Abstract
Glioma is the most frequent type of malignant tumor in the central nervous system, accounting for about 80% of primary malignant brain tumors, usually with a poor prognosis. A number of studies have been conducted on the molecular abnormalities in glioma to further understand its pathogenesis, and it has been found that lncRNAs (long non-coding RNA) play a key role in angiogenesis, tumor growth, infiltration and metastasis of glioma. Since specific lncRNAs have an aberrant expression in brain tissue, cerebrospinal fluid as well as peripheral circulation of glioma patients, they are considered to be potential biomarkers. This review focuses on the biological characteristics of lncRNA and its value as a biomarker for glioma diagnosis and prognosis. Moreover, in view of the role of lncRNAs in glioma proliferation and chemoradiotherapy resistance, we discussed the feasibility for lncRNAs as therapeutic targets. Finally, the persisting deficiencies and future prospects of using lncRNAs as clinical biomarkers and therapeutic targets were concluded.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is the most common and aggressive primary tumor of central nervous system, accounting for about 80% of primary malignant brain tumors [1]. In the 2016 World Health Organization (WHO) classification, gliomas are categorized into diffuse astrocytoma, oligodendroglioma and glioblastoma based on histological characteristics [2]. Glioblastoma is the most frequent and malignant subtype, with a median survival of only 14.1 months [3]. Innovatively, molecular features are incorporated into the diagnostic criteria of glioma according to the classification, including isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion. Therefore, a better understanding of the genetic and molecular pathogenesis of glioma could contribute to more effective therapies. In recent years, emerging fields like genomics, transcriptomics and proteomics have brought an explosion of information about glioma. Accordingly, the research on glioma biomarkers is developing rapidly [4], and long non-coding RNA (lncRNA) have been paid more and more attention.
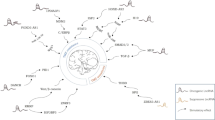
LncRNAs are a class of non-coding RNAs with longer than 200 nucleotides, involved in transcriptional, post-transcriptional and epigenetic levels of gene regulation [5, 6]. The way in which lncRNAs interacts with other biomolecules and modulates gene expression may be roughly fall into five categories (Fig. 1). (1) Signals. Under different stimulation, lncRNAs are specifically transcribed and participate in signal transduction. Some lncRNAs have regulatory function in the signaling pathways after being transcribed, while others simply act as by-products in the regulatory pathways [7]. (2) Molecular decoys. This kind of lncRNAs directly bind to DNA-binding proteins or other transcription factors, thus blocking the action of this signaling pathway and regulating downstream gene transcription[8]. In particular, lncRNAs may act as effective natural microRNA sponges that regulate gene expression by competitively binding microRNAs which known as competitive endogenous RNAs (ceRNAs) [9]. (3) Molecular guides. LncRNAs bind to those proteins and chromatin-modifying complexes, which usually identified as transcription factors, and then recruit them to specific sequences in the chromatin [10]. (4) Scaffolds. Multiple related transcription factors bind to single lncRNA molecule to initiate cross talk and integration between different signaling pathways. LncRNA scaffolds can co-regulate gene transcription temporally and spatially, which are conducive to the body and cells to produce feedback to external signals quickly [11].
Four action modes of lncRNA mechanism. Four action modes of lncRNA mechanism. A Signals. LncRNAs are specifically transcribed under the stimulation of upstream signals and participate in signaling pathway as signal transduction molecules. B Decays. LncRNAs directly bind to transcription factors or miRNAs, thereby blocking the action and signaling pathway of the molecules and regulating downstream gene transcription. C Guides. LncRNAs are combined with proteins and chromatin-modifying complexes, which are then localized to specific DNA sequences. D Scaffolds. Multiple related transcription factors bind to the same lncRNA and jointly regulate gene transcription or inhibition
Recent evidence indicates that aberrant lncRNA expression plays an important role in glioma pathogenesis [12], such as biogenesis, proliferation, angiogenesis and treatment resistance. When compared with mRNAs. the expression level of lncRNAs is higher in brain tissues than in other tissues [13], so lncRNAs may be more suitable biomarkers for glioma. In this review, we explore the potential use of lncRNAs as diagnostic and prognostic biomarkers, as well as their possible application in clinical treatment of glioma.
LncRNAs as biomarkers for glioma
lncRNA and glioma diagnosis
The current approach to diagnose glioma are mainly based on neuroimaging and biopsy. With the introduction of molecular parameters in the diagnosis of glioma [2], biopsy is of great significance for the treatment of glioma patients. However, the invasive procedure of tissue acquisition itself brings risks to the patients, and there is no way to know the subsequent mutation of tumor. Furthermore, the focal sampling of a lesion may not fully capture the intratumoral heterogeneity [14]. By contrast, liquid biopsy, referring to the replacement of surgical biopsy specimens with fluid samples such as blood and cerebrospinal fluid (CSF), may be the answer to these challenges. Liquid biopsy offers the promise of diagnosis and mutational analysis of glioma in a non-invasive manner [15], being available as an effective supplement to existing solid biopsy. Liquid biopsies usually detect circulating tumor cells, extracellular vesicles, circulating tumor DNA (ctDNA) and circulating tumor RNA (ctRNA). As one of them, lncRNA is receiving more and more attention.
The best-known usage of lncRNA in cancer diagnosis is Prostate cancer antigen 3 (PCA3) in prostate cancer. PCA3 is a specific lncRNA with an increased expression in more than 90% prostate tumors [16], which is already clinically used for prostate cancer detection and has been approved by the US Food and Drug Administration (FDA) [17]. Compared with prostate cancer, the most important characteristic of glioma is the existence of the blood–brain barrier, hinders the migration of glioma biomarkers to peripheral blood. Moreover, due to the presence of a large number of RNA enzymes in the blood, the half-life of free lncRNAs in the plasma is only 3 h and are easily decomposed. Nevertheless, lncRNAs can still exist stably in the peripheral blood attributed to the fact that exosomes serve as carriers for most lncRNAs [18]. The concentration of lncRNAs in exosomes can be even higher than that in derived cells [19]. Therefore, lncRNAs in peripheral blood and CSF could be suitable biomarkers for the diagnosis and prognosis of glioma.
At the molecular level, the lncRNA expression show considerable variation whether between glioma and normal tissue or among different glioma subtypes. Zhang et al. analyzed a cohort of gene expression data from the Gene Expression Omnibus (GEO) and found 129 lncRNAs, which showed a more than two-fold difference between gliomas and normal brain tissues [20]. Another analysis of glioma data based on The Cancer Genome Atlas (TCGA) showed that lncRNAs are extensively induced or repressed in both glioblastoma and low-grade glioma (LGG) [21]. Some differentially expressed lncRNAs were reported to have the potential function as biomarkers for glioma diagnosis (Table1).
It is worth mentioning that serum lncRNA HOX transcript antisense RNA (HOTAIR) can perform a diagnostic biomarker for glioblastoma, with a sensitivity of 86.1% and a specificity of 87.5%. According to the data in TCGA, glioblastoma can be classified into classical, mesenchymal, neural, and proneural subtypes based on gene expression [32]. Research has found that HOTAIR expression varied in different subtypes, which was markedly increased in the classical and mesenchymal subtypes compared with the neural and proneural subtypes [33]. Similarly, HOXA11-AS expression also showed great difference among the four glioblastoma subtypes. Specifically, the expression in the classical and mesenchymal subtypes was higher than that in the neural and proneural subtypes [34]. These results suggested that the expression level of some lncRNAs is closely related to tumor classification, and may even affect the malignant behavior of tumors.
lncRNA and glioma prognosis
Treatment for gliomas includes surgical resection, radiation and chemotherapy. The Response Assessment in Neuro-Oncology (RANO) criteria based on magnetic resonance imaging (MRI) is considered to be the gold standard for treatment efficacy evaluation [35]. The main problem in measuring treatment effectiveness is pseudoprogression [36, 37], which is caused by the response of brain tissue to chemotherapy and radiotherapy. The imaging feature of pseudoprogression is defined as increased enhancement and edema on MRI, hard to differ from tumor progression. This condition may be due to edema and increased vascular permeability caused by treatment-related local inflammation. Differential diagnosis is necessary because a combination of chemotherapy and radiotherapy may induce pseudoprogression in about 30% patients [38, 39]. Unfortunately, There is currently no effective radiological technique to distinguish pseudoprogression from tumor recurrence [40]. Several studies showed that lncRNA biomarkers may aid in the determination of glioma recurrence. For instance, lncRNA family with sequence similarity 225 member B (FAM225B) upregulates in recurrent glioblastoma, and is identified to be an independent prognostic factor for recurrent glioblastoma [41].
The multi-level involvement of lncRNAs in tumor biological process, makes them potential choices as prognostic biomarkers of glioma. Multiple lncRNAs have been confirmed to be closely related to the clinicopathological data and prognosis of glioma patients. A meta-analysis including 14 eligible studies and 1415 glioma patients indicated that lncRNA expression not only significantly correlated with overall survival (OS) in glioma patients but also associated with tumor diameter, tumor grade, and Karnofsky Performance Status Scale (KPS) [42]. According to another meta-analysis study, the expression of urothelial carcinoma-associated (UCA1) was positively associated with tumor size, while high MALAT1 expression could predict short OS [43]. Compared with single lncRNA, lncRNA signature displayed stronger predictive effect for prognosis (Table 2). Most of these studies were based on lncRNAs in brain tissue, but some studies investigated the relationship between lncRNAs in serum and glioma prognosis. For example, Shen et al. found the expression level of HOTAIR and growth arrest-specific transcript 5 (GAS5) in serum were associated with survival, recurrence and progression in glioblastoma [44].
LncRNAs as therapeutic targets for glioma
lncRNA and signaling pathways
As mentioned above, the expression of lncRNAs in glioma differs greatly from that in normal tissues. Studies have confirmed that lncRNAs involved in various signaling pathways and play a role in diverse biological behaviors of glioma, such as proliferation, migration and invasion. Accordingly, lncRNAs can serve as therapeutic targets through regulating these signaling pathways (Fig. 2).
LncRNAs regulating important signaling pathways in glioma. lncRNAs can not only upregulate or activate the receptors on the surface of the cell membrane but also affect key moleculesin the signaling pathways, influecing the biological behavior of glioma. mTOR mammalian target of rapamycin, GSK-3 glycogen synthase kinase-3, LEF lymphoid enhancer factor; TCF, T-cell factor
LncRNAs in PI3K/Akt signaling pathway
Phosphoinositide 3-kinase (PI3K) can be activated by some growth factor receptors such as epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), fibroblast growth factor receptor (FGFR) as well as the insulin-like growth factor receptor (IGFR). The abnormal activation of PI3K and its downstream signaling pathways affects the apoptosis of cells, and has been suggested to be involved in the growth, metabolism, invasion and angiogenesis of glioma [59].
lncRNA LPP antisense RNA-2 (LPP-AS2) is a lncRNA, which is found upregulated in glioblastoma. It functions as a ceRNA and decoys for miR-7-5p to upregulate the expression of EGFR and activate the downstream PI3K/AKT/c-MYC pathway. Besides, LPP-AS2 is both directly and transcriptionally regulated by c-MYC, forming a positive feedback loop and promote tumorigenesis [60]. Similarly, lncRNA small nucleolar RNA host gene 16 (SNHG16) binds to miR-373-3p to regulate EGFR expression [61].
Phosphatase and tension homolog (PTEN) is a major tumor suppressor gene, which depletes levels of.
phosphoinositol 3,4, 5-triphosphate (PIP3) and downregulate PI3K [59]. Studies found lncRNA brain cytoplasmic RNA 1 (BCYRN1) [62], and DiGeorge Syndrome Critical Region Gene 5 (DGCR5) [63] function as ceRNAs, regulate PTEN expression and therefore play a tumor-suppressive role via PI3K/Akt pathway.
LncRNAs in Wnt/β-catenin signaling pathway
Wnt is a group of secreted glycoproteins. When Wnt signal is lost, cytoplasmic β‐catenin acts as an intercellular adhesion protein and degraded via the ubiquitination pathway. Once Wnt signal is activated, β‐catenin translocates from cytoplasm to nucleus, functions as a transcriptional coactivator and then regulates the expressions of target genes, such as c‐Myc and cyclin D1 [64]. Compared to normal tissues, expression of β-catenin is significantly higher in glioma tissues, which is also associated with higher histological malignancy grade and worse prognosis [65].
LncRNAs plays various roles in Wnt/β-catenin signaling pathway. LncRNA solute carrier family 8 member A1 antisense RNA 1 (SLC8A1-AS1) is highly upregulated in glioma tissues. It promotes proliferation, colony formation, migration, and invasion through activating Wnt/β‐catenin signaling [66]. Consistently, lncRNA ADAM Metallopeptidase with Thrombospondin Type 1 Motif 9 Antisense RNA 1 (ADAMTS9-AS1) deficiency attenuates glioma cell proliferation and induced glioma cell apoptosis proliferation and migration of glioma cells by suppressing Wnt/β‐catenin pathway [67]. Moreover, knockdown of lncRNA RP5-821D11.7 (lncRNA-RP5) negatively affects glioma proliferation, colony formation, migration and reduces epithelial-mesenchymal transition (EMT) through Wnt/β-catenin cascade [68]. Conversely, lncRNA ST7 antisense RNA 1 (ST7-AS1) expression is reduced in glioma tissues. ST7-AS1 overexpression downregulates polypyrimidine tract-binding protein 1 (PTBP1) expression, suppresses Wnt/β-catenin pathway and inhibits glioma progression, which might be a promising therapeutic target [69].
LncRNAs in Notch signaling pathway
Notch signaling consists of transmembrane receptors, transmembrane ligands and DNA binding proteins. Notch is hydrolyzed by proteases and releases Notch protein fragment such as Notch intracellular domain (NICD), and then binds to transcription factor CSL (CBF1, Suppressor of Hairless, Lag-1) to regulate downstream gene expression of both normal cells and tumor cells [70]. The most important physiological implication of the Notch pathway in glioma lies in its maintenance of glioma stem cells (GSCs). Activation of the Notch pathway can produce more NICD and then induce GSC differentiation and contribute to intra-tumor heterogeneity [71]. Long intergenic non-protein coding RNA 1410 (LINC01410) motivates Notch signaling pathway and accelerates the progression of glioma by sponging miR-506-3p and promoting NOTCH2 receptor [72]. Prostate cancer-up-regulated long noncoding RNA 1(PlncRNA-1) promotes cell proliferation and inhibits cell apoptosis via modulation of Notch pathway, indicating that lncRNAs play vital roles in the regulatory of Notch signaling pathway and have potential of becoming therapeutic targets.
lncRNA and therapeutic resistance
In all treatments of glioma like surgical resection, radiation and chemotherapy, therapeutic resistance is an inevitable problem. The therapeutic tolerance of gliomas is closely related to GSCs. GSCs have features of pluripotency and self-renewal and are capable of producing a variety of cell types that constitute the bulk of tumor [73]. After surgical resection, GSCs remaining are considered to be the main cause of glioma recurrence [74]. Similarly, CSCs are believed to be responsible for temozolomide (TMZ) resistance and become a source of new tumor cells [75]. CD133-expressing GSCs are enriched in glioma after radiation therapy, which were found to promote radiation resistance through various pathways such as the DNA damage checkpoint [76], Notch [77] and NF-κB [78].
There are a number of lncRNAs involving the self-renewal, proliferation and differentiation of GSCs. Fritah et al. analyzed the transcriptome changes of GSCs under the standard chemotherapy by TMZ and found that TMZ induced the expression of a large number of lncRNAs. The researchers also integrated thousands of molecular associations in databases and generate a gene regulatory network. 22 lncRNAs were extracted from the network, which involve in regulatory loops of drug response and have prognostic value in gliomas [79].
Notch signaling has received a lot of attention in promoting GSC self-renewal and suppressing GSC differentiation. It was found that Notch signaling regulates lncRNA taurine-upregulated gene 1(TUG1) expression in glioma, while TUG1 helps maintain the stemness of GSCs through antagonizing miR-145. Additionally, TUG1 targeting treatment induces GSC differentiation and inhibits cell proliferation in vivo [80]. Moreover, Several r lncRNAs such as SOX2 overlapping transcript (SOX2OT) [81], LINK00152 [82], TP73-AS1 [83] and HOTAIRM1 [84] also play critical roles in the malignant behavior of GSCs and considered to be potential therapeutic targets for glioma.
TMZ, as a kind of alkylating agent, is the first-line therapy for glioma. Unlike many other chemotherapeutic drugs, TMZ can readily cross the blood–brain barrier [85]. Its metabolic products can methylate the guanine residues, The methylated guanine, which cannot be repaired by DNA mismatch repair (MMR), and leads to replication-associated double-stranded DNA breaks, G2/M cell cycle arrest, and eventl apoptosis [86].
Unfortunately, temozolomide only extends survival by two months [87], and gliomas are always resistant to TMZ. The most classic mechanism of resistance to TMZ therapy is upregulation of the enzyme methylguanine-DNA methyltransferase (MGMT), which directly repairs methylated guanine. Several lncRNAs involved in the regulation of MGMT expression. Oncogene transforming growth factor beta1 (TGF-β1) is able to upregulate lncRNA H19 and HOXD-AS2, decrease miR-198 expression, and then promote temozolomide resistance and MGMT expression. [88]. Temozolomide-associated lncRNA in glioblastoma recurrence (lnc-TALC) can also increase MGMT expression by mediating the acetylation of H3K9, H3K27 and H3K36 in MGMT promoter regions through the c-Met/Stat3/p300 axis [89].
Extensive studies reported alterations in the DNA mismatch repair (MMR) system also conferred resistance to temozolomide [90, 91]. One research found that lncRNA X-inactive Specific Transcript (XIST) coregulates MMR and MGMT pathways at the same time. XIST directly targets miR-29c to regulate one of the key MMR proteins, MSH6, and downregulates MGMT expression simultaneously [92].
Given to their extensive involvement in transcriptional and post-transcriptional regulatory, lncRNAs may also regulate drug-resistance pathways independent from MGMT. LncRNA small nucleolar RNA host gene 12 (SNHG12) acts as a sponge for miR-129-5p, leading to anti-apoptosis and G1/S transition via the MAPK/ERK pathway [93]. CACS2 is a tumor-suppressive lncRNA that inhibits the proliferation of glioma cells and amplifies TMZ-induced repression of cell proliferation [94]. SET-binding factor 2 antisense RNA1 (SBF2-AS1) [95] and MALAT1 [96] also enhances chemoresistance to temozolomide.
As stated above, the resistance of glioblastoma to radiotherapy is similarly complex. Multiple signaling pathways collectively maintain the intrinsically-radioresistant glioma stem cell populations. [97]. In other tumors, lncRNAs meditate radioresistance through various mechanisms including repair of DNA damage, cell cycle arrest, apoptosis, CSCs regulation, EMT, and autophagy [98]. Liu et al. use clustered regularly interspaced short palindromic repeat interference (CRISPRi) to screen out nine lncRNAs sensitizing cells to radiation, named as lncRNA Glioma Radiation Sensitizers (lncGRS) [99]. The expression of lncRNA antisense hypoxia-inducible factor (AHIF) in glioblastoma augments under radiation, conferring the ability of vitality, invasion and radiation resistance on tumor cells. More importantly, the ability can be transmitted between tumor cells through exosomes [100]. Lnc-RI acts as a ceRNA by competitively binding to miR-193a-3p, stabilizes RAD51 mRNA and increases spontaneous DNA double-strand break (DSBs) repair levels [101]. Furthermore, TPTEP1 competitively interacting with mir-106a-5p to upregulate MaPK14 expression, thereby activating the P38 MaPK signaling pathway, and suppressing glioma stemness and radioresistance [102]. LINK-RA1 can stabilize the level of H2B K120 monoubiquitylation (H2Bub1), thus inhibiting the activation of autophagy and contributing to the radioresistance of glioma cells [103] (Fig. 3).
Role of lncRNAs in resistance to radiotherapy and chemotherapy in glioma. Role of lncRNAs in resistance to radiotherapy and chemotherapy in glioma. LncRNAs can mediate drug resistance of glioma through regulating the production of MGMT and the MMR system respectively. In radiotherapy, lncRNAs contribute to radiotherapy resistance by participating in double-strand break (DSBs) repair. The self-renewal of GSC is regulated by multiple lncRNAs, which have substantial effects in treatment resistance
Conclusions and future prospects
Since been discovered in the last century, lncRNAs were once thought to be transcriptional noise for a long time. With the report [104] of HOTAIR, the regulation of lncRNAs on gene expression has become a focus of research. In the field of glioma, researchers have explored many lncRNAs with diagnostic and therapeutic potential. Glioma lncRNAs can be transported by exosomes, cross the blood–brain barrier and easily detected in peripheral circulation. Moreover, lncRNAs have the features of disease specificity and cell type specificity.
Although substantial experimental evidence and bioinformatics analysis suggest that lncRNAs have great potential as biomarkers in glioma, translating basic research into clinical practice still faces many difficulties, which is also the direction for future investigation. Firstly, to date, none of the lncRNA specificity is sufficiently high to pass the requirements of the Tumor Marker Utility Grading System Levels of Evidence/NCCN for clinical application [105]. Secondly, technical standards for the extraction of lncRNA samples need to be established, and there is an absence of clinical research to obtain biological correlation, sensitivity, specificity, etc. Lastly, the concentration of lncRNA in peripheral blood is too low, and there lacks a rapid, economical and efficient approach for detection. Real-time polymerase chain reaction (RT-PCR) is the gold standard for RNA level measurement, but it requires expensive equipment and is highly sensitive to genomic DNA contamination [106]. Microarray-based method is not easily interfered by contamination, but does not involve the amplification of samples, resulting in low sensitivity [107]. New detection methods like nanosensors are highly anticipated. It's also required to determine whether the dysregulated lncRNA expression can be used as a predictor of tumorigenesis rather than just prognosis.
It’s exciting to make therapeutic targeting of lncRNAs in the clinic a reality. Nevertheless, better understanding of the off-target effects of nucleic acid therapeutics and potential toxicity is needed. CRISPR-Cas9 is another exciting technology that can be used to target lncRNAs, however, further research is needed to fully understand its effects and application.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
References
Ohgaki H, Kleihues P (2005) Epidemiology and etiology of gliomas. Acta Neuropathol 109(1):93–108
Louis DN et al (2016) The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Prados MD et al (2009) Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol 27(4):579–584
Kros JM et al (2015) Circulating glioma biomarkers. Neuro Oncol 17(3):343–360
Bonasio R, Shiekhattar R (2014) Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48:433–455
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43(6):904–914
Guttman M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227
Guenther MG et al (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130(1):77–88
Tay Y, Rinn J, Pandolfi PP (2014) The multilayered complexity of ceRNA crosstalk and competition. Nature 505(7483):344–352
Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330(6004):612–616
Spitale RC, Tsai MC, Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6(5):539–543
Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21(11):1253–1261
Derrien T et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789
Patel AP et al (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344(6190):1396–1401
Zachariah MA et al (2018) Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol 20(9):1155–1161
Hessels D et al (2003) DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 44(1):8–15 (discussion 15-6)
Groskopf J et al (2006) APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem 52(6):1089–1095
Noerholm M et al (2012) RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer 12:22
Arita T et al (2013) Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res 33(8):3185–3193
Zhang X et al (2012) Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis 48(1):1–8
Reon BJ et al (2016) Expression of lncRNAs in Low-Grade Gliomas and Glioblastoma Multiforme: An In Silico Analysis. PLoS Med 13(12):e1002192
Sun Y, Jing Y, Zhang Y (2021) Serum lncRNA-ANRIL and SOX9 expression levels in glioma patients and their relationship with poor prognosis. World J Surg Oncol 19(1):287
Li X, Zhang H, Wu X (2019) Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene 686:1–7
Mei JC, Yan G, Mei SQ (2020) Diagnostic and prognostic potentials of long noncoding RNA ELF3-AS1 in glioma patients. Dis Mark 2020:8871746
Wu X et al (2021) LncRNA GAS8-AS1 downregulates lncRNA NEAT1 to inhibit glioblastoma cell proliferation. Brain Behav 11(6):e02128
Tan SK et al (2018) Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer 17(1):74
Zhao WH et al (2019) Association between expression of HOTAIR and invasiveness of gliomas, and its predictive value. Adv Clin Exp Med 28(9):1179–1183
Hua X et al (2019) LINK-A lncRNA participates in the pathogenesis of glioma by interacting with survivin. Exp Ther Med 18(3):1581–1586
Chen L et al (2020) Overexpression of LncRNA PSMG3-AS1 distinguishes glioblastomas from sarcoidosis. J Mol Neurosci 70(12):2015–2019
Fang J, Huang J (2019) Clinical significance of the expression of long non-coding RNA PVT1 in glioma. Cancer Biomark 24(4):509–513
Yuan Q et al (2020) Analysis of long noncoding RNA ZNF667-AS1 as a potential biomarker for diagnosis and prognosis of glioma patients. Dis Markers 2020:8895968
Verhaak RG et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1):98–110
Zhang JX et al (2013) HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol 15(12):1595–1603
Wang Q et al (2016) A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett 373(2):251–259
Chinot OL et al (2013) Response assessment criteria for glioblastoma: practical adaptation and implementation in clinical trials of antiangiogenic therapy. Curr Neurol Neurosci Rep 13(5):347
Wen PY et al (2010) Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 28(11):1963–1972
Brandsma D et al (2008) Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol 9(5):453–461
de Wit MC et al (2004) Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology 63(3):535–537
Taal W et al (2008) Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer 113(2):405–410
Kruser TJ, Mehta MP, Robins HI (2013) Pseudoprogression after glioma therapy: a comprehensive review. Expert Rev Neurother 13(4):389–403
Li J et al (2020) FAM225B Is a Prognostic lncRNA for Patients with Recurrent Glioblastoma. Dis Markers 2020:8888085
Li J et al (2020) Prognostic and clinicopathological significance of long non-coding RNA in glioma. Neurosurg Rev 43(1):1–8
Zhou Q et al (2018) lncRNAs as potential molecular biomarkers for the clinicopathology and prognosis of glioma: a systematic review and meta-analysis. Gene 668:77–86
Shen J et al (2018) Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog 57(1):137–141
Lin JZ, Lin N, Zhao WJ (2020) Identification and validation of a six-lncRNA prognostic signature with its ceRNA networks and candidate drugs in lower-grade gliomas. Genomics 112(5):2990–3002
Yang X et al (2021) Identification of an epithelial-mesenchymal transition-related lncRNA prognostic signature for patients with glioblastoma. Sci Rep 11(1):23694
Li X, Meng Y (2019) Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer 19(1):813
Maimaiti A et al (2021) Identification and validation of a novel eight mutant-derived long non-coding RNAs signature as a prognostic biomarker for genome instability in low-grade glioma. Aging (Albany NY) 13(11):15164–15192
Maimaiti A et al (2021) Identification and validation of an individualized prognostic signature of lower-grade glioma based on nine immune related long non-coding RNA. Clin Neurol Neurosurg 201:106464
Huang K et al (2021) Development and Validation of an Mesenchymal-Related Long Non-Coding RNA Prognostic Model in Glioma. Front Oncol 11:726745
Qiu X et al (2021) Development and Validation of an Immune-Related Long Non-coding RNA Prognostic Model in Glioma. J Cancer 12(14):4264–4276
Pan YB et al (2020) Prognostic and predictive value of a long non-coding rna signature in glioma: a lncRNA expression analysis. Front Oncol 10:1057
Wen J et al (2020) Identification and verification on prognostic index of lower-grade glioma immune-related LncRNAs. Front Oncol 10:578809
Zheng J et al (2021) A prognostic ferroptosis-related lncRNAs signature associated with immune landscape and radiotherapy response in glioma. Front Cell Dev Biol 9:675555
Wang Y et al (2020) Identification of a glycolysis-related LncRNA signature to predict survival in diffuse glioma patients. Front Oncol 10:597877
Zhao J, Wang L, Wei B (2020) Identification and validation of an energy metabolism-related lncRNA-mRNA signature for lower-grade glioma. Biomed Res Int 2020:3708231
Gao WZ et al (2018) Identification of a multidimensional transcriptome signature for survival prediction of postoperative glioblastoma multiforme patients. J Transl Med 16(1):368
Song L et al (2019) Genome-wide identification of lncRNAs as novel prognosis biomarkers of glioma. J Cell Biochem 120(12):19518–19528
Shahcheraghi SH et al (2020) Wnt/beta-catenin and PI3K/Akt/mTOR signaling pathways in glioblastoma: two main targets for drug design: a review. Curr Pharm Des 26(15):1729–1741
Zhang X et al (2020) Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J Exp Clin Cancer Res 39(1):196
Zhou XY et al (2020) lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics 112(1):1021–1029
Mu M et al (2020) LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene 39(45):6879–6892
He Z et al (2020) LncRNA DGCR5 plays a tumor-suppressive role in glioma via the miR-21/Smad7 and miR-23a/PTEN axes. Aging (Albany NY) 12(20):20285–20307
He L et al (2019) Wnt/β-catenin signaling cascade: a promising target for glioma therapy. J Cell Physiol 234(3):2217–2228
Denysenko T et al (2016) WNT/β-catenin signaling pathway and downstream modulators in low- and high-grade glioma. Cancer Genomics Proteomics 13(1):31–45
He L et al (2021) Knockdown of long non-coding RNA SLC8A1-AS1 attenuates cell invasion and migration in glioma via suppression of Wnt/β-catenin signaling pathways. Brain Res Bull 176:112–120
Zhou C et al (2021), LncRNA ADAMTS9-AS1 knockdown suppresses cell proliferation and migration in glioma via down-regulating Wnt/β-catenin signaling pathway. Bosn J Basic Med Sci
Younis M, Shaikh S, Shahzad KA (2021) Long non-coding RNA RP5–821D11.7 promotes proliferation, migration, and epithelial-mesenchymal transition in glioma and glioma stem-like cells. Prog Biophys Mol Biol, 2021.
Sheng J et al (2021) p53-targeted lncRNA ST7-AS1 acts as a tumour suppressor by interacting with PTBP1 to suppress the Wnt/β-catenin signalling pathway in glioma. Cancer Lett 503:54–68
Wu Y, Qian Z (2019) Long non-coding RNAs (lncRNAs) and microRNAs regulatory pathways in the tumorigenesis and pathogenesis of glioma. Discov Med 28(153):129–138
Parmigiani E, Taylor V, Giachino C (2020) Oncogenic and Tumor-Suppressive Functions of NOTCH Signaling in Glioma. Cells 9(10):15
Zhao X, Shen F, Yang B (2021) LncRNA LINC01410 Induced by MYC Accelerates Glioma Progression via Sponging miR-506-3p and Modulating NOTCH2 Expression to Motivate Notch Signaling Pathway. Cell Mol Neurobiol
Piccirillo SG, Vescovi AL (2007) Brain tumour stem cells: possibilities of new therapeutic strategies. Expert Opin Biol Ther 7(8):1129–1135
Nduom EK, Hadjipanayis CG, Van Meir EG (2012) Glioblastoma cancer stem-like cells: implications for pathogenesis and treatment. Cancer J 18(1):100–106
Chen J et al (2012) A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488(7412):522–526
Bhat KPL et al (2013) Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24(3):331–346
Bao S et al (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444(7120):756–760
Wang J et al (2010) Notch promotes radioresistance of glioma stem cells. Stem Cells 28(1):17–28
Fritah S et al (2020) Temozolomide-Induced RNA Interactome Uncovers Novel LncRNA Regulatory Loops in Glioblastoma. Cancers (Basel) 12(9):15
Katsushima K et al (2016) Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun 7:13616
Su R et al (2017) Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer 16(1):171
Yu M et al (2017) Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer 16(1):110
Zhang B et al (2021) Long non-coding RNA TP73-AS1 is a potential immune related prognostic biomarker for glioma. Aging (Albany NY) 13(4):5638–5649
Xia H et al (2020) Long Noncoding RNA HOTAIRM1 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells Through Regulation of HOX Gene Expression. Neurotherapeutics 17(2):754–764
Schreck KC, Grossman SA (2018) Role of temozolomide in the treatment of cancers involving the central nervous system. Oncology (Williston Park) 32(11):555–560
Strobel H et al (2019) Temozolomide and Other Alkylating Agents in Glioblastoma Therapy. Biomedicines 7(3):9
Stupp R et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996
Nie E et al (2021) TGF-beta1 modulates temozolomide resistance in glioblastoma via altered microRNA processing and elevated MGMT. Neuro Oncol 23(3):435–446
Wu P et al (2019) Lnc-TALC promotes O(6)-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat Commun 10(1):2045
Cahill DP et al (2007) Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 13(7):2038–2045
Stark AM et al (2015) Expression of DNA mismatch repair proteins MLH1, MSH2, and MSH6 in recurrent glioblastoma. Neurol Res 37(2):95–105
Du P et al (2017) LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci Rep 37(5):47
Lu C et al (2020) DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer 19(1):28
Liao Y et al (2017) LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J Cell Biochem 118(7):1889–1899
Zhang Z et al (2019) Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer Res 38(1):166
Kim S-S et al (2018) Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res 46(3):1424–1440
Ou A, Yung WKA, Majd N (2020) Molecular Mechanisms of Treatment Resistance in Glioblastoma. Int J Mol Sci 22(1):45
Zhu J et al (2019) Molecular mechanisms of lncRNAs in regulating cancer cell radiosensitivity. Biosci Rep 39(8):9
Liu SJ et al (2020) CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol 21(1):83
Dai X et al (2019) AHIF promotes glioblastoma progression and radioresistance via exosomes. Int J Oncol 54(1):261–270
Shen L et al (2018) LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res 46(2):717–729
Tang T et al (2020) lncRNA TPTEP1 inhibits stemness and radioresistance of glioma through miR106a5pmediated P38 MAPK signaling. Mol Med Rep 22(6):4857–4867
Zheng J et al (2020) Linc-RA1 inhibits autophagy and promotes radioresistance by preventing H2Bub1/USP44 combination in glioma cells. Cell Death Dis 11(9):758
Rinn JL et al (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129(7):1311–1323
Febbo PG et al (2011) NCCN task force report: evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw 9(5):S1-32 (quiz S33)
Chen M et al (2021) A novel biosensor for the ultrasensitive detection of the lncRNA biomarker MALAT1 in non-small cell lung cancer. Sci Rep 11(1):3666
Szilágyi M et al (2020) Circulating cell-free nucleic acids: main characteristics and clinical application. Int J Mol Sci 21(18):56
Acknowledgements
We acknowledge the National Natural Science Foundations of China, Natural Science Foundations of Heilongjiang, the “Chunhui Plan” of the Ministry of Education, Distinguished Young Foundations of the First Affiliated Hospital of Harbin Medical University and subject "The effects of Nrf2/HO-1 signaling pathway inhibition on antiglioma effect of arsenious acid" for financial support.
Funding
This work was supported by the National Natural Science Foundations of China (81971135); Natural Science Foundations of Heilongjiang (YQ2020H014); the “Chunhui Plan” of the Ministry of Education (HLJ2019009); Distinguished Young Foundations of the First Affiliated Hospital of Harbin Medical University (HYD2020JQ0014); "The effects of Nrf2/HO-1 signaling pathway inhibition on antiglioma effect of arsenious acid"(JDFYK-2019002).
Author information
Authors and Affiliations
Contributions
XX and YL had the idea for the article and drafted the work; IG performed the literature search; YL helped to draw the figures; Rui Liu helped to polish up the revised manuscript; NW and GY critically revised the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors confirm that this article content has no conflict of interest.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
Not applicable.
Humans and animal rights
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, X., Liang, Y., Gareev, I. et al. LncRNA as potential biomarker and therapeutic target in glioma. Mol Biol Rep 50, 841–851 (2023). https://doi.org/10.1007/s11033-022-08056-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08056-y