, we utilized E. coli ATCC 25,922 standard strain and S. aureus 29,213 for Gram-negative and Gram-positive bacteria, respectively. Following an overnight incubation at 37 °C, the size of inhibition zone was measured and interpreted as sensitive, intermediate and resistant.
Phenotypic screening of ESBLs, MBLs, and CRE in E. coli and K. pneumoniae
E. coli and K. pneumoniae isolates related to NI were used for phenotypic ESBLs, metallo-β-lactamases (MBLs) and CRE tests. The initial screening was performed with five antibiotics, including cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefpodoxime (10 μg), and aztreonam (30 μg) (TAV). In each antibiotic, the inhibition zone resistance was examined by confirmatory tests according to CLSI 2020 [22]. Briefly, ceftazidime (30 μg) and ceftazidime/clavulanate (30/10 μg; TAV) discs were used as the combination disk test. Sensitivity zone diameter of at least 5 mm in the composite disk compared to ceftazidime was considered as ESBLs. K. pneumoniae ATCC 700,603 was employed as the control strain. To determine MBLs, the two discs of imipenem (10 μg) with a distance of 15 mm were placed on MHA plate that was inoculated by 0.5-McFarland standard. One imipenem disc was impregnated with 10 μl of 0.5 M ethylenediamine tetraacetic acid (EDTA). The plates were incubated at 35 °C for 16–18 h. In the combination disk with EDTA, the sensitivity zone diameter of 7 mm and higher was considered as MBLs.
Phenotypic screening of CRE resistance in E. coli and K. pneumoniae
The modified carbapenem inactivation method was used to detect CREs resistance in E. coli and K. pneumoniae. After incubation of the meropenem disc in bacterial suspension for 2 h, the disk was replaced on MHA (Merck Co.) and inoculated with E. coli ATCC 25,922 as the standard strain. CRE activity was distinctive by no zone on MHA (Merck Co.) [22].
Molecular detection of ESBLs, MBLs, and CRE genes
Following the bacterial DNA extraction by boiling method [23], polymerase chain reaction (PCR) was performed for specific primers shown in the Table 1. Genes encoding ESBLs (blaCTX-M, blaOXA-11, and blaSHV), MBLs (blaNDM, blaVIM, and blaIMP), and CRE (blaKPC, blaOXA-48, and blaOXA-23,) were detected.
Statistical analysis
Statistical analysis was conducted using the “IBM SPSS statistics 22” software (IBM analytics; USA). Statistical significance of variables was determined by chi-square and Fisher's exact tests. The results were presented as descriptive statistics in terms of relative frequency. A p-value of ≤ 0.05 was considered statistically significant.
Results
Patient characteristics and prevalence of NIs
A total of 602 patients in the ICU 1 (n = 239; 70.39%) and ICU 2 (n = 363; 30.605) were recruited and analyzed for six months. The average length of hospital stay on the admission was 22 days. The lowest stay was two days, while the maximum stay was 217 days. According to the positive cultures, 71 patients, including 44 (61.97%) men and 27 (38.03%) women with the mean age of 49.53 years (± 23.06 standard deviation), showed NIs. The patients in the age range of 14–29 years had the most hospital infections. Overall, the prevalence of NIs in ICU1 was estimated as, 17.99% and 7.77% in ICU 2. The prevalence of NI was significantly different in both ICUs. However, the rate of this infection in ICU 1 was higher than that of ICU 2 (p < 0.001). The detailed information of the patients is represented in Table 2.
Bacteria causing NIs
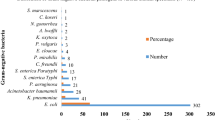
In total, 71 patients had positive culture. Of 103 bacterial isolates identified, 79 isolates (76.70%) were Gram-negative, and 24 (23.30%) were Gram-positive. The most prevalent Gram-negative bacteria were E. coli (n = 29; 28.16%), A. baumannii (n = 15; 14.56%), and K. pneumoniae (n = 13; 12.62%), whereas the most common bacterium was S. aureus (n = 12; 11.66%), as shown in Fig. 1. Regarding the type of clinical infection, respiratory system (52; 50.48%) was the most common organ involved, followed by urinary tract system (n = 26; 25.25%), bloodstream (n = 22; 21.36%), and wound (n = 3; 2.91%), as depicted in Table 3. A. baumannii and K. pneumoniae were the most common bacteria isolated from the respiratory tract system, and E. coli was the most prevalent infectious agent of urinary tract system.
Antimicrobial susceptibility pattern
Antimicrobial susceptibility pattern in Gram-negative bacteria showed the highest resistance to third-generation cephalosporins, including ceftazidime (n = 69; 87.34%), cefotaxime and cefpodoxime (n = 59; 74.69%), and ceftriaxone (n = 58; 73%). Moreover, the highest susceptibility was observed for amikacin (n = 25; 34.61%), followed by imipenem and meropenem (n = 27; 34.17%). Among Gram-negative bacteria, A. baumannii showed the highest resistance to most of antimicrobials. Gram-positive bacteria showed the highest resistance to penicillin (n = 22; 91.67%) and cefoxitin (n = 19; 79.17%), and the highest susceptibility to tigecycline (n = 24; 100%), followed by trimethoprim/sulfamethoxazole (n = 23; 84. 95%) and linezolid (n = 22; 91.67%), as illustrated in Table 4. Multidrug resistance is defined as resistance of a microorganism to at least one agent in three or more antimicrobial categories. Overall, the prevalence of MDR bacteria was 58.25% (n = 60). E. cloacae (n = 4; 100%), S. aureus (n = 11; 91.66%), and A. baumannii (n = 13; 86.66%) were the most commonly MDR bacteria isolated. The ratio of the number of MDR isolates to the total number of isolates was statistically significant (p < 0.001;
See supplementary file 1 for more details).).
Phenotypic detection of ESBLs and CREs in E. coli and K. pneumoniae isolates
A total of 42 isolates, including 29 E. coli and 13 K. pneumoniae isolates related to NI, were obtained in ICUs 1 and 2 and used for phenotypic tests. Of 42 isolates of E. coli and K. pneumoniae, 32 (76.19%) isolates, i.e. 22 (75.86%) E. coli and 10 (76.93%) K. pneumoniae, were ESBL producers. Also, 10 (23.81%) isolates were non-ESBLs isolates. Based on the phenotypic detection of CREs, 6 (14.28%) isolates, including 2 (6.89%) E. coli and 4 (30.77%) K. pneumoniae isolates, were identified as CRE producers (Table 5).
Molecular detection of ESBLs, MBLs, CRE genes and PCR product sequencing
The blaCTX-M gene encoding ESBLs was detected in 29 (90.62%) isolates, including 22 (100%) E. coli and 7 (70%) K. pneumoniae isolates. The blaSHV was identified in 7 (21.87%) isolates of E. coli (1; 4.54%) and K. pneumoniae (n = 6; 60%). The blaOXA-11 encoding ESBLs was found in 10 (31.25%) isolates of E. coli (n = 7; 31.81%) and K. pneumoniae (n = 3; 30%). The blaNDM gene was detected in 4 (66.66%) K. pneumoniae isolates, and the blaOXA-23 and blaOXA-48 genes were observed in 3 (50%) K. pneumoniae and only 1 (16.66%) E. coli isolate, respectively. However, the blaVIM, blaKPC, and blaIMP genes were not found in any isolates (Table 6). PCR sequencing products were performed for five blaNDM and one blaOXA-23 gene products, and 98% nucleotide similarity was observed in both genes. In addition, four blaNDM gene products were identified as blaNDM-1.
Coexistence of ESBL- and CRE-encoding genes in E. coli and K. pneumoniae isolates
PCR confirmed the coexistence of blaCTX-M and blaSHV in five isolates and blaCTX-M and blaOXA-11 genes in 10 isolates, and three genes (blaCTX-M, blaSHV, and blaOXA-11) encoding ESBLs were harbored by two K. pneumoniae isolates. In addition, the coexistence of two genes, blaNDM-1 and blaOXA-23, were observed in three isolates of K. pneumoniae. Also, three K. pneumoniae isolates carried four genes, including blaCTX-M, blaSHV, blaNDM, and blaOXA-23, and two K. pneumoniae isolates harbored five genes, including blaOXA-11, blaCTX-M, blaSHV, blaNDM -1 and blaOXA-23.
(See supplementary file 2 for more details).).
Discussion
NIs are one of the major global health problems involved almost all hospitalized patients at risk for these infections [10]. In is study, the prevalence of NIs in both ICUs was reported as 11.79%, which was 17.99% in ICU 1 and 7.77% in ICU 2. Previous studies conducted in Ahvaz and Gorgan cities of Iran have reported 10% and 13% NI prevalence in ICU, respectively [24, 25]. The rate of NI prevalence obtained in our study for ICU was lower than the estimates provided by studies in Zahedan (76.9%) and Birjand (27.2%) cities. Several reasons for differences in the prevalence of NIs by diverse studies in Iran might be variations in the study periods and type of units under investigation, as well as unreported real rate of infections. In the present study, NIs caused by fungal infections were not studied due to the limited number of samples compared to other investigations conducted in Iran [26, 27], Northern India (30.4%) [28], and China (26.07%) [29]. The length of patient stay is a risk factor and associated with the increased incidence of NIs. There was a difference in the number of hospitals and that of patients in prior studies [28, 29].
The most common infections in our study were respiratory tract infections, followed by urinary tract infection. In a previous study comparing NIs in the internal ICU with those of surgery ICU demonstrated that respiratory tract infection and UTI were the most common infections in the two ICUs, respectively. In other investigations in India and Germany, respiratory tract infection was the most common infected organ. Patients in the ICU require supportive treatment; therefore, mechanical ventilation and urinary catheterization are widely used, which can be risk factors for the infection of the patients in this unit [-31].
In the current study, Gram-negative bacteria were more identified than Gram-positive bacteria in positive cultures. Similar to our result, Nouri et al. [32] reported Gram-negative bacteria as the main causes of NIs. We also found that the most frequent Gram-negative bacteria were E. coli and A. baumannii, and Gram-positive bacterium was S. aureus. This outcome is in agreement with an earlier study in Hamadan showing E. coli and K. pneumoniae as the most prevalent Gram-negative bacteria and S. aureus as the most frequent Gram-positive bacterium [32].
Another study has also reported K. pneumoniae, Acinetobacter species, and P. aeruginosa as the most prevalent Gram-negative bacteria and S. aureus the most prevalent Gram-positive bacterium. Gram-negative bacteria have the greatest diversity, thus achieving high adaptability in the environment. These bacteria are flora in the human digestive system and skin and simply transfer from equipment and personnel to patients. S. aureus is a flora of the skin and nasal and can easily spread [7, 32, 33]. The prevalence of antibiotic-resistant bacteria has become a public health concern worldwide. Increased antimicrobial resistance among microorganisms causing NIs is associated with high mortality in hospitalized patients.
Antimicrobial susceptibility testing in Gram-negative bacteria showed the highest susceptibility to amikacin, followed by imipenem and meropenem. Also, the lowest susceptibility rates were related to ceftazidime, cefotaxime, and cefpodoxime, respectively. The result of our study showing that A. baumannii had the most resistance to all antimicrobial categories were consistent with other findings in Iran and Uganda [7, 34].
In Gram-positive bacteria, tigecycline, trimethoprim/sulfamethoxazole, and linezolid had the highest efficiency. Also, the highest resistance was related to penicillin, which supports Tolera et al.’ investigation in Ethiopia [10]. The prevalence of MRSA in our study was higher than a previous study conducted in Gauteng Academic Hospital in South, which seems worrying [35]. These differences in high level of antibiotic resistance could be due to regional and genetic variations of species and also genetic differences between individuals. However, it is thought that factors such as long-term hospitalization of patients, continuous use of broad-spectrum antibiotics during hospitalization, cultural diversity, irregular consumption of drug, and self-treatment are the most important factors affecting the high-level resistance of antibiotics [36, 37].
In the present study, the phenotypical prevalence of ESBLs was observed in 32 (76.19%) E. coli and K. pneumoniae isolates, which in Hasani’s study, the prevalence rate of ESBLs was 87.8% [37]. However, different prevalence has been reported in Iran and other countries. In Bialvaei’s study, the rates of ESBLs production were 27.27% in E. coli and 25.9% in K. pneumoniae [38].
According to a systematic review and meta-analysis, the prevalence of ESBLs was reported as 43.2% in Iran, which is higher than the rate stated in developed countries such as France (1.5%) and Germany (3.3%) [39]. In our study, sample collection was carried out in ICUs. In this unit the use of broad-spectrum antibiotics is very high, which can be a reason for the high level of ESBLs. The investigation of ESBL genes showed high prevalence of the blaCTX-M and low prevalence of blaSHV. Similarly, previous studies have reported the prevalence of 92.5% and 100% for blaCTX-M in E. coli and K. pneumoniae isolates, respectively [37, 40].
The low prevalence of blaSHV was confirmed by a former study conducted in Khuzestan (3.5%) and Tehran cities (10%) of Iran [41, 42]. Phenotypical tests exhibited that 6 (14.28%) isolates of E. coli and K. pneumoniae harbored MBLs and CRE. This result is in line with a previous study showing that the overall rates of carbapenem resistance were 24% and 5% in K. pneumoniae and E. coli isolates, respectively [43].
In molecular detection, the most common carbapenemase genes included blaNDM-1 and blaOXA-23, which former gene was mostly prevalent in K. pneumoniae, and the latter gene was mainly detected in K. pneumonia isolates. Shahcheraghi et al., identified the first blaNDM-1 among K. pneumonia isolates in Iran [44]. Fazeli et al., [45] and Moghadampour et al., [46] have also found blaNDM-1 gene among K. pneumonia isolates, which is consistent with Huang’s finding conducted in China [41, 45,47,47]. In our study, the genes blaIMP, blaVIM, and blaKPC were not detected, while blaNDM, blaOXA-48, and blaKPC were the genes not identified in Kiaei et al.’s study in Keramn [48]. In this study, the prevalence of blaNDM and blaOXA-48 were similar or almost similar to Jalalvand’s study in Tehran city [49].
K. pneumoniae and E. coli are the most important pathogens of NIs and can be associated with increased mortality and mortality related to carbapenem resistance worldwide. The available antibiotics for carbapenem-resistant isolates are limited. Furthermore, isolates carrying the blaNDM-1 gene are MDR and resistant to almost all beta-lactam antibiotics, fluoroquinolones, and aminoglycosides. Therefore, the emergence and spread of these isolates is a great challenge and should be taken into consideration [50, 51]. Unsupervised and indiscriminate use of antibiotics has always been important factors in creating resistance and reducing treatment options. Thus, application of a specific guideline like antimicrobial stewardship program and establishment of surveillance systems to analyze antibiotic resistance pattern can decline trends of bacterial resistance and NIs in the ICU. This study had some limitations. The small sample size and single center study were among these limitations. Other limitations included lack of access to patient records, including the type of NIs, antibiotic prescription history, and risk factors for antibiotic-resistant bacteria. Moreover, the identification of bacterial with culture and biochemical tests and the time of the study may be potential sources of bias and possible confounder of this study, respectively. To generalize the results to the entire study area, it is suggested that further studies be conducted periodically with a larger sample and more centers.
Conclusion
In the present study, the most NIs was identified in the ICU 1. Moreover, the Gram-negative bacteria E. coli, A. baumannii, and K. pneumoniae with high levels of resistance were introduced as the most common bacteria causing NIs in the ICU. The interesting point of our study was that we, for the first time, detected blaOXA-11, blaOXA-23, and blaNDM-1 genes in E. coli and K. pneumoniae isolates in Ilam city of Iran. These genes showed high resistance to carbapenems, which is a significant issue that needs to be taken into account. Therefore, identifying endemic pthogenic bacteria, source of infection, determining antibiotic susceptibility pattern in ICU, finding and implementing an appropriate strategy for diagnosing, prescribing medication, and monitoring infection control could be effective in reducing the rate of such resistance, NIs, and also patients’ mortality.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
References
Mohammadi M, Vaisi Raiegan A, Jalali R, Ghobadi A, Salari N, Barati H (2019) The prevalence of nosocomial infections in Iranian hospitals. J Babol Univ Medical Sci 21:39–45
Hogeveen M, Heijting I, Jansen S, Hopman J, Overbeek MGD, Tostman A, Bekker V. (2021) Nosocomial infections in neonatal care: a scoping review protocol of published surveillance case definitions. https://osf.io/preprints/w9ce2/
Khammar M, Hassanzadeh S, Tara F, Siahsar M, Tahmasbi F, Keikha M, Ghazvini K (2021) A-4year Study on Antimicrobial Susceptibility Trends of Nosocomial Infections in a Mashhad Referral Hospital, Mashhad. Rev Clin Med, Iran, p 8
Nimer NA (2022) Nosocomial infection and antibiotic-resistant threat in the middle east. Infect Drug Resist. https://doi.org/10.2147/IDR.S351755
Rajabi M, Abdar ME, Rafiei H, Aflatoonia MR, Abdar ZE (2016) Nosocomial infections and epidemiology of antibiotic resistance in teaching hospitals in south east of Iran. Glob J Health Sci 8:190
Centers for disease control and prevention [CDC] Campaign to prevent antimicrobial resistance in healthcare settings: why a campaign? Atlanta GCfDCaP. (2001). https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5115a5.htm
Agaba P, Tumukunde J, Tindimwebwa J, Kwizera A (2017) Nosocomial bacterial infections and their antimicrobial susceptibility patterns among patients in Ugandan intensive care units: a cross sectional study. BMC Res Notes 10:1–12
Leone M, Bouadma L, Bouhemad B, Brissaud O, Dauger S, Gibot S, Hraiech S, Jung B, Kipnis E, Launey Y (2018) Hospital-acquired pneumonia in ICU. Anaesth Crit Care Pain Med 37:83–98
Khan A, Miller WR, Arias CA (2018) Mechanisms of antimicrobial resistance among hospital-associated pathogens. Expert Rev Anti Infect Ther 16:269–287
Tolera M, Abate D, Dheresa M, Marami D (2018) Bacterial nosocomial infections and antimicrobial susceptibility pattern among patients admitted at Hiwot Fana Specialized University Hospital. Adv Med, Eastern Ethiopia. https://doi.org/10.1155/2018/2127814
Nwafia IN, Ohanu ME, Ebede SO, Ozumba UC (2019) Molecular detection and antibiotic resistance pattern of extended-spectrum beta-lactamase producing Escherichia coli in a Tertiary Hospital in Enugu. Nigeria Ann Clin Microbiol Antimicrob 18:1–7
Peralta G, Sanchez MB, Garrido JC, De Benito I, Cano ME, Martínez-Martínez L, Roiz MP (2007) Impact of antibiotic resistance and of adequate empirical antibiotic treatment in the prognosis of patients with Escherichia coli bacteraemia. J Antimicrob Chemother 60:855–863
Goltz J (2022) Investigating Factors Associated with Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae Colonization and/or Infection in Humans: A One Health Approach: University of Guelph. https://atrium.lib.uoguelph.ca/xmlui/handle/10214/26906
Urase T, Okazaki M, Tsutsui H (2020) Prevalence of ESBL-producing Escherichia coli and carbapenem-resistant Enterobacteriaceae in treated wastewater: a comparison with nosocomial infection surveillance. J Water Health 18:899–910
Surgers L, Boyd A, Boelle P-Y, Lalande V, Jolivot P-A, Girard P-M, Arlet G, Cambier C, Homor A, Decre D (2017) Clinical and microbiological determinants of severe and fatal outcomes in patients infected with Enterobacteriaceae producing extended-spectrum β-lactamase. Eur J Clin Microbiol Infect Dis 36:1261–1268
Xiao T, Yang K, Zhou Y, Zhang S, Ji J, Ying C, Shen P, Xiao Y (2019) Risk factors and outcomes in non-transplant patients with extended-spectrum beta-lactamase-producing Escherichia coli bacteremia: a retrospective study from 2013 to 2016. Antimicrob Resist Infect Control 8:1–11
Maechler F, Schwab F, Hansen S, Fankhauser C, Harbarth S, Huttner BD, Diaz-Agero C, Lopez N, Canton R, Ruiz-Garbajosa P (2020) Contact isolation versus standard precautions to decrease acquisition of extended-spectrum β-lactamase-producing Enterobacterales in non-critical care wards: a cluster-randomised crossover trial. Lancet Infect Dis 20:575–584
Ding Y, Wang Y, Hsia Y, Sharland M, Heath PT (2019) Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China. Ann Clin Microbiol Antimicrob 18:1–8
Campion M, Scully G (2018) Antibiotic use in the intensive care unit: optimization and de-escalation. J Intensive Care Med 33:647–655
Garner J, Jarvis W, Emori T, Horan T, Hughes J (1991) CDC definitions for nosocomial infections 1988. Z Arztl Fortbild 85:818–827
Cheesbrough M (2005) District laboratory practice in tropical countries, part 2. Cambridge university press, Cambridge
Weinstein MP, Lewis JS (2020) The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes. J Clin Microbiol 58:e01864-e11819
Yamagishi J, Sato Y, Shinozaki N, Ye B, Tsuboi A, Nagasaki M, Yamashita R (2016) Comparison of boiling and robotics automation method in DNA extraction for metagenomic sequencing of human oral microbes. PLoS ONE 11:e0154389
Golsha R, Ashoori N, Tajik M, Sohrabi A, Montazeri M (2020) Prevalence of nosocomial infections in intensive care units in shahid Sayyad-E-Shirazi Hospital of Gorgan During 2016-2018. Tabari Biomed Stu Res J. https://doi.org/10.18502/tbsrj.v2i2.3761
Birgani AG, Asadpoor S (2008-2009) Nosocomial infections in intensive care unit of Ahvaz Arya Hospital. Modern Care Journal. 2011;8(2): 58–93.
Tabatabaei SM, Pour FB, Osmani S. (2015) Epidemiology of hospital-acquired infections and related anti-microbial resistance patterns in a tertiary-care teaching hospital in Zahedan, Southeast Iran. Int J Infect. 2(4). e29079 https://doi.org/10.17795/iji-29079.
Zare-Bidaki M, Allahyari E, Nikoomanesh F, Ebrahimzadeh A (2021) A comparative analysis of nosocomial infections between internal and surgical intensive care units of university hospitals in Birjand, Iran from 2016 to 2017: a retrospective study. J Basic Res Med Sci 8:32–41
Boora S, Singh P, Verma A, Chauhan A, Lathwal A, Mathur P (2021) Point-prevalence survey for the hospital-acquired infections in intensive care units of trauma center in a tertiary care hospital of Northern India. J Lab Physicians 14(2):115–118
Wang J, Liu F, Tartari E, Huang J, Harbarth S, Pittet D, Zingg W (2018) The prevalence of healthcare-associated infections in mainland China: a systematic review and meta-analysis. Infect Control Hosp Epidemiol 39:701–709
Mitharwal SM, Yaddanapudi S, Bhardwaj N, Gautam V, Biswal M, Yaddanapudi L (2016) Intensive care unit-acquired infections in a tertiary care hospital: an epidemiologic survey and influence on patient outcomes. Am J Infect Control 44:e113–e117
Ott E, Saathoff S, Graf K, Schwab F, Chaberny IF (2013) The prevalence of nosocomial and community acquired infections in a university hospital: an observational study. Dtsch Arztebl Int 110:533
Nouri F, Karami P, Zarei O, Kosari F, Alikhani MY, Zandkarimi E, Zarandi ER, Taheri M (2020) Prevalence of common nosocomial infections and evaluation of antibiotic resistance patterns in patients with secondary infections in Hamadan. Iran Infect Drug Resist 13:2365
Rahimi-Bashar F, Karami P, Khaledi A, Dehghan A, Seifrabie MA, Yaghoobi MH (2018) Evaluation of the prevalence of nosocomial infection in different wards of Be’sat hospital of Hamedan. Avicenna J Clin Microbiol Infect 5:31–35
Shoaei S, Sali S, Yousefi H (2017) Incidence and resistance patterns of nosocomial infections in labbafi nejad hospital admitted patients during 2012–2014. Int J Infect Dis 3:78–81
Fortuin-de Smidt MC, Singh-Moodley A, Badat R, Quan V, Kularatne R, Nana T, Lekalakala R, Govender NP, Perovic O (2015) Staphylococcus aureus bacteraemia in Gauteng academic hospitals. South Africa Int J Infect Dis 30:41–48
Pournajafi A, Mahmoudi A (2020) Prevalence of extended-spectrum beta-lactamases production in Escherichia coli isolated from urinary tract infection samples in Zanjan hospitals. South Asian Res J Pharm Sci, Iran
Hasani A, Mohammadzadeh A, Kafil HS, Rezaee MA, Hasani A, Aghazadeh M (2015) Characterization of TEM-, SHV-, CTX-and AmpC-type β-lactamases from cephalosporin resistant Escherichia coli isolates from Northwest of Iran. J Pure Appl Microbiol 9:3401–3406
Bialvaei AZ, Kafil HS, Asgharzadeh M, Aghazadeh M, Yousefi M (2016) CTX-M extended-spectrum β-lactamase-producing Klebsiella spp, Salmonella spp, Shigella spp and Escherichia coli isolates in Iranian hospitals. Braz J Microbiol 47:706–711
Jabalameli L, Beigverdi R, Ranjbar HH, Pouriran R, Jabalameli F, Emaneini M (2021) Phenotypic and genotypic prevalence of extended-spectrum β-Lactamase-Producing Escherichia coli: a systematic review and meta-analysis in Iran. Microb Drug Resist 27:73–86
Saisi H, Makobe C, Kangongo M, Kariuki S (2019) Prevalence of CTXM, SHV, TEM AND OXA genes among extended-spectrum beta-lactamase producing Klebsiella pneumoniae from Mukuru Slum. Kenya Adv Microbiol 9:853–862
Zadeh SB, Shakib P, Zolfaghari MR, Sheikh AF (2021) Prevalence of Escherichia coli and Klebsiella pneumoniae, Producing Extended-Spectrum Beta-Lactamase (ESBLs) from Clinical Specimen in Khuzestan. Gene, Cell and Tissue, Iran, p 8
Shahbazi S, Karam MRA, Habibi M, Talebi A, Bouzari S (2018) Distribution of extended-spectrum β-lactam, quinolone and carbapenem resistance genes, and genetic diversity among uropathogenic Escherichia coli isolates in Tehran. Iran J Globa Antimicrob Resist 14:118–125
Nasiri MJ, Mirsaeidi M, Mousavi SMJ, Arshadi M, Fardsanei F, Deihim B, Davoudabadi S, Zamani S, Hajikhani B, Goudarzi H (2020) Prevalence and mechanisms of carbapenem resistance in Klebsiella pneumoniae and Escherichia coli: a systematic review and meta-analysis of cross-sectional studies from Iran. Microb Drug Resist 26:1491–1502
Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, Nasiri S, Owlia P, Nikbin VS, Imani Fooladi AA (2013) First report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb Drug Resist 19:30–36
Fazeli H, Norouzi-Barough M, Ahadi A, Shokri D, Solgi H (2015) Detection of New Delhi Metallo-Beta-Lactamase-1 (NDM-1) in carbapenem-resistant Klebsiella pneumoniae isolated from a university hospital in Iran. Hippokratia 19:205
Moghadampour M, Rezaei A, Faghri J (2018) The emergence of bla OXA-48 and bla NDM among ESBL-producing Klebsiella pneumoniae in clinical isolates of a tertiary hospital in Iran. Acta Microbiol Immunol Hung 65:335–344
Huang X, Cheng X, Sun P, Tang C, Ni F, Liu G (2018) Characteristics of NDM-1-producing Klebsiella pneumoniae ST234 and ST1412 isolates spread in a neonatal unit. BMC Microbiol 18:1–6
Kiaei S, Moradi M, Hosseini-Nave H, Ziasistani M, Kalantar-Neyestanaki D (2019) Endemic dissemination of different sequence types of carbapenem-resistant Klebsiella pneumoniae strains harboring blaNDM and 16S rRNA methylase genes in Kerman hospitals, Iran, from 2015 to 2017. Infect Drug Resist 12:45
Jalalvand K, Shayanfar N, Shahcheraghi F, Amini E, Mohammadpour M, Babaheidarian P (2020) Evaluation of phenotypic and genotypic characteristics of carbapnemases-producing enterobacteriaceae and its prevalence in a referral hospital in Tehran city. Iran J Pathol 15:86
Eyvazi S, Hakemi-Vala M, Hashemi A, Bejestani FB, Elahi N. (2018) Emergence of NDM-1-producing Escherichia coli in Iran. Arch Clin Infect Dis 13(4):e62029
Gao B, Li X, Yang F, Chen W, Zhao Y, Bai G, Zhang Z (2019) Molecular epidemiology and risk factors of ventilator-associated pneumonia infection caused by carbapenem-resistant enterobacteriaceae. Front Pharmacol 10:262
Acknowledgements
We would like to thank the Clinical Microbiology Research Center, Ilam University of Medical Sciences, Ilam, Iran, for their cooperation. The authors also acknowledge the Vice Chancellor for Research Affairs, Ilam University of Medical Sciences, for their executive and financial support.
Author information
Authors and Affiliations
Contributions
NS and HK contributed to the study conception and design. Data collection and analysis were performed by MH and SK. The first draft of the manuscript was written by MH and SK, and all the authors commented on the previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflict of interest in this study.
Ethical approval
The study protocol was approved by the local ethics committee of Ilam University of Medical Sciences Iran (ethical code: IR.MADILAM.REC.1400.003). Written informed consent was received from each patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hashemian, M., Kazemian, H., Kaviar, V.H. et al. Nosocomial infections and antimicrobial susceptibility patterns among patients admitted to intensive care unit of Imam Khomeini hospital in Ilam, Iran. Mol Biol Rep 50, 5565–5574 (2023). https://doi.org/10.1007/s11033-023-08476-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08476-4