Abstract
Cryptococcus species is an opportunistic yeast pathogen and classified into different molecular types according to typing techniques including multilocus sequence typing (MLST). The study aimed to investigate the genotypes of environmental Cryptococcus isolates using MLST and the relationship between the in vitro antifungal susceptibility and sequence types of isolates. Genotyping Cryptococcus isolates was performed by the MLST method at seven nuclear loci. Antifungal susceptibility was determined by using CLSI broth micro-dilution method for amphotericin B, fluconazole, itraconazole, voriconazole, flucytosine, and luliconazole. Seven sequence types (ST) were detected using MLST analysis, with the most frequent (50%) ST77, followed by ST4 (16.7%) among 30 C. neoformans isolates. All antifungals demonstrated excellent activity against isolates, except for itraconazole and amphotericin B that were non-wild type against 53.3% and 10% of isolates, respectively. Although seven sequence types belonging to C. neoformans isolates were detected, ST77 was the main sequence type in Ahvaz. Also, non-wild type isolates were only found against itraconazole and amphotericin B.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Cryptococcus contains haploid encapsulated yeasts that belong to the division Basidiomycota, and they can cause life-threatening infections in immunocompromised individuals [1,2,3]. This genus was initially classified based on capsular epitopes and serological tests into four serotypes (A to D) [1], two species (Cryptococcus neoformans and C. gattii), and three varieties, including C. neoformans var. grubii (serotype A), C. neoformans var. neoformans (serotype D), C. gattii var. gattii (serotype B and C) [4]. According to the revised taxonomy in 2015, this genus has been now categorized into seven species include C. neoformans, C. deneoformans, C. gattii, C. deuterogattii, C. bacillisporus, C. tetragattii, and C. decagattii [5]. Out of them, C. neoformans (previously C. neoformans var. grubii) is the most common species and has a worldwide geographic distribution. It is frequently isolated from pigeon excreta, and less commonly from other sources such as soil, tree, and woody debris [6,7,8].
Since 2009, the multi-locus sequence typing (MLST) technique has been adopted by the International Society for Human and Animal Mycology (ISHAM) working group as a standard method for studying fungal genetic diversity, molecular epidemiology, and ancestral evolution. MLST and PCR-based typing techniques caused extensive and essential changes in the classification of C. neoformans [9], so that C. neoformans includes three molecular types VNI, VNII, and VNB for C. neoformans and VN IV for C. deneoformans [5, 10].
Both amphotericin B and 5-flucytosine (5FC) are considered as the gold standard for treating cryptococcal infections. In addition, azoles, especially fluconazole, are also used in the disease's maintenance and prevention phase [11]. However, resistance to azoles and 5FC has been reported both in vitro and in vivo studies, which can be due to clinical resistance and differences in molecular types [12, 13]. In the present study, we analyzed C. neoformans isolates by MLST using seven genetic loci and our genotypes’ association with other countries’ genotypes was evaluated. The antifungal susceptibility of isolates was evaluated against six antifungals. Due to the lack of information about C. neoformans sensitivity to luliconazole, its sensitivity results were compared with other routine drugs.
Materials and methods
Strains of Cryptococcus neoformans
In this study, 30 isolates of C. neoformans (C. neoformans var. grubii) with guano pigeon origin were investigated. All isolates were previously isolated using niger seed agar from pigeon droppings samples from private houses and pet-shops in different areas in Ahvaz (a city in the southwest of Iran, capital of Khuzestan province) [14]. Isolates were identified using molecular methods and sequences data deposited in GenBank (Accession numbers; LC535977-80, LC535983-90, LC535993-5, LC536010, LC536012-3, LC536015-6, LC537134, LC537136-9, LC537153, LC545844-5, LC536002, LC536004) [14]. All isolates were kept in distilled water at room conditions in the department of Medical Mycology, Ahvaz Jundishapur University of medical sciences.
DNA extraction
DNA extraction of isolates was performed by the described method of Makimura et al., with some modification [15]. Briefly, isolates were subcultured on Sabouraud dextrose agar (SDA) (Biolife, Italia) at 35 °C overnight. Yeast colonies were collected in the sterile microtubes contain 50 mg glass beads (Sigma—Aldrich, USA) and 300 µL lysis buffer and stored at − 20 °C for 24 h. Then microtubes contents were homogenized by a SpeedMill PLUS Homogenizer (Analytikjena, Germany). Supernatants were removed and DNAs extracted using phenol-chloroform-isoamyl alcohol (Sigma—Aldrich, Germany) and kept at − 20 °C.
MLST genotyping
For MLST genotyping, six unlinked genes including CAP59, GPD1, LAC1, PLB1, SOD1, URA5, and the IGS1 non-coding region were used, as described by the ISHAM consensus MLST scheme for the C. neoformans species complexes (http://mlst.mycologylab.org/). The PCR conditions for CAP59, GPD1, SOD1, URA5 and the IGS1 loci were performed as described by Meyer et al. [16]. The PCR conditions for the LAC1 locus were modified, as 94 °C 4 min; 30 cycles: 94 °C 30 s, 61.5 °C 30 s, 72 °C 1 min. Also, the conditions of 12 cycles: 65 °C 30 s with a 2 °C step-down every two cycles 95 °C 2 min; 95 °C 30 s, 65 °C 30 s, 72 °C 1 min; followed by 20 cycles: 95 °C 30 s, 65 °C 30 s, 72 °C 1 min were used for PLB1 locus. Amplification was performed using Red master mix (Amplicon, Denmark) and PCR products sequenced using the primers presented by Meyer et al. [16]. All sequencing results from each gene were aligned using the Clustal W algorithm at MEGA V.6.06 software available at http://megasoftware.net after being manually edited using the software Chromas v. 2.6.6 available at http://technelysium.com.au/ChromasPro.html.
All 210 sequences (7 loci for 30 isolates) were deposited at GenBank and accession numbers were obtained, accession numbers, LC598352–LC598381 for CAP59; accession numbers, LC598535–LC598564 for URA5; accession numbers, LC598565–LC598594 for SOD1; accession numbers, LC598595–LC598624 for PLB1; accession numbers, LC598625–LC598654 for LAC1; accession numbers, LC598655–LC598684 for IGS1; accession numbers LC598685–LC598714 for GPD1. Then, allele type (AT) and sequence types (ST) were identified by loading and comparing sequences on the C. neoformans MLST database, a global database of sequence information from C. neoformans available at http://mlst.mycologylab.org/.
Phylogenetic analysis
The UPGMA algorithm was implemented in the BioNumeric program to confirm evolutionary relationships in all sequenced types, show allelic profiles, and compare allelic profiles' drug sensitivity. The allelic profiles of the isolates were used to generate a minimum spanning tree including isolate from China, Thailand, Japan, Brazil, India, Hong Kong, Indonesia, Kuwait, Qatar and Italy. Data for these countries were extracted from published papers [9, 17,18,19].
Nucleotide diversity
In this study several indexes such as number of polymorphic sites (S), nucleotide diversity (p), number of haplotypes (h), haplotype diversity (Hd), and average number of nucleotide differences (k) were calculated by DNAsp 6.12.03 software available at (http://www.ub.edu/dnasp/) to assess the extent of DNA polymorphisms. Also, three tests, Tajima's D, Fu & Li's D, and Fu & Li's F for neutrality were calculated using this software. A negative or positive D value suggests purifying selection/population size expansion or balancing selection/a decrease in population size, respectively. The phenomenon of recombination inside the C. neoformans isolates was calculated using Watterson's estimate per sequence (Өs) at DNAsp 6.12.03 and genetic disequilibrium method by Fstat software available at https://www2.unil.ch/popgen/softwares/fstat.htm.
Antifungal susceptibility
Antifungal susceptibility tests were performed against amphotericin B (Sigma—Aldrich, Germany), fluconazole (Serva, USA), itraconazole (Sigma—Aldrich, Germany(, voriconazole (Sigma—Aldrich, Germany), 5FC (Sigma—Aldrich, Germany), and luliconazole (APIChem Technology, China) based on broth microdilution of clinical and laboratory standards institute (CLSI) M27 4th protocol [20]. Antifungals were prepared in dimethyl sulfoxide (DMSO, Merck, Germany) in a two-fold dilution series including 0.00781–16 μg/ml for amphotericin B, 0.01562–32 μg/ml for fluconazole, 0.00195–4 μg/ml for itraconazole and flucytosine, 0.00097–2 μg/ml for voriconazole, and 0.000244–0.5 μg/ml for luliconazole.
A yeast suspension equivalent to 0.5 McFarland in sterile saline solution (0.85%) was prepared from a 48 h culture of C. neoformans. Then, a concentration of 1–5 × 103 cells/mL was prepared from each yeast suspension in RPMI 1640 (Gibco, UK). 100 μL of yeast suspension and 100 µL of serial dilutions of each antifungal were added into each microplate well. A drug-free well (positive control) and yeast-free (negative control) were included in the test. Microplates were incubated at 35 °C for 48–72 h and then the minimum inhibitory concentrations (MIC) of each antifungal were detected for isolates. The MIC that inhibited 50% (MIC50) and 90% (MIC90), and MICGeometric (GM) were also calculated. There are not defined breakpoints for used antifungals against Cryptococcus species. As a result, epidemiological cut-off values (ECVs) were calculated by M59 guideline as follows: ≥ 0.5 μg/ml for amphotericin B, ≥ 8 μg/ml for fluconazole and flucytosine, and ≥ 0.25 μg/ml for itraconazole and voriconazole [21].
Results
Multilocus sequence type
Using the MLST analysis, seven sequence types including ST77, ST4, ST31, ST55, ST57, ST58, and ST324 were detected among 30 C. neoformans isolates. The most prevalent genotype was ST77 in 15 (50%) isolates, followed by ST4 in 5 (16.7%) isolates. The allelic profile of all sequence types is shown in Fig. 1. All sequence types were previously found in other countries such as China (ST31, ST57), India (ST31, ST4, ST77), Thailand (ST31, ST4), Brazil (ST31, ST77), German (ST77, ST58), Japan (ST31, ST4), Italy (ST55) and Indonesia and Hong Kong (ST4) [9, 18, 19].
Phylogenetic analysis
According to UPGMA phylogenetic algorithm, two separate clusters were detected among our C. neoformans VNI isolates based on similarity in allelic profiles. The first cluster consisted of 2 sequence types and three isolates, whereas the second cluster supported five sequence types and 27 isolates. There was also a 46.8% similarity between the two clusters. The highest similarity (85.7%) was seen between ST77 and ST31 which differ only in two nucleotides at the IGS1 locus. In contrast, ST324 in cluster 1 had the highest divergence compared to other sequence types (Fig. 1).
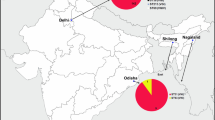
The MStree analysis shows that most of our sequence types are similar to Chinese, Italian, and Brazilian sequence types. However, they are genetically descended from India and, to a lesser extent, from Kuwait. Sequence type 324 and sequence type 58 are ancestrally close to sequence types from Italy with a distant genetic affinity with ancestral sequence types in India (Fig. 2).
Minimum spanning trees showing sequence type relationships among the different geographic regions and our data Cryptococcus neoformans isolates. The circle size and the branch thickness indicate the number of isolates and the evolutionary relationship between the isolates, respectively. Different color represents different geographic areas. (The type of STs and the ST numbers added in supplementary file)
Nucleotide diversity
In this study, 18 allele types for seven MLST gene regions were detected based on the MLST database. These allele types are as follows; 5 allele types for LAC1, 3 allele types for PLB1, 3 allele types for IGS1, 3 allele types for URA5, 2 allele types for CAP59, 1 allele type for GPD1, and 1 allele type for SOD1 loci (Fig. 1). The highest polymorphism was observed in LAC1 (46 sites polymorphisms), whereas two loci, GPD1 and SOD1, were monomorphic. According to three indexes h, Hd, and π, the highest and lowest genetic diversity was observed in the LAC1 gene region (h = 7, Hd = 0.680, π = 0.01329) and CAP59 (h = 2, Hd = 0.129, π = 0.00023) respectively (Table 1).
According to Tajima's D test, D value in all loci except GPD1 and IGS1 was negative and significant (≤ 0.05). It is evidence of purifying selection or population size expansion in these five loci. In comparison, evidence of balancing selection or a decrease in population size was observed in two other loci. The recombination phenomenon was analyzed using three tests, theta Watterson (Өs), Pairwise Homoplasy Index (PHI), and the minimum number of recombination events (Rm) (Table 1). Based on these tests, three recombinations have occurred within our study population with the most recombinant, and mutation in the LAC1 locus (Rm = 1 and Өs = 8.330). In contrast, recombination in the concatenated dataset (intergenic) was not observed in our population (PHI, P = 1). Furthermore, the linkage disequilibrium test between pairs of loci did not show a statistical significance for any loci except LAC1 and IGS1. These results could be evidence of recombination within our isolates and rejection of the null hypothesis (Table 1).
Antifungal susceptibility testing
The susceptibility of 30 C. neoformans isolates to six antifungal agents is presented in Table 2. Based on defined ECVs by CLSI M59 guidelines, all isolates were found to be wild-type phenotype (WT) to fluconazole, voriconazole, and flucytosine. It is found that 53.3% and 10% isolates were non-wild type phenotype (non-WT) to itraconazole and amphotericin B, respectively. Although there was no relationship between sequence types and the non-WT phenotype to itraconazole and amphotericin B, it is important to note that the non-WT phenotypes were observed in four sequence types, ST58, ST31, ST77, and ST4 (Fig. 1).
Discussion
Unfortunately, despite the development of genotypic methods, very limited studies have been conducted on the genotypic diversity of Cryptococcus spp. in clinical and environmental isolates in some geographical regions. Most clinical available reports in Iran are as case reports [2, 22, 23], and environmental epidemiological studies are limited to formal identification and/or consequent PCR and sequencing of the ITS or IGS1 regions or PCR–RFLP [24, 25], and AFLP method by Pakshir et al. [26]. Molecular techniques have shown that there is no geographical difference in the distribution of Cryptococcus complex species, however, VNI and VGI are the predominant molecular types for C. neoformans and C. gattii, respectively [27]. Therefore, in the present study, we analyzed the MLST characteristics of 30 C. neoformans isolates by the ISHAM-MLST consensus scheme.
Our results showed moderate genetic variation by the existence of seven sequence types from 30 isolates in Ahvaz, Iran, with the dominance of ST77. ST77 is widespread in other geographical regions of the world. Most environmental isolates of C. neoformans in the Andrade–Silva study belonging to ST77 [28]. Prakash et al. in molecular epidemiology in India, reported ST77 as the second most common sequence type [3]. Also, this sequence type was included in half of the environmental isolates from Brazil [6], reflecting the fact that ST77 has adapted to many geographic niches, both environmental and clinical and can cause infection due to recovering this sequence type from environmental and clinical samples. ST4 was the second most frequent sequence type in our study. This sequence type was reported in Thailand, Japan, India, Hong Kong, Indonesia, Kuwait, Qatar [19]. Although no sequence type from Iran has been reported so far, a study showed the presence of ST4, ST5, ST23, ST69, ST93, ST174, ST175, ST185, and ST192 in Kuwait and ST4, ST5, ST31 in Qatar (neighbor countries of Iran) [17]. Only two of these sequence types (ST4 and ST31) overlap with what was obtained in this study. The most common sequence types (ST31, ST77, ST55, and ST57) were directly originated from the Indian isolates. ST324 and ST58 have a genetic distance from other isolates, and originated from Italian isolates. ST4 is similar to Kuwait, Qatari, Japanese, and Indonesian isolates. A glance at these limited data suggests that there is likely abundant genetic diversity of C. neoformans in Iran and all isolates have Indian ancestor.
Three recombination events were found in concatenated sequences using Rm analysis and a pairwise genotypic disequilibrium test detected recombination phenomenon among LAC1 and IGS1 loci. The nucleotide diversity in the LAC1 locus can be attributed to the adaptation of the fungus to environmental conditions (cold and heat) [9]. Similarly, Cogliati et al. illustrated that environmental isolates of Cryptococcus have a higher nucleotide diversity at the LAC1 locus than clinical isolates [9]. The results of the PHI test also did not show a significant difference, which refers to the clonal nature of our C. neoformans population structure. Finally, taken together, these results may indicate recombination within our isolates and/or suggest non-meiotic reproduction [19].
From 2018, ECVs have been described for several antifungals drug against C. neoformans in the CLSI guideline. According to this guideline, all our isolates (100%) were found to be WT (susceptible) to fluconazole, voriconazole, and flucytosine. Moreover, 53.3% and 10% of isolates were non-WT (resistant) to itraconazole and amphotericin B, respectively. Although our results were consistent with Rocha et al. in amphotericin B and fluconazole susceptibility [29], in contrast, we demonstrated resistance to itraconazole in 53.3% isolates. Furthermore, Chen et al. found similar results with us for the three antifungals fluconazole, voriconazole, and 5FC, but they found that all isolates were sensitive to amphotericin B and itraconazole in contrast with our findings that 10% and 53.3% of isolates were non-WT to amphotericin B and itraconazole, respectively [18].
Conclusions
Overall, the differences in ECVs were not associated with sequence types. However, except for ST77 and ST4, most sequence types were limited to one or two isolates, and the possibility of correct evaluation and correlation of sequence type with drug profile was weak. Luliconazole is a new imidazole, and no ECVs or breakpoints have been reported in either the global CLSI or EUCAST protocols. Although the MIC range of luliconazole was considerably lower than other studied azoles, the MICs obtained for C. neoformans were much higher than the MICs obtained for others studied filamentous fungi and Candida spp., so far [30,31,32].
References
Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG (2006) Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172(4):2223–2238. https://doi.org/10.1534/genetics.105.046672
Hashemi R, Majidi A, Tabatabaey A, Motamed H (2014) Fatal disseminated cryptococcus infection in an immunocompetent patient. Arc Clin Infect Dis 9(3):e20246. https://doi.org/10.5812/archcid.20246
Prakash A, Sundar G, Sharma B, Hagen F, Meis JF, Chowdhary A (2020) Genotypic diversity in clinical and environmental isolates of Cryptococcus neoformans from India using multilocus microsatellite and multilocus sequence typing. Mycoses 63(3):284–293. https://doi.org/10.1111/myc.13041
Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, Abeln EC et al (2001) Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147(4):891–907. https://doi.org/10.1099/00221287-147-4-891
Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR et al (2017) The case for adopting the “species complex” nomenclature for the etiologic agents of Cryptococcosis. mSphere. https://doi.org/10.1128/mSphere.00357-16
Ferreira-Paim K, Andrade-Silva L, Fonseca FM, Ferreira TB, Mora DJ, Andrade-Silva J et al (2017) MLST-based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV patients in Southeastern Brazil. PLoS Negl Trop Dis 11(1):e0005223. https://doi.org/10.1371/journal.pntd.0005223
Cogliati M, Zamfirova RR, Tortorano AM, Viviani MA, Network FC (2013) Molecular epidemiology of Italian clinical Cryptococcus neoformans var. grubii isolates. Med Mycol 51(5):499–506. https://doi.org/10.3109/13693786.2012.751642
Alves GSB, Freire AKL, Bentes AdS, Pinheiro JFdS, de Souza JVB, Wanke B et al (2016) Molecular typing of environmental Cryptococcus neoformans/C. gattii species complex isolates from Manaus, Amazonas, Brazil. Mycoses 59(8):509–515. https://doi.org/10.1111/myc.12499
Cogliati M, Desnos-Ollivier M, McCormick-Smith I, Rickerts V, Ferreira-Paim K, Meyer W et al (2019) Genotypes and population genetics of Cryptococcus neoformans and Cryptococcus gattii species complexes in Europe and the mediterranean area. Fungal Genet Biol 129:16–29. https://doi.org/10.1016/j.fgb.2019.04.001
Munoz M, Camargo M, Ramirez JD (2018) Estimating the intra-taxa diversity, population genetic structure, and evolutionary pathways of Cryptococcus neoformans and Cryptococcus gattii. Front Genet 9:148. https://doi.org/10.3389/fgene.2018.00148
Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ et al (2010) Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis 50(3):291–322. https://doi.org/10.1086/649858
Beale MA, Sabiiti W, Robertson EJ, Fuentes-Cabrejo KM, O’Hanlon SJ, Jarvis JN et al (2015) Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across Southern Africa. PLoS Negl Trop Dis 9(6):e0003847. https://doi.org/10.1371/journal.pntd.0003847
Hong N, Chen M, Xu N, Al-Hatmi AM, Zhang C, Pan WH et al (2019) Genotypic diversity and antifungal susceptibility of Cryptococcus neoformans isolates from paediatric patients in China. Mycoses 62(2):171–180. https://doi.org/10.1111/myc.12863
Moslem M, Fatahinia M, Kiasat N, Zarei Mahmoudabadi A (2020) Predominance of Cryptococcus neoformans var. grubii in Ahvaz, molecular identification and evaluation of virulence factors. Jundishapur J Microbiol 13(11):e112408. https://doi.org/10.5812/jjm.112408
Makimura K, Murayama SY, Yamaguchi H (1994) Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol 40(5):358–364. https://doi.org/10.1099/00222615-40-5-358
Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC et al (2009) Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47(6):561–570. https://doi.org/10.1080/13693780902953886
Thanh LT, Phan TH, Rattanavong S, Nguyen TM, Duong AV, Dacon C et al (2018) Multilocus sequence typing of Cryptococcus neoformans var. grubii from Laos in a regional and global context. Med Mycol 57(5):557–565. https://doi.org/10.1093/mmy/myy105
Chen Y-H, Yu F, Bian Z-Y, Hong J-M, Zhang N, Zhong Q-S et al (2018) Multilocus sequence typing reveals both shared and unique genotypes of Cryptococcus neoformans in Jiangxi province, China. Sci Rep 8(1):1495. https://doi.org/10.1038/s41598-018-20054-4
Khayhan K, Hagen F, Pan W, Simwami S, Fisher MC, Wahyuningsih R et al (2013) Geographically structured populations of Cryptococcus neoformans variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS ONE 8(9):e72222. https://doi.org/10.1371/journal.pone.0072222
CLSI (2017) Reference method for broth dilution antifungal susceptibility testing of yeasts; CLSI standard M27, 4th edn. Clinical and Laboratory Standards Institute, Wayne
CLSI (2018) Epidemiological cutoff values for antifungal susceptibility testing; CLSI supplement M59, 2nd edn. Clinical and Laboratory Standards Institute, Wayne
Gharabaghi MA, Allameh SF (2014) Primary pulmonary cryptococcosis. Case Rep. https://doi.org/10.1136/bcr-2014-203821
Badali H, Alian S, Fakhim H, Falahatinejad M, Moradi A, Mohammad Davoudi M et al (2015) Cryptococcal meningitis due to Cryptococcus neoformans genotype AFLP 1/VNI in Iran: a review of the literature. Mycoses 58(12):689–693. https://doi.org/10.1111/myc.12415
Mirhendi S, Kordbacheh P, Kazemi B, Samiei S, Pezeshki M, Khorramizadeh M (2001) A PCR-RFLP method to identification of the important opportunistic fungi: Candida species, Cryptococcus neoformans, Aspergillus famigatus and Fusarium solani. Iran J Pub Health 30(3–4):103–106
Afshari SAK, Shokohi T, Aghili R, Badali H (2012) Epidemiology and molecular characterization of Cryptococcus neoformans isolated from pigeon excreta in Mazandaran province, northern Iran. J Med Mycol 22(2):160–166. https://doi.org/10.1016/j.mycmed
Pakshir K, Fakhim H, Vaezi A, Meis JF, Mahmoodi M, Zomorodian K et al (2018) Molecular epidemiology of environmental Cryptococcus species isolates based on amplified fragment length polymorphism. J Mycol Med 28(4):599–605. https://doi.org/10.1016/j.mycmed.2018.09.005
Mihara T, Izumikawa K, Kakeya H, Ngamskulrungroj P, Umeyama T, Takazono T et al (2013) Multilocus sequence typing of Cryptococcus neoformans in non-HIV associated cryptococcosis in Nagasaki, Japan. Med Mycol 51(3):252–260. https://doi.org/10.3109/13693786.2012.708883
Andrade-Silva LE, Ferreira-Paim K, Ferreira TB, Vilas-Boas A, Mora DJ, Manzato VM et al (2018) Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PLoS ONE 13(3):e0193237. https://doi.org/10.1371/journal.pone.0193237
Rocha DFS, Cruz KS, da Silva Santos CS, Menescal LSF, da Silva Neto JR, Pinheiro SB et al (2018) MLST reveals a clonal population structure for Cryptococcus neoformans molecular type VNI isolates from clinical sources in Amazonas, Northern-Brazil. PLoS ONE 13(6):e0197841. https://doi.org/10.1371/journal.pone.0197841
Abastabar M, Rahimi N, Meis JF, Aslani N, Khodavaisy S, Nabili M et al (2016) Potent activities of novel imidazoles lanoconazole and luliconazole against a collection of azole-resistant and-susceptible Aspergillus fumigatus strains. Antimicrob Agents Chemother 60(11):6916–6919. https://doi.org/10.1128/AAC.01193-16
Taghipour S, Kiasat N, Shafiei S, Halvaeezadeh M, Rezaei-Matehkolaei A, Mahmoudabadi AZ (2018) Luliconazole, a new antifungal against Candida species isolated from different sources. J Med Mycol 28(2):374–378. https://doi.org/10.1016/j.mycmed.2017.11.004
Moslem M, Mahmoudabadi AZ (2020) The high efficacy of luliconazole against environmental and otomycosis Aspergillus flavus strains. Iran J Microbiol 12(2):170
Acknowledgements
This study was part of a Ph.D. thesis (Maryam Moslem) supported by Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. We would like to thank the medical mycology department, Ahvaz Jundishapur University of Medical Sciences, for their support.
Funding
The Infectious and Tropical Diseases Research Centre, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences supported this study (OG: 9836).
Author information
Authors and Affiliations
Contributions
Study concept and design, AZM and MF; sampling, isolation, conducting the experiments, MM; data analysis, MM, and NK; drafting of the manuscript, MM; Critical editing, AZM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no potential conflicts of interest.
Ethical approval
The ethical committee approved this project of Ahvaz Jundishapur University of Medical Sciences (IR.AJUMS.REC.1398.648).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Moslem, M., Fatahinia, M., Kiasat, N. et al. Genotypic diversity of Iranian Cryptococcus neoformans using multilocus sequence typing (MLST) and susceptibility to antifungals. Mol Biol Rep 48, 4201–4208 (2021). https://doi.org/10.1007/s11033-021-06433-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06433-7