Abstract
A role of Retinol Binding Protein-4 (RBP4) in insulin resistance is widely studied. However, there is paucity of information on its receptor viz., Stimulated by Retinoic Acid-6 (STRA6) with insulin resistance. To address this, we investigated the regulation of RBP4/STRA6 expression in 3T3-L1 adipocytes exposed to glucolipotoxicity (GLT) and in visceral adipose tissue (VAT) from high fat diet (HFD) fed insulin-resistant rats. 3T3-L1 adipocytes were subjected to GLT and other experimental maneuvers with and without vildagliptin or metformin. Real-time PCR and western-blot experiments were performed to analyze RBP4, STRA6, PPARγ gene and protein expression. Adipored staining and glucose uptake assay were performed to evaluate lipid and glucose metabolism. Oral glucose tolerance test (OGTT) and Insulin Tolerance Test (ITT) were performed to determine the extent of insulin resistance in HFD fed male Wistar rats. Total serum RBP4 was measured by quantitative sandwich enzyme-linked immunosorbent assay kit. Adipocytes under GLT exhibited significantly increased RBP4/STRA6 expressions and decreased insulin sensitivity/glucose uptake. Vildagliptin and metformin not only restored the above but also decreased the expression of IL-6, NFκB, SOCS-3 along with lipid accumulation. Furthermore, HFD fed rats exhibited significantly increased serum levels of RBP4 along with VAT expression of RBP4, STRA6, PPARγ, IL-6. These molecules were significantly altered by the vildagliptin/ metformin treatment. We conclude that RBP4/STRA6 pathway is primarily involved in mediating inflammation and insulin resistance in adipocytes and visceral adipose tissues under glucolipotoxicity and in insulin resistant rats.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retinol binding protein-4 (RBP4) is a liver and adipocyte specific transport protein, playing a major role in mediating insulin resistance and inflammation [1]. Increased mRNA and protein levels of RBP4 along with higher expression of gluconeogenic enzymes has been reported in insulin resistant GLUT4-KO mice models [2]. Molecular basis of these mechanisms was not fully understood; however, it was shown later that RBP4 mediate its action via its membrane receptor viz., Stimulated by retinoic acid 6 (STRA6), which activates JAK2/STAT5 pathway and thereby inhibit insulin signalling [3, 4]. Study on another RBP4 receptor, RBPR2 suggests a possible role of it in mediating metabolic actions of RBP4 in liver and fat tissues [5]. In vivo studies using STRA6 null mice also showed that STRA6 contributes to high fat diet-induced insulin resistance [3]. Another study reported that STRA6 null mice exhibits decreased body weight, fat mass, leptin and insulin levels which might be attributed to the increased insulin sensitivity in STRA6−/− mice [6]. One study has also suggested that STRA6 is very crucial for adipogenesis, and cells depleted with the STRA6 lost their adipogenicity even in the presence of RBP4 [7]. This implies that STRA6 may be necessary for RBP4 to mediate insulin resistance and STRA6 activation is linked to inhibition of insulin receptor responses in white adipose tissue.
“Asian Indian Phenotype” refers to the unique clinical and bio-chemical abnormalities in Indians that includes high insulin resistance and abdominal adiposity despite lower body mass index leading to increased vulnerability for the development of diabetes and cardiovascular disorders [8,9,10]. One of our earlier studies demonstrated increased circulatory levels of RBP4 linked to insulin resistance, obesity, impaired glucose tolerance (IGT) as well as type 2 diabetes mellitus (T2DM) [11]. While there are studies claiming the involvement of RBP4 and STRA6 in insulin resistance and glucose homeostasis, there is still lot more to be explored regarding their mechanistic action(s) and what downstream molecules takes part in this process. Also, there is a lack of knowledge on the effect of the existing anti-diabetic drugs specifically—Metformin (an insulin sensitizer) and Vildagliptin (an insulin secretagogue) on RBP4/STRA6 alterations. Earlier, both these drugs were shown to inhibit transcriptional activities of PPARα & PPARγ [12] and RBP4 promoter was shown to be activated by the transcription factors like cAMP, PPARα and PPARγ [13]. Therefore, in the present study, we are investigating the regulation of RBP4/STRA6 expression in 3T3-L1 adipocytes exposed to glucolipotoxicity and in visceral adipose tissue (VAT) from high fat diet (HFD) fed insulin-resistant rats in the absence and presence of vildagliptin or metformin.
Materials and methods
3T3-L1 fibroblasts culture, differentiation and treatment regimen
3T3-L1 mouse fibroblasts were procured from ATCC (USA) and were induced to differentiate into mature adipocytes by standard procedures [14] and we have used cells with passage number less than 20. Briefly, cells were grown to 80% confluence in DMEM (Gibco, Thermofisher Scientific, USA) containing 10% fetal bovine serum (FBS). Differentiation was induced by incubating the cells in fresh DMEM containing 1 µM dexamethasone, 10 µg/ml insulin (Millipore Sigma, USA), 0.5 mM isobutylmethylxanthine (IBMX) (Millipore Sigma, USA), and 10% FBS for 3 days. Cells were then maintained for another 6 days in DMEM with 10% FBS, and 10 µg/ml insulin. The cells used for experimentation were more than 80% differentiated as determined visually.
For glucolipotoxicity (GLT) treatment, cells were serum starved for 4 h and then exposed to high glucose (25 mM) and palmitate (500 µM) for 6 h. Palmitate was prepared by complexing with albumin. Briefly, sodium palmitate was dissolved in pre-heated NaOH (0.1 M) and then complexed with 10% BSA at 55 °C as described elsewhere [15, 16]. Cells were treated with vildagliptin (100 nM) and metformin (1 mM) as reported previously [17, 18] (Glenmark Pharmaceuticals, Mumbai, India) along with GLT. For the experiments performed with PPARγ inhibitor-GW9962, cells were treated with vildagliptin and GW9962 (10 µM) separately for 6 h.
Quantitative real-time PCR
Total RNA was prepared from the adipocytes using TRIzol reagent (TaKaRa, Japan) according to the manufacturer’s instructions. Complementary DNA was prepared from the isolated RNA by reverse transcribing 1 µg of total RNA using reverse transcriptase enzyme (Thermoscientific, Waltham, USA), and random hexamer primers (IDT, USA). Real time quantitative PCR was performed on a Light-cycler (Roche, Basel, Switzerland) with appropriate cycle conditions using SYBR Premix Ex Taq II (TaKaRa, Japan) and using appropriate sets of forward and reverse primers (Supplementary Tables 1 and 2). Relative quantification of gene transcripts was calculated using 2−ΔΔCt method and normalized to β-actin transcripts. Negative controls were run as well to ensure the absence of contamination.
Western blot analysis
Cells were harvested from the culture plates using RIPA lysis buffer (Thermoscientific, Waltham, USA) supplemented with protease and phosphatase inhibitors. Equal amount of protein (30 µg of protein) was loaded per lane and were separated by SDS-PAGE; transferred to Polyvinylidene difluoride (PVDF) membrane (Millipore, Burlington, USA); and incubated with specific primary (overnight) and secondary antibodies (1 h) dissolved in TBST. Primary antibodies for phospho-AKT (ser473), and AKT were purchased from Cell signaling (Danvers, USA); RBP4 and STRA6 were procured from Enzo Life Science (Farmingdale, USA) and Novus Biologicals (Littleton, USA), respectively. adiponectin, TNFα, IL-6, MCP-1, GLUT-4 were purchased from Santa Cruz (Dallas, USA). PPARγ was purchased from Abcam, Cambridge. HRP-conjugated secondary antibodies were purchased from Santa Cruz (Dallas, USA). Bands were visualized using enhanced chemiluminescent reagent using BioRad gel dock instrument (BioRad, Hercules, USA). Images of the band intensity were quantified using Image J software. Data are represented as relative protein expression i.e., the ratio of test protein and β-actin.
Adipored staining
Differentiated 3T3-L1 cells were treated with GLT in the presence and absence of vildagliptin or metformin for 24 h. Then the cells were washed twice with PBS and fixed with 4% paraformaldehyde for 30 min followed by three times PBS wash and permeabilized using 0.25% triton X-100. Following permeabilization, cells were washed again with PBS, and incubated with adipored (Lonza, Basel, Switzerland) or 15 min in the dark. Finally, cells were washed with PBS and mounted on the slides using DAPI-mounting medium (Fluoroshield with DAPI, MilliporeSigma, St. Louis, USA). The fluorescent images were obtained using a Confocal Laser Scanning microscope (Carl-Zeiss LSM 700, Oberkochen, Germany) with a × 60 objective.
Glucose uptake assay
Equal number of cells were propagated in 12-well plates (2 × 104 cells/well) and differentiated with standard protocol. Cells were treated with GLT for 6 h with and without vildagliptin or metformin, prior to the start of glucose uptake experiment. 2-NBDG glucose uptake was performed based on the method described previously [19]. Briefly, cells were washed with Krebs–Ringer Buffer (KRB) without glucose (supplemented with 0.1% BSA and CaCl2), and stimulated with 100 nM insulin for 30 min in appropriate wells except control. Cells were then treated with 40 µM 2- 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl) Amino)-2-Deoxyglucose (NBDG, BioVision, San Francisco, USA) in the same media for 1 h. After incubation, free 2-NBDG was washed off with KRB buffer from the cells. Finally, cells were then lysed with lysis buffer (20 mM Tris, 1% Sodium deoxycholate, 40 mM KCL) and gently homogenized with 100% DMSO. The contents of the lysed cells were transferred to 96-well fluorescence plate and read at excitation wavelength of 485 nm and emission wavelength of 535 nm.
Animal study
All animal experiments were carried out in compliance with National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) as well as with the regulations specified by the Committee for the Purpose of Control and Supervision on Experiments on Animals (CPCSEA, Govt. of India) and approved from the Institutional Animal Ethics Committee. Male Wistar rats (10–12 weeks of age) were divided into five groups (n = 6 each) and blood glucose were measured at the beginning of the study. The groups comprised of rats fed with (1) Normal Pellet diet (NPD), (2) High fat diet (HFD; 65% Kcal from fats), (3) HFD + Vildagliptin (3 mg/kg of body weight) (Prophylactic), (4) HFD + Vildagliptin (Therapeutic) (3 mg/kg of body weight). (5) HFD + Metformin (Therapeutic) (150 mg/kg of body weight). Dosage was selected as per the previous study [20]. Prophylactic treatment started along with HFD treatment, before the onset of insulin resistance and was given for 24 weeks Therapeutic treatment started after the onset of insulin resistance in Wistar rats and was given for 16 weeks. NPD and HFD diets were custom-made and procured from the National Institute of Nutrition (NIN), Hyderabad, India. At 24th week, fasting blood samples were collected to estimate the biochemical markers and then the animals were euthanized and visceral adipose tissue (VAT) were excised and stored at − 80 °C. Gene expression and protein expression studies were performed as explained above. Biochemical assays were performed by standard protocols.
Insulin tolerance test (ITT) and oral glucose tolerance test (OGTT)
Animals were characterized as insulin resistant by insulin tolerance test (ITT) and as glucose intolerant by oral glucose tolerance test (OGTT) [21]. Both ITT and OGTT were performed 60 days after HFD treatment. For ITT, rats were fasted for 4 h, and insulin (Insulatard-0.5 U/kg/BW) was administered intraperitoneally followed by the measurement of blood glucose concentration at different time points. For OGTT, rats were fasted for 16 h followed by oral administration of glucose (2.0 g/kg/BW). The blood was collected from the tail vein and blood glucose concentration was measured at different time points (30, 60 and 120 min) by one touch glucose analyzer (USA).
RBP4 estimation
Total serum RBP4 (sum of all the RBP4 isoforms including holo-RBP4 and apo-RBP4 present in the circulation) was measured by quantitative sandwich enzyme-linked immunosorbent assay (ELISA) (Cusabio, China) according to the manufacturer’s instructions. The values are expressed in ng/ml. The intra- and inter-assay coefficients of variation were < 5% and < 10%, respectively.
Statistical analysis
Differences were evaluated by one-way analysis of variance (ANOVA) using GraphPad Prism, Version 5 (GraphPad Software Inc., CA, USA) and a p value < 0.05 was considered as statistically significant.
Results
Vildagliptin and metformin down-regulate GLT-induced RBP4 and STRA6 expression in 3T3-L1 adipocytes: involvement of PPARγ
We first studied the effects of GLT on RBP4, STRA6, and PPARγ gene expression in 3T3-L1 adipocytes. We found that GLT significantly increased RBP4, STRA6, and PPARγ mRNA expression levels while vildagliptin or metformin treatment reduced their expressions in the GLT treated adipocytes (Fig. 1A–C). Consistent with the mRNA expression data, protein expression of RBP4, STRA6, and PPARγ were also increased in GLT treated cells, while vildagliptin and metformin treatment significantly (p < 0.05) reduced these modulations made by GLT (Fig. 1D, E).
mRNA expression patterns of RBP4 (A), STRA6 (B), & PPARγ (C) in 3T3-L1 adipocytes under different experimental conditions. Representative protein expression blot for RBP-4, STRA-6, and PPARγ along with β-Actin (D). Cumulative protein expression data of RBP4, STRA6, and PPARγ (E). *p < 0.05, compared to their corresponding Control., #p < 0.05, compared to their corresponding GLT. F and G 3T3-L1 cells were differentiated and treated with GW9962 (10 µM) or Vildagliptin (100 nM) for 6 h. Representative protein expression blot for PPAR-γ, RBP-4 and STRA-6 along with β-Actin (F). Cumulative protein expression data of PPARγ, RBP-4, and STRA-6 (G). Results are expressed as mean ± SEM of minimum three independent determinations. *p < 0.05, compared to their corresponding Control
Since both vildagliptin and metformin treatment was effectively found to reduce both PPARγ and RBP4 protein expression in 3T3-L1 adipocytes in the presence of GLT, we next examined whether PPARγ instigate any effect on RBP4 expression. We used specific PPARγ inhibitor-GW996 and observed that GW996 significantly reduced (p < 0.05) both PPARγ and RBP4 protein expression in 3T3-L1 adipocytes compared to untreated cells (Fig. 1F, G), suggesting that downregulation of RBP4 expression by vildagliptin could be by inhibiting PPARγ. We also observed that both GW996 and vildagliptin were able to down-regulate STRA6 protein expression; however, the effect of GW996 on STRA6 could not reach statistical significance (Fig. 1F, G).
Effect of vildagliptin and metformin on GLT induced inflammation in 3T3-L1 adipocytes
Since RBP4 is linked to inflammatory network, we also studied several inflammatory signatures along with RBP4. We observed that there was significant increase in the pro-inflammatory mediators viz., TNFα (2A), IL-6 (2B), NFκB (2C), and SOCS-3 (2D) by the GLT treatment. Treatment with vildagliptin and metformin significantly (p < 0.05) reduced GLT induced mRNA expression of these pro-inflammatory markers. (Fig. 2A–D). Consistent with mRNA data, protein expression profiles of TNFα and IL-6 were also up-regulated in GLT treated cells (Fig. 2E). Treatment with vildagliptin or metformin significantly reduced (p < 0.05) protein expression of TNFα whereas IL-6 protein expression was significantly reduced (p < 0.05) only by metformin treatment (Fig. 2E, F).
mRNA expression patterns of TNFα (A), IL-6 (B), NFκB (C), and SOCS-3 (D) in 3T3-L1 adipocytes in different experimental conditions. Protein expression patterns of TNFα, and IL-6 along with β-Actin (E). Cumulative protein expression data of TNFα and IL-6 in 3T3-L1 adipocytes under different experimental conditions (F). Results are expressed as mean ± SEM of minimum three independent determinations. *p < 0.05, compared to their corresponding Control
Effect of vildagliptin and metformin on glucose uptake and lipid accumulation in 3T3-L1 adipocytes
While insulin stimulated glucose uptake was impaired in adipocytes subjected to GLT treatment, both vildagliptin and metformin significantly increased the insulin stimulated glucose uptake in adipocytes even in the presence of GLT (Fig. 3A). While metformin was well known for its insulin sensitising effects in adipocytes [17], vildagliptin role was still not tested in this regard. We observed that vildagliptin synergistically enhanced pAKT expression in the presence of insulin in 3T3-L1 adipocytes (Fig. 3B, C) which emphasizes that vildagliptin stimulation of glucose uptake in 3T3-L1 adipocytes might be mediated by increased pAKT expression.
Insulin stimulated glucose uptake in 3T3-L1 adipocytes (A). *p < 0.05, compared to control; #p < 0.05, compared to INS; @p < 0.05, compared to GLT + INS. Results are expressed as mean ± SEM of minimum three independent experiments. Representative protein blot of pAKT (B) and cumulative protein expression data of pAKT (C) in 3T3-L1 adipocytes under different experimental conditions *p < 0.05, compared to − ve Insulin, #p < 0.05, compared to + ve Insulin, δp < 0.05, compared to Vilda + Insulin. mRNA expression patterns of PGC-1α (D) in 3T3-L1 adipocytes under different experimental conditions. Adipored staining of 3T3-L1 adipocytes under different experimental conditions (E). Mean fluorescence intensity (F). *p < 0.05, compared to control; #p < 0.05, compared to GLT. Results are expressed as mean ± SEM of minimum three independent experiments
Apart from inflammation, impaired lipid accumulation is another factor for the induction of insulin resistance in adipocytes. PPARγ and PGC-1α are two important genes that promote fatty acid synthesis and lipid accumulation in adipocytes. Both PPARγ and its co-activator PGC-1α gene expression were increased in the presence of GLT, while vildagliptin or metformin treatment significantly (p < 0.05) reduced both PPARγ and PGC-1α gene expression (Figs. 1C and 3D) in 3T3-L1 adipocytes. Besides these, vildagliptin or metformin was also found to significantly (p < 0.05) reduce lipid accumulation as evident from the adipored staining in 3T3-L1 adipocytes under GLT treatment (Fig. 3E, F).
Clinical characteristics of the study animals and assessment of insulin tolerance test (ITT) and oral glucose tolerance test (OGTT)
Clinical and biochemical status of the study groups is provided in Supplementary Table 3. Significant elevation (p < 0.05) of body weight, fasting plasma glucose (FPG), total cholesterol, and serum triglyceride levels were observed in HFD fed insulin resistant rats compared to normal pellet diet (NPD) group. Prophylactic treatment with vildagliptin which is concurrently started with HFD significantly reduced serum cholesterol and triglyceride levels, however, no change was observed in FPG levels in prophylactic vildagliptin treatment group. Therapeutic treatment with vildagliptin significantly reduced FPG and serum triglyceride levels compared to HFD group, whereas metformin treatment significantly reduced cholesterol levels. No treatment has significantly altered HDL cholesterol levels. A significant reduction (p < 0.05) in body weight was observed in all the treatment groups compared to the HFD group.
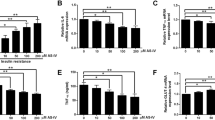
To study the changes in insulin sensitivity, we measured the dynamic characteristics of blood glucose after insulin injection in rats fed with NPD (or) HFD for 12 weeks by [20] insulin tolerance test (ITT). Compared to NPD animals, glucose disposal curve depicts a sharp increase (p for trend, < 0.05) in glucose values in HFD animals when injected with the same concentration of insulin confirming the loss of insulin sensitivity (Fig. 4A).
Blood glucose values during insulin tolerance test (ITT) in NPD and HFD condition at different time points (all values are mean ± SEM) (A). Glucose excursion pattern measured during oral glucose tolerance test (OGTT) in NPD and HFD condition under the experimental condition at different time points (all values are mean ± SEM) (B). C Serum levels of RBP4 in rats treated with different drugs. Data presented as mean ± SEM. *p < 0.001, compared to NPD; #p < 0.05, compared to HFD
Oral glucose tolerance test (OGTT) was employed to determine the glycaemic responses to exogenously administered glucose in rats fed with NPD (or) HFD diet before starting the treatment regime. Compared to the NPD group, rats fed with HFD exhibited more glucose levels at all the time points during OGTT (p for trend, < 0.05), demonstrating that HFD-fed animals become glucose-intolerant (Fig. 4B).
In vitro relevance of our findings central to the role of RBP4/STRA6 are also demonstrated in high-fat fed diabetic rats from different treatment groups. Total serum RBP4 levels were significantly increased (p < 0.05) in HFD fed rats compared to the NPD group (Fig. 4C). A significant reduction (p < 0.05) in RBP4 levels were observed in the prophylactic vildagliptin treatment group compared to HFD group. Therapeutic vildagliptin and metformin treated groups were also exhibited reduced serum RBP4 levels compared to the HFD group; albeit this could not reach statistical significance.
Expression levels of RBP4, PPARγ and other pro-inflammatory genes in the visceral adipose tissue (VAT) of the study rats
mRNA expression of RBP4, STRA6, TNFα, IL-6, PPARγ, FASN, ATGL, and HSL were found to be significantly (p < 0.05) up-regulated in VAT of HFD fed insulin resistant, glucose intolerant rats (Fig. 5A–H). Prophylactic vildagliptin administration significantly (p < 0.05) reduced RBP4, TNFα, IL-6, PPARγ, ATGL, & HSL (data not significant) gene expression in VAT of HFD fed rats. Therapeutic treatment with vildagliptin and metformin significantly (p < 0.05) reduced the expression of RBP4, STRA6, TNFα, and IL-6 gene expression in VAT of HFD fed rats. Therapeutic treatment with vildagliptin is more beneficial in regulating PPARγ and FASN gene expression and controlling lipid accumulation in VAT from HFD fed rats, whereas therapeutic metformin treatment regulated lipolysis more efficiently in HFD fed rats as evident by the reduced expression of ATGL and HSL expression in VAT.
mRNA expression patterns of RBP4 (A), STRA6 (B), TNFα (C), IL-6 (D), PPARγ (E), FASN (F), ATGL (G), & HSL (H) normalized to β-Actin in study animals. *p < 0.05, compared to NPD; #p < 0.05, compared to HFD. I and J Visceral adipose tissue specific protein expression blots of RBP4, PPARγ, GLUT4, IL-6 along with β-Actin (I). Cumulative Protein expression data of RBP4, PPARγ, GLUT-4, and IL-6 of minimum six study animals (J). Results are expressed as mean ± SEM. *p < 0.05, compared to their corresponding NPD; #p < 0.05, compared to their corresponding HFD
Consistent with mRNA expression, HFD-fed animals also showed increased protein expression of RBP4, PPARγ, and IL-6 in VAT samples. Both prophylactic & therapeutic vildagliptin and metformin treatment reduced RBP4 protein expression albeit without statistical significance (Fig. 5I). Increased PPARγ protein expression in VAT from HFD-fed rats was significantly reduced by all the test agents. IL-6 protein expression was also significantly reduced (p < 0.05) by both therapeutic vildagliptin and metformin treatment in HFD-fed rats (Fig. 5I, J). Protein expression of GLUT4 was significantly (p < 0.05) down-regulated in HFD-fed animals whereas all test agents significantly (p < 0.05) up-regulated and normalized GLUT4 protein expression indicating insulin sensitizing role of vildagliptin and metformin in VAT from the HFD fed rats (Fig. 5I, J).
Discussion
The following are the nut-shell findings of our study: (1) 3T3-L1 adipocytes exposed to glucolipotoxicity exhibited increased RBP4/STRA6 expression and decreased insulin sensitivity as evident from decreased levels of pAKT and glucose uptake. Vildagliptin and Metformin treatment significantly restored the above signaling alterations. (2). HFD fed insulin resistant rats exhibited significantly increased serum levels of RBP4 as well as their VAT showed significant increase in the expression levels of RBP4, STRA-6 and PPARγ along with augmentation of inflammatory markers and impaired levels of GLUT4. Interestingly, all the above mediators of inflammation and insulin resistance were significantly reduced in VAT from the rats treated with either vildagliptin or metformin. The differences in mRNA and protein expression states in certain molecular signatures could be due to post-transcription/post-translational modifications.
Elevated levels of RBP4 have been shown linked to insulin resistance in both animal models and humans [1, 22, 23] with the involvement of JAK-STAT pathway as well as PPARγ in mediating insulin resistance by RBP4 [2]. Increased levels of RBP4 have been shown associated with insulin resistance, obesity and T2DM in different ethnic populations [24, 25]. A recent prospective study also reported association of serum RBP4 levels and risk of incident T2DM in individuals with prediabetes [26]. Interestingly, it has been identified that STRA6 is a membrane receptor of RBP4, which is responsible for mediating downstream effects of RBP4. However, RBP4 has been shown to mediate insulin resistance by STRA6-independent [27, 28] or STRA6-dependent pathways [29, 30]. Genetic studies showed that single nucleotide polymorphisms in STRA6 are associated with T2DM [31], raising the possibility that STRA6 plays an important role in metabolic disorders like type 2 diabetes. In addition, STRA6 has been proposed to play a role in insulin resistance on the basis of data from adipose specific STRA6 knockout mouse [5]. Our in vitro work supports this as 3T3-L1 adipocytes treated with GLT exhibited increased expression of both RBP4 and STRA6.
Since, elevated RBP4 and STRA6 are associated with altered metabolic states and insulin resistance, lowering RBP4 and STRA6 would be therapeutically beneficial. We studied the effect of anti-diabetic drugs, vildagliptin which is an insulin secretagogue and metformin, an insulin sensitizer. Both these drugs are very effective in treating hyperglycaemia, but their role in reducing obesity associated inflammation and lipid accumulation especially in adipocytes is poorly understood. Interestingly in our study, both vildagliptin and metformin treatment down-regulates GLT induced RBP4 mRNA and protein expression, thus alleviating insulin resistance in 3T3-L1 adipocytes.
PPARγ is an important adipogenic transcription factor involved in promoting lipid accumulation and differentiation of adipocytes [32]. Heterozygous PPARγ-deficient mice are protected from HFD induced or aging-induced adipocyte hypertrophy, obesity, and insulin resistance [33]. Interestingly, PPARγ is also known to act as a potent transcriptional activator of RBP4 gene promoter [11]. We observed that in addition to the action as a RBP4 repressor, both vildagliptin and metformin also reduced PPARγ gene expression in adipocytes. This indicates that PPARγ might play a role in regulation of RBP4 expression in 3T3-L1 adipocytes. On co-treatment of PPARγ inhibitor-GW9962 with vildagliptin in our experiment, we observed that GW9962 treatment down-regulated RBP4 expression in 3T3-L1 adipocytes. This implies that vildagliptin down-regulation of RBP-4 expression could be partly mediated by PPARγ in adipocytes; however, this need to be verified in gene-silencing/KO experiments.
RBP4 is also considered as a pro-inflammatory signature, as increased expression of RBP4 has been shown associated with activation of T-cell as well as Antigen presenting cells (APCs) [34, 35]. RBP4 also stimulates expression of pro-inflammatory cytokines by activating c-JNK and TLR4 pathways in human and mouse macrophages [28]. However, a study reported by Sell & Eckel, shows that RBP4 production by human adipocytes were down-regulated by exogenous treatment of TNF-α [29]. In this context, we observed that both vildagliptin and metformin are effective in reducing inflammation in 3T3-L1 adipocytes by reducing transcription of TNFα, IL-6 and NFκB molecules. A recent study has also reported the beneficial effects of metformin in lowering age-related inflammation, where it has been shown how metformin could activate autophagy to enhance the activity of anti-inflammatory regulatory T-cells (Tregs) and thereby mitigate inflammation [36]. This study is consistent with our observation where the levels of RBP4 and other inflammatory cytokines were significantly reduced by metformin and this might have been mediated by adipocytes associated Tregs cells. The reduction in inflammation along with the reduced expression of RBP4, has positive effects on insulin sensitivity in adipocytes, although the exact down-stream molecules that could mediate this process are not yet fully identified.
In previous studies, it has been shown that RBP4/STRA6 up-regulation was associated with the reduced expression of pAKT and phosphorylated insulin receptor (pIR) both in adipocytes and hepatocytes [1, 2]. In the present study, we reported that vildagliptin has a synergistic effect on insulin stimulated pAKT expression in 3T3-L1 adipocytes and this might be originated from the reduced expression of RBP4 under vildagliptin treatment. The increased pAKT expression is also consistent with the increased glucose uptake under vildagliptin treatment regime, emphasizing an insulin sensitizing role of vildagliptin in 3T3-L1 adipocytes.
Lipid metabolism which comprehends lipogenesis and lipolysis is an important aspect of adipose tissue physiology. An improvement in lipid homeostasis is associated with both reduction in inflammation and increased insulin sensitivity [37,38,39]. We reported reduced accumulation of lipid droplets in mature 3T3-L1 adipocytes treated with vildagliptin or metformin. Reduction in lipid accumulation is also accompanied by decreased mRNA expression of PPARγ as well as PGC-1α in vildagliptin and metformin treated cells. Thus, our study in 3T3-L1 adipocytes emphasizes that vildagliptin and metformin could beneficially alter RBP4/STRA6 signalling in reducing inflammation and improving lipid and glucose homeostasis.
Consistent with our in vitro 3T3-L1 adipocytes work, similar findings were seen in our pre-clinical HFD fed insulin resistant rats. As HFD is known to mediate insulin resistance in animal models [19], we also observed that HFD fed rats displayed loss of insulin sensitivity and become glucose intolerant. Moreover, HFD fed rats also displayed increased circulatory levels of RBP4, higher expression of visceral adipose tissue specific RBP4 and PPARγ along with increased expression of pro-inflammatory cytokines compared to VAT from normal pellet diet (NPD) fed animals. Previous studies have also corroborated this finding that RBP4 circulatory levels are elevated in HFD fed insulin resistant mice, and links elevated RBP4 levels to increased hepatic PEPCK expression [21].
Prophylactic vildagliptin treatment seems to be more effective than therapeutic vildagliptin as prophylactic treatment significantly reduced both RBP4 mRNA as well as circulatory RBP4 levels in the HFD fed animals. In the therapeutic treatment groups, in-spite of a significant reduction of PPARγ in VAT, RBP4 reduction was seen as a trend without statistical significance. This shows that, RBP4 might be differentially regulated independent of PPARγ in VAT of HFD fed rats, and the reduction in circulatory RBP4 in HFD fed animals might be acting through liver, as a major fraction of serum RBP4 comes from the liver secretion. This can be a limitation of our study that we couldn’t able to test the role of liver-specific RBP4 in regulating insulin resistance and type 2 diabetes, but will be an exciting avenue for us to test this hypothesis in future.
In addition to above observations, treatment with vildagliptin and metformin significantly reduced VAT specific expression of pro-inflammatory cytokines TNFα, and IL-6 which was found to be up-regulated during HFD induction of insulin resistance and glucose intolerance. Vildagliptin or Metformin is also found to significantly up-regulate GLUT4 expression in VAT of HFD-fed rats indicating the insulin sensitizing effects of these drugs and our observation were also consistent with the previous reports [40, 41]. Reduction in adipose tissue inflammation along with RBP4 expression as well as regulation of lipid homeostasis by vildagliptin is our study is also an important observation. Very recently, Circulating RBP4 levels were shown inversely and independently correlated with β-cell function across the spectrum of glycemia [42]. Therefore, it would be worth to explore whether the beneficial insulin secretagogue action of vildagliptin could be operated by regulation of RBP4 in pancreatic beta cells.
Our study emphasizes that RBP4/STRA6 pathway as one of the players in mediating inflammation and insulin resistance in adipocytes under glucolipotoxicity as well as in VAT from the HFD fed insulin resistant rats. One of the major significant finding that has emerge from the study that both vildagliptin and metformin resists several signatures of inflammation and improves insulin sensitivity by regulating lipid accumulation and glucose uptake in 3T3-L1 adipocytes as well as in VAT from the HFD-fed rats. However, we could not establish any mechanistic insights that can prove RBP4/STRA6 induce insulin resistance via inflammatory responses, as this requires generation of robust knockout animal and cell line models through advance genome editing techniques.
Conclusion
In conclusion, as RBP4 is a known molecule for mediating inflammation and insulin resistance in insulin responsive tissues, we suggest that the beneficial regulation of RBP4/STRA6 signaling by vildagliptin and metformin extends their newer modes of action which needs to be harnessed by future investigations of clinical setting.
Data availability
Data is available and will be provided on reasonable request.
Code availability
Not applicable.
Abbreviations
- RBP4:
-
Retinol Binding Protein-4
- STRA6:
-
Stimulated by retinoic Acid 6
- GLT:
-
Glucolipotoxicity
- VAT:
-
Visceral Adipose Tissue
- NPD:
-
Normal Pellet Diet
- HFD:
-
High Fat Diet
- JAK:
-
Janus Kinase
- STAT:
-
Signal Transducer and activator of Transcription
- PPARα/γ:
-
Peroxisome proliferator-activated receptor alpha/gamma
- IBMX:
-
Isobutyl methylxanthine
- cAMP:
-
Cyclic adenosine 3ʹ,5ʹ-cyclic monophosphate
References
Berry DC, Jin H, Majumdar A, Noy N (2011) Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci USA 108:4340–4345. https://doi.org/10.1073/pnas.1011115108
Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM et al (2005) Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 436:356–362. https://doi.org/10.1038/nature03711
Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O’Byrne SM, DeSantis D et al (2013) The STRA6 receptor is essential for retinol-binding protein induced insulin resistance but not for maintaining vitamin a homeostasis in tissues other than the eye. J Biol Chem 288:24528–24539. https://doi.org/10.1074/jbc.M113.484014
Berry DC, Noy N (2012) Signaling by vitamin A and retinol-binding protein in regulation of insulin responses and lipid homeostasis. Biochim Biophys Acta 1821:168–176. https://doi.org/10.1016/j.bbalip.2011.07.002
Alapatt P, Guo F, Komanetsky SM, Wang S, Cai J, Sargsyan A, Rodríguez Díaz E, Bacon BT, Aryal P, Graham TE (2013) Liver retinol transporter and receptor for serum retinol-binding protein (RBP4). J Biol Chem 288:1250–1265. https://doi.org/10.1074/jbc.M112.369132
Zemany L, Kraus BJ, Norseen J, Saito T, Peroni OD, Johnson RL et al (2014) Downregulation of STRA6 in adipocytes and adipose stromovascular fraction in obesity and effects of adipocyte-specific STRA6 knockdown in vivo. Mol Cell Biol 34:1170–1186. https://doi.org/10.1128/mcb.01106-13
Muenzner M, Tuvia N, Deutschmann C, Witte N, Tolkachov A, Valai A et al (2013) Retinol-binding protein 4 and its membrane receptor STRA6 control adipogenesis by regulating cellular retinoid homeostasis and retinoic acid receptor activity. Mol Cell Biol 33:4068–4082. https://doi.org/10.1128/mcb.00221-13
Anjana RM, Pradeepa R, Deepa M, Datta M, Sudha V, Unnikrishnan R et al (2011) Prevalence of diabetes and prediabetes (impaired fasting glucose and/or impaired glucose tolerance) in urban and rural India: Phase I results of the Indian council of medical research-INdia DIABetes (ICMR-INDIAB) study. Diabetologia 54:3022–3027. https://doi.org/10.1007/s00125-011-2291-5
Mohan V, Amutha A, Ranjani H, Unnikrishnan R, Datta M, Anjana RM et al (2013) Associations of β-cell function and insulin resistance with youth-onset type 2 diabetes and prediabetes among Asian Indians. Diabetes Technol Ther 15:315–322. https://doi.org/10.1089/dia.2012.0259
Staimez LR, Weber MB, Ranjani H, Ali MK, Echouffo-Tcheugui JB, Phillips LS et al (2013) Evidence of reduced β-cell function in Asian Indians with mild dysglycemia. Diabetes Care 36:2772–2778. https://doi.org/10.2337/dc12-2290
Pandey GK, Balasubramanyam J, Balakumar M, Deepa M, Anjana RM, Abhijit S et al (2015) Altered circulating levels of retinol binding protein 4 and transthyretin in relation to insulin resistance, obesity, and glucose intolerance in Asian Indians. Endocr Pract 21:861–869. https://doi.org/10.4158/EP14558.OR
Sozio MS, Lu C, Zeng Y, Liangpunsakul S, Crabb DW (2011) Activated AMPK inhibits PPAR-α and PPAR-γ transcriptional activity in hepatoma cells. Am J Physiol 301:739–747. https://doi.org/10.1152/ajpgi.00432.2010
Rosell M, Hondares E, Iwamoto S, Gonzalez FJ, Wabitsch M, Staels B et al (2012) Peroxisome proliferator-activated receptors-α and -γ and cAMP-mediated pathways, control retinol-binding protein-4 gene expression in brown adipose tissue. Endocrinology 153:1162–1173. https://doi.org/10.1210/en.2011-1367
Scott MA, Nguyen VT, Levi B, James AW (2011) Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev 20:1793–1804. https://doi.org/10.1089/scd.2011.0040
Pandey GK, Vadivel S, Raghavan S, Mohan V, Balasubramanyam M, Gokulakrishnan K (2019) High molecular weight adiponectin reduces glucolipotoxicity-induced inflammation and improves lipid metabolism and insulin sensitivity via APPL1-AMPK-GLUT4 regulation in 3T3-L1 adipocytes. Atherosclerosis 288:67–75. https://doi.org/10.1016/j.atherosclerosis.2019.07.011
Lenin R, Maria MS, Agrawal M, Balasubramanyam J, Mohan V, Balasubramanyam M (2012) Amelioration of glucolipotoxicity-induced endoplasmic reticulum stress by a chemical chaperone in human THP-1 monocytes. Exp Diabetes Res 2012:356487. https://doi.org/10.1155/2012/356487
Ishii M, Shibata R, Kondo K, Kambara T, Shimizu Y, Tanigawa T et al (2014) Vildagliptin stimulates endothelial cell network formation and ischemia-induced revascularization via an endothelial nitric-oxide synthase-dependent mechanism. J Biol Chem 289:27235–27245. https://doi.org/10.1074/jbc.M114.557835
Qin ZY, Zhang M, Dai YM, Wang YM, Zhu GZ, Zhao YP et al (2014) Metformin prevents LYRM1-induced insulin resistance in 3T3-L1 adipocytes via a mitochondrial-dependent mechanism. Exp Biol Med 239:1567–1574. https://doi.org/10.1177/1535370214537746
Rajaram N, Frees AE, Fontanella AN, Zhong J, Hansen K, Dewhirst MW et al (2013) Delivery rate affects uptake of a fluorescent glucose analog in murine metastatic breast cancer. PLoS ONE 8:e76524. https://doi.org/10.1371/journal.pone.0076524
Apaijai N, Chinda K, Palee S, Chattipakorn S, Chattipakorn N (2014) Combined vildagliptin and metformin exert better cardioprotection than monotherapy against ischemia-reperfusion injury in obese-insulin resistant rats. PLoS ONE 9:e102374. https://doi.org/10.1371/journal.pone.0102374
Balakumar M, Raji L, Prabhu D, Sathishkumar C, Prabu P, Mohan V et al (2016) High-fructose diet is as detrimental as high-fat diet in the induction of insulin resistance and diabetes mediated by hepatic/pancreatic endoplasmic reticulum (ER) stress. Mol Cell Biochem 423:93–104. https://doi.org/10.1007/s11010-016-2828-5
Graham TE, Yang Q, Blüher M, Hammarstedt A, Ciaraldi TP, Henry RR et al (2006) Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med 354:2552–2563. https://doi.org/10.2337/db06-0616
Cheng J, Li Y, Wu G, Zheng J, Lu H, Shi X et al (2014) Ectopic expression of RBP4 impairs the insulin pathway and inguinal fat deposition in mice. J Physiol Biochem 70:479–486. https://doi.org/10.1007/s13105-014-0326-3
Wang Y, Sun L, Lin X, Yuan JM, Koh WP, Pan A (2019) Retinol binding protein 4 and risk of type 2 diabetes in Singapore Chinese men and women: a nested case-control study. Nutr Metab 16:3. https://doi.org/10.1186/s12986-018-0329-0
Kwanbunjan K, Panprathip P, Phosat C, Chumpathat N, Wechjakwen N, Puduang S et al (2018) Association of retinol binding protein 4 and transthyretin with triglyceride levels and insulin resistance in rural thais with high type 2 diabetes risk. BMC Endocr Disord 18:26. https://doi.org/10.1186/s12902-018-0254-2
Fan J, Yin S, Lin D, Liu Y, Chen N, Bai X et al (2019) Association of serum retinol-binding protein 4 levels and the risk of incident type 2 diabetes in subjects with prediabetes. Diabetes Care 42:1574–1581. https://doi.org/10.2337/dc19-0265
Farjo KM, Farjo RA, Halsey S, Moiseyev G, Ma J-X (2012) Retinol-binding protein 4 induces inflammation in human endothelial cells by an NADPH oxidase-and nuclear factor kappa b-dependent and retinol-independent mechanism. Mol Cell Biol 32:5103–5115. https://doi.org/10.1128/mcb.00820-12
Norseen J, Hosooka T, Hammarstedt A, Yore MM, Kant S, Aryal P et al (2012) Retinol-binding protein 4 inhibits insulin signaling in adipocytes by inducing proinflammatory cytokines in macrophages through a c-Jun N-terminal kinase- and toll-like receptor 4-dependent and retinol-independent mechanism. Mol Cell Biol 32:2010–2019. https://doi.org/10.1128/mcb.06193-11
Sell H, Eckel J (2012) Regulation of retinol binding protein 4 production in primary human adipocytes by adiponectin, troglitazone and TNF-alpha. Diabetologia 50:2221–2223. https://doi.org/10.1007/s00125-007-0764-3
Chen CH, Hsieh TJ, Der LK, Lin HY, Lee MY, Hung WW et al (2012) Increased unbound retinol-binding protein 4 concentration induces apoptosis through receptor-mediated signaling. J Biol Chem 287:9694–9707. https://doi.org/10.1074/jbc.M111.301721
Nair AK, Sugunan D, Kumar H, Anilkumar G (2010) Case-control analysis of SNPs in GLUT4, RBP4 and STRA6: association of SNPs in STRA6 with type 2 diabetes in a south indian population. PLoS ONE 5:e11444. https://doi.org/10.1371/journal.pone.0011444
Zhang L, Sun W, Duan X, Duan Y, Sun H (2019) Promoting differentiation and lipid metabolism are the primary effects for DINP exposure on 3T3-L1 preadipocytes. Environ Pollut 255:113154. https://doi.org/10.1016/j.envpol.2019.113154
Gumbilai V, Ebihara K, Aizawa-Abe M, Ebihara C, Zhao M, Yamamoto Y et al (2016) Fat mass reduction with adipocyte hypertrophy and insulin resistance in heterozygous PPARγ mutant rats. Diabetes 65:2954–2965. https://doi.org/10.2337/db15-1422
Moraes-Vieira PM, Yore MM, Dwyer PM, Syed I, Aryal P, Kahn BB (2014) RBP4 activates antigen-presenting cells, leading to adipose tissue inflammation and systemic insulin resistance. Cell Metab 19:512–526. https://doi.org/10.1016/j.cmet.2014.01.018
Moraes-Vieira PM, Castoldi A, Aryal P, Wellenstein K, Peroni OD, Kahn BB (2016) Antigen presentation and T-cell activation are critical for RBP4-induced insulin resistance. Diabetes 65:1317–1327. https://doi.org/10.2337/db15-1696
Bharath LP, Agrawal M, McCambridge G, Nicholas DA, Hasturk H, Liu J, Jiang K, Liu R, Guo Z, Deeney J, Apovian CM, Snyder-Cappione J, Hawk GS, Fleeman RM, Pihl RMF, Thompson K, Belkina AC, Cui L, Proctor EA, Kern PA, Nikolajczyk BS (2020) Metformin enhances autophagy and normalizes mitochondrial function to alleviate aging-associated inflammation. Cell Metab 32:44-55.e6. https://doi.org/10.1016/j.cmet.2020.04.015
Lazra Y, Falach A, Frenkel L, Rozenberg K, Sampson S, Rosenzweig T (2015) Autocrine/paracrine function of globular adiponectin: inhibition of lipid metabolism and inflammatory response in 3t3-l1 adipocytes. J Cell Biochem 116:754–766. https://doi.org/10.1002/jcb.25031
Kanuri BN, Kanshana JS, Rebello SC, Pathak P, Gupta AP, Gayen JR et al (2017) Altered glucose and lipid homeostasis in liver and adipose tissue pre-dispose inducible NOS knockout mice to insulin resistance. Sci Rep 7:41009. https://doi.org/10.1038/srep41009
Yang R, Wang L, Xie J, Li X, Liu S, Qiu S et al (2018) Treatment of type 2 diabetes mellitus via reversing insulin resistance and regulating lipid homeostasis in vitro and in vivo using cajanonic acid A. Int J Mol Med 42:2329–2342. https://doi.org/10.3892/ijmm.2018.3836
Lee JO, Lee SK, Kim JH, Kim N, You GY, Moon JW et al (2012) Metformin regulates glucose transporter 4 (GLUT4) translocation through AMP-activated protein kinase (AMPK)-mediated Cbl/CAP signaling in 3T3-L1 preadipocyte cells. J Biol Chem 287:44121–44129. https://doi.org/10.1074/jbc.M112.361386
Akarte AS, Srinivasan BP, Gandhi S (2012) Vildagliptin selectively ameliorates GLP-1, GLUT4, SREBP-1c mRNA levels and stimulates β-cell proliferation resulting in improved glucose homeostasis in rats with streptozotocin-induced diabetes. J Diabetes Complicat 26:266–274. https://doi.org/10.1016/j.jdiacomp.2012.03.013
Huang R, Yin S, Ye Y, Chen N, Luo S, Xia M et al (2020) Circulating retinol binding protein 4 is inversely associated with pancreatic β-cell function across the spectrum of glycemia. Diabetes Care 43:1258–1265. https://doi.org/10.2337/dc19-2432
Acknowledgements
Authors acknowledge research grant supports received from the Department of Biotechnology (DBT), Govt. of India (Grant No. BT/PR4082/MED/30/684/2011) and the Madras Diabetes Research Foundation—Intramural Research Funding (MIRF). Gautam Kumar Pandey acknowledges the Council of Scientific & Industrial Research (CSIR), India for his financial assistance (Senior Research Fellowship). Dr. Kuppan Gokulakrishnan acknowledges the DBT/Wellcome Trust India Alliance for their support.
Author information
Authors and Affiliations
Contributions
KG conceptualized the work; KG and GKP designed & executed the study methods, analyzed data, interpreted the results and drafted the manuscript. GKP performed experiments and cell-culture related work. SC helped in confocal microscopy. SS and PD helped in performing animal study. KG, NM, MB, and VM edited the manuscript, critically reviewed and helped in drafting the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests. All authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gokulakrishnan, K., Pandey, G.K., Sathishkumar, C. et al. Augmentation of RBP4/STRA6 signaling leads to insulin resistance and inflammation and the plausible therapeutic role of vildagliptin and metformin. Mol Biol Rep 48, 4093–4106 (2021). https://doi.org/10.1007/s11033-021-06420-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06420-y