Abstract
Entomopathogenic fungi are an important factor in the natural regulation of arthropod populations. Moreover, some can exist as an endophyte in many plant species and establish a mutualistic relationship. In this study, we have investigated the endophytic growth of Beauveria bassiana within different tissues of Phaseolus vulgaris in the presence and absence of Tetranuychus urticae. After the colonization of the B. bassiana within the internal tissues of P. vulgaris. The susceptibility of T. urticae appeared to depend on the life stage where high, moderate, and low mortalities were recorded among adults, nymphs, and eggs, respectively. In addition, this study provided, for the first time, molecular insight into the endophytic growth of B. bassiana by analyzing the expression of several genes involved in the development of the entomopathogenic fungi at 0-, 2-, and 7- days post-inoculation. B. bassiana displayed preferential tissue colonization within P. vulgaris that can be put into the following order based on the detection rate: leaf > stem > root. After analyzing the development-implicated genes (degrading enzymes, sugar transporter, hydrophobins, cell wall synthesis, secondary metabolites, stress management), the most remarkable finding is the detection of behavioral change between parasitic and endophytic Beauveria during post-penetration events. This study elucidates the tri-trophic interaction between fungus-plant-herbivore.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The control of herbivores has largely relied upon the use of chemical pesticides [1]. The secondary effects of these molecules include, but are not limited to, (i) insect resistance development (ii) toxicity to non-target organisms and (iii) residue in food crops [2]. Therefore, biologists are endeavoring to find alternatives to the harmful conventional pesticides [3]. Beauveria bassiana (Bals.) Vuill. (Hypocreales: Cordycipitaceae), a soil-borne cosmopolitan entomopathogenic fungus is one of the most famous biopesticides and its aerial conidia consist the basis of several commercially available mycoinsecticides [4]. Beauveria bassiana has been previously described as a generalist pathogen targeting a broad physiological and ecological range of insects and mites [5,6,7,8,9,10,11]. Generally, inundatively applied fungal cells will adhere, germinate, possibly differentiate, penetrate the integument of arthropods, and grow vegetatively as blastospores within their hemocoel [12]. A study conducted by Butt et al. [13] highlighted the genes implicated in the developmental processes of entomopathogenic fungi. The authors described in detail some genes involved in each key stage of the pathogenesis providing an insight into the entomopathogenic fungi-arthropod interaction. B. bassiana genes Cdep1, Bbchit1, and Cyp5337A1 encode proteases, chitinases, and lipases that take part in the degradation of the arthropod cuticle [14]. Moreover, a study conducted by Zhang et al. [15] demonstrated the vital role of the fungal hydrophobins, one of the most principal components of the aerial conidia cell wall. These proteins, encoded by the genes Hyd1 and Hyd2, are involved in hydrophobicity, attachment, and virulence of B. bassiana. The composition of B. bassiana cell wall also includes chitin and glucan, encoded by AY743592 and AY743593, respectively [16]. Beauvericin and bassianolide, the most famous secondary metabolites, are encoded by the genes Bbbeas and Bbbsls, respectively [17, 18]. The cyclooligomer depsipeptides are thought to be important in the pathogenesis of entomopathogenic fungi. A study conducted by Xiao et al. [19] demonstrated that BBA_08728 is responsible for encoding the sugar transporters in B. bassiana. These membrane proteins are believed to have the ability to transport glucose as well as plant cell wall-derived sugars. Furthermore, a mitogen-activated protein kinase (MAPK)-encoding gene, Bbhog1, contributes to stress management and virulence of B. bassiana [20]. However, increased interest in the potential of the endophytic growth of entomopathogenic fungi was recently noted [21]. This new role of the entomopathogenic fungi is being explored for the management of herbivores [22,23,24], plant pathogens [25], and its part in plant growth promotion [23, 26, 27]. The endophytic interaction is relative to plant as well as the fungi species and this association could be advantageous, neutral, or even disadvantageous [28]. Recent studies conducted by Jaber and Ownley [29,30,31], and Vega [24] showed the different endophytic entomopathogenic fungi (EEPF) that can reduce the herbivory of a broad range of pests and enhance the growth of various plants. Numerous studies reported antagonistic activity of B. bassiana, growing internally within the host plant tissues, against plant pests such as Aphis gossypii Glover (Hemiptera: Aphididae) [32, 33], Helicoverpa armigera Hübner (Lepidoptera: Noctuidae) [28], Liriomyza huidobrensis Blanchard (Diptera: Agromyzidae) [34]. Moreover, a review conducted by 30,31 declared the EEPF safe to auxiliary insects. Also, multi-kingdom associations may be established simultaneously between EEPF-herbivore-plant allowing nutrient exchange between all parties. The EEPF can translocate insect-derived nitrogen to the associated crop without causing any symptoms [35], in parallel, the plant can provide carbon to its symbiotic partner [36]. Even though the colonization of EEPF is considered a promising form of natural pest control, studies analyzing the patterns and dynamics of endophytic growth are lacking. A study conducted by Wagner and Lewis [37] briefly described the endophytic growth of entomopathogenic fungi within plant tissues. The authors reported similarities in the adhesion, germination, and penetration of EPIF into the plants when compared to arthropods. However, our understanding of this colonization is still very limited,hence, the limited utilization of EEPF as biopesticides. To elucidate the relationship between EEPF-plant-herbivore, Phaseolus vulgaris L., the common bean, and Tetranychus urticae Koch, the two-spotted spider mite, are outstanding options. Phaseolus vulgaris is one of the most important crops on the economic scale [38]. Also, the endophytic growth of B. bassiana within the internal tissues of P. vulgaris has been previously demonstrated [28]. Tetranychus urticae is a major pest feeding on a broad range of crops including the common bean. The two-spotted mite begins to feed shortly after emergence from the underside of leaves by piercing the epidermal tissue of the host plant withdrawing the contents via their stylets [39]. The latest reviews showed that T. urticae can thrive on 3,800 plant species belonging to 140 families [40]. Favorable outbreak conditions for T. urticae have been created by modern farming systems, fertilizers, and overexploitation of synthetic pesticides Dagli and Tunc [41]. The control of spider mite populations is still largely relying upon chemical acaricides such as organophosphates, neonicotinoids, and inhibitors of complex II and III of mitochondrial electrons [42]. Due to its rapid resistance development, the currently available acaricides are insufficient to suppress the populations of T. urticae in many areas worldwide [42]. The objective of this study is to reveal the expression dynamics of genes involved in key developmental processes of B. bassiana. In their review of 2016, Valero-Jiménez et al. highlighted the vital role of the aforementioned genes during the parasitism of B. bassiana. The expression analysis of those genes elucidated the genetics behind the virulence of entomopathogenic fungi towards insects. However, the role played by those genes during the internal colonization of entomopathogenic fungi is still poorly understood. Here, we investigate (i) the antagonistic activity of a Beauveria sp. to all developmental stages of T. urticae when present as endophyte within the tissues of P. vulgaris and (ii) the transcription profile of genes Cdep1, Bbchit1, Cyp5337A1, Hyd1, Hyd2, Bbbeas, Bbbsls, BBA_08728, and Bbhog1 of B. bassiana when present as an endophyte within the tissues of P. vulgaris in the presence and absence of T. urticae.
Material and methods
Source of entomopathogenic fungi
Beauveria bassiana strain LTB01 (accession number: TEF-α = EU177813, RBP2 = MK908095, BLOC = MK884877, ITS = DQ984676) was used in this study. The strain was initially originated from the corpses of Cephalcia tannourinensis (Hymenoptera: Pamphiliidae) Chevin, and preserved in the laboratories of the American University of Beirut. B. bassiana strain LTB01 was grown on Potato Dextrose Agar (PDA) medium (amended with 0.5 g/L streptomycin) in dark conditions at 27 °C for 14 days. The conidia that developed were disassociated from the medium via a sterile flamed loop after adding 20 ml of sterile distilled water. To remove hyphal cells, the suspension was filtered through 8 layers of cheesecloth. The concentration of the conidia suspension was evaluated using a Neubauer-improved hemocytometer (BLAUBRAND®, Germany). Several dilutions were performed on the mother suspension to obtain the concentration of 2 × 108 conidia/ml. Tween 80 (0.1% of the volume) (Sigma-Aldrich, USA) was added to avoid the formation of a cluster of propagules. The conidial viability was assessed according to the protocol described by Goettel and Inglis [43]. In brief, a random volume of the spore suspension was inoculated on the top of the PDA petri dish that was transferred into an incubator in dark conditions at 27 °C for 48 h. The conidial germination was evaluated under an optical microscope (at ×40 magnification) where all conidia with an apparent germ tube (at least their size) were considered viable. A total of 5 PDA Petri dishes were assessed for conidial viability with an average of 500 conidia per plate. The LTB01 strain recorded viability of 94% ± 1.4 of the conidia.
Plant material
Seeds of the common bean were surface sterilized by submerging them in 96% ethanol and 1% sodium hypochlorite for 10 s and 3 min, respectively. The sterilizing process was repeated 3 times to remove any contaminant microorganisms. The entire surface of sterilized seeds was rinsed with distilled water and rubbed up against PDA Petri dishes, incubated at 27 °C, and observed for microbial contamination. The seeds that caused microbial growth were discarded while the rest were kept until further use.
Surface sterilized seeds were germinated and grown in pots containing a sterile mixture of soil and sand at a 2:1 ratio and transferred into a controlled greenhouse (20–30 °C, 80% RH, under natural light). The greenhouse was divided into 10 compartments each containing 20 plants of common bean. The compartments were sprayed alternately with (i) spore suspension of B. bassiana strain LTB01 (2 × 108 conidia/ml) + 0.1% Tween (treatment compartment) (ii) sterile water + 0.1% Tween (control compartment) making it a total of 5 technical replicates (5 compartments) within a greenhouse. The above-mentioned compartments were separated with a buffer zone, consisted of 2 non-sprayed compartments, to avoid cross-contamination. This experiment was also repeated in 3 biological replicates (3 different greenhouses in 3 different testing time points; 25 °C with a photoperiod of 16/8 h.) making it a total of 15 compartments of each treatment. The leaves (adaxial surface) of 14-days old plants were manually sprayed with 5 ml of spore suspensions before being covered with a plastic bag for 24 h to maintain humidity. The quantity of inoculum on leaves was estimated at 5.9 μl/cm2 (S.E. = 0.6), obtained by measuring the surface of 20 P. vulgaris leaves using a Delta-T Leaf Area Meter Mk 2 (Delta-T Devices Ltd., Cambridge, UK) and obtaining the weight of spore suspension on the leaves. No additional chemical insecticides or acaricides were applied during the experiments.
Assessment of endophytic growth
Sterilization method
Seven days post-inoculation, the endophytic growth of B. bassiana was evaluated in P. vulgaris plant tissues (leaf, root, stem). To ascertain the internal development of B. bassiana within the plant tissues, thorough surface sterilization was performed as follows: plant tissues were first washed with 0.1% Tween solution for 3 min to break hydrophobic bonds between conidia and plant cells [15]. Afterward, the plant tissues were submerged in 96% ethanol and 1% sodium hypochlorite for 10 s and 3 min respectively before rinsing in sterile water. The sterilizing process was repeated 3 times to remove any viable fungal epiphytes from the external surfaces. To ensure the success of the sterilization method, a pilot study was priorly conducted that consists of subjecting fungal cells directly to the sterilization procedure and confirming the total unviability of the cells. To re-confirm the sterilization process efficacy, aliquots from the final rinse step and surface sterilized tissues were inoculated onto PDA Petri dishes and observed for microbial contamination.
Endophytic growth detection by re-isolation
The endophytic growth was assessed according to the method described by Posada et al. [44]. In brief, surface-sterilized plant tissues were dried on sterile towels, then dissected using a sterile scalpel into 5 mm fragments. The tissues were directly inoculated on top of PDA Petri dishes (amended with 0.5 g/L streptomycin) cultured at 27 °C for 14 days. Five plants were randomly chosen from each of the 5 compartments within a greenhouse (25 bean plants/greenhouse) making a total of 75 plants for 3 biological replicates (3 different greenhouses in 3 different testing time points). Three fragments were dissected from each plant part (leaves, root, and stem) making it a total of 9 fragments from each plant,therefore, 675 plant tissue samples (225 leaves, roots, and stem) were evaluated. The endophytic growth was assessed on every sample based on the presence or absence of fungal cultures (number of colonized fragments).
Detection of fungal DNA
The endophytic growth was also confirmed using molecular detection according to the method described by Rehner et al. [45]. In brief, plant tissues (100 mg), selected as described above, were ground under liquid nitrogen and the DNA of the mixture was extracted using DNeasy kit (QIAGEN, GmbH, Hilden, Germany) following the manufacturer’s instructions. The internal transcribed spacer (ITS) were amplified on a thermocycler (CFX96 Real-Time System, BIO-RAD, USA) using the primers pairs ITS1F (5′ CTTGGTCATTTAGAGGAAGTAA 3′) [46] ITS4 (5′TCCTCCGCTTATTGATATGC 3′) [47] and BB.fw (5′ GAACCTACCTATCGTTGCTTC 3′) BB.rv (5′ ATTCGAGGTCAACGTTCAG 3′) [48] for the two-step nested PCR-approach. To check the success and specificity of the PCR, all amplicons were separated on gel electrophoresis (1.5% agarose) then purified using the QUIAquick PCR purification kit (QIAGEN, GmbH, Hilden, Germany) following the manufacturer's instructions. To determine the identity of the DNA, all purified amplicons were sequenced using the ABI3500 instrument.
Evaluation of the endophytic activity against two-spotted spider mite (TSSM)
The strain of T. urticae was originally collected from P. vulgaris and reared under laboratory conditions (25 °C, 16/8 h L:D) without exposure to chemical molecules. To limit the variation in susceptibility due to differences in activities associated with developmental stages, single-age populations of the mites were prepared as described by Chandler et al. [6]. After confirmation of the endophytic growth of B. bassiana, 30 mites belonging to each life stage (eggs, nymphs, adults) of the mite were inoculated on the adaxis of the first pair of 3-week-old P. vulgaris leaves using a fine brush. Five plants from each of the 5 compartments within a greenhouse were used (25 bean plants/greenhouse). This experiment was repeated in 3 biological replicates (3 different greenhouses) making it a total of 75 plants; thus, 2250 individuals of each developmental stage of the mite were tested for their mortalities till 7 days post-inoculation (DPI). The sampling of the infected mites was performed as described by Batta [49] with slight modifications. In brief, dead individuals were surface sterilized, and then transferred to PDA Petri dishes (amended with 0.5 g/L streptomycin) which were incubated at 27 °C to promote fungal growth. The sampling of the infected mites continued for 14 days after initial transfer into the Petri dishes. Additionally, the macroscopic and microscopic characteristics of fungal colonies were observed.
Genetic expression of endophyte B. bassiana
The expression analysis focused on eleven genes believed to play an essential role in key developmental stages of B. bassiana. The leaves of 14-days old plants were treated with 2 × 108 spore suspensions as described above. After 24 h, 30 motile stages (larvae and adults) of T. urticae were allowed to feed on the treated leaves of P. vulgaris.
The RNA extraction was performed on leaf fragments collected from plants at 0, 2- and 7-days post-inoculation of spore suspensions using Zymo Research, USA kit according to the manufacturer's instructions. Until further use, purified RNA was stored at − 80 °C. Two μg of purified RNA were transcribed, using BIORAD's kit, into cDNA using the manufacturer's instructions. SYBR® Green rt-PCR (real-time PCR) Sigma- Aldrich was used to obtain rt-PCR products. rt-PCR reactions were performed in a CFX96 Real-Time System (BIO-RAD) using the specific primer pairs (Table 1). The PCR reactions, including the cDNA dilutions and primer concentrations, were adjusted to obtain approximatively similar gene amplification efficiencies reactions were optimally set to master mix (25 μl) containing: 12.5 μl SYBR® Green rt-PCR (Sigma-Aldrich), 150 nM or 250 nM forward and reverse primers (Macrogen, Korea), and 2 μl of cDNA. PCR thermal conditions consisted of one cycle of 50 °C for 2 min, one cycle of 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The melting curve analysis (single peak) was considered as an indicator of the amplification specificity. Moreover, negative controls (no template) were run for each gene to detect primer dimerization and unspecific amplification. To minimize intra-experiments, the rt-PCR analysis was performed in triplicate (technical replicates), and the mean of 3 biological replicates (3 different plant tissue/cDNA samples in 3 different testing time points) was calculated for the gene expression analysis. The amplification efficacy for each primer set was calculated as E = 10[−1/slope]; slopes of standard curves were generated by plotting the log of cDNA versus Ct values acquired over a range of 10- fold dilution to the mother solution (Table 1). The average value collected at 0 days post-inoculation was considered as calibrator and average values collected at 2- and 7- days post-inoculation were considered as test values and time required for the entomopathogenic fungi penetration and endophytic growth respectively. The relative differences in the expression level of genes in different samples were calculated using the Livak and Schmittgen [50] method (2−ΔΔCq). Cyclophilin A was chosen as a reference gene (housekeeping gene) due to its expression stability in Beauveria spp. [51].
Statistical analyses
The data obtained by the re-isolation, detection of fungal DNA experiment were statistically analyzed by one-way Analysis of Variance (ANOVA). The antagonist activity of EEPF/plant against T. urticae was analyzed by Kaplan Meier survival curves using SPSS (version 25) software. The statistical differences between data obtained with each treatment and the control for each experiment were measured by the Log-rank test expressed by Chi-2 results and P-values. The data obtained by rt-PCR was analyzed by two-way ANOVA (days post-inoculation and presence/absence of T. urticae) using SPSS Statistics for Windows, version 25.0 (SPSS Inc., Chicago, Ill., USA). The Tukey test was used at the 5% threshold for the separation of means.
Results
Evaluation of endophytic growth by re-isolation
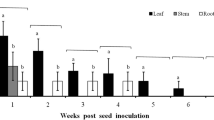
The endophytic growth of B. bassiana in P. vulgaris plant tissues tested positive. The fungal development detected from dishes inoculated with various plant tissues (leaf = 48.4 ± 1.9%; stem = 32.4 ± 2%; root = 3.96 ± 0.7%) was significantly different (F = 153.461 df = 2, P < 0.05). No B. bassiana endophytic growth was detected within any tissue of the control plants.
Detection of fungal DNA
The ITS of entomopathogenic fungi was considered as evidence of endophytic growth. The amplification of fungal DNA detected from the treatment plant tissue extracts (leaf = 27.06 ± 4.9%; stem = 15.06 ± 1.9; root = 0 ± 0%) was also significantly different (F = 19.61 df = 2, P < 0.05). No amplification was detected from the control plant tissue extracts. The amount of fungal DNA at the different plant tissues can be put into the following order according to their cycle threshold values (cycles ± S.E.): root (no DNA amplification) < stem (Ct = 34.92 ± 0.2) < leaf (Ct = 36.34 ± 0). The same melting temperature was recorded in this study indicating a specific and unique amplification. After conducting sequence alignment, the amplicon revealed to be 100% identical with the previously described ITS of B. bassiana LTB01 strain (accession number: DQ984676) [54].
Pathogenicity of EEPF colonized P. vulgaris against all developmental stages of T. urticae
The activity of Beauveria, when present as endophyte within the tissues of P. vulgaris, was daily observed on the survival (adults and nymphs) and the hatching (eggs) of T. urticae for 7 DPIDPI. Significantly high, moderate, and low effects were observed against the adults (χ2 = 818.354; df = 1; P < 0.0001), nymphs (χ2 = 345.766; df = 1; P < 0.0001) and eggs (χ2 = 173.496; df = 1; P < 0.0001) of T. urticae respectively (Fig. 1). The antagonistic activity of EEPF/plant against the different developmental, based on their chi-square value, can be put into the following order: adults > nymphs > eggs; therefore, the survival capacity seems to be relative to the developmental stage of the mite. Furthermore, the mycosis percentage recorded in this study was 3.7 (± 0.2), 2.9 (± 0.1), and 0 (± 0) % for the adults, nymphs, and eggs of T. urticae respectively.
Curves representing survival (of a adults and b nymphs) and hatching (of c eggs) of T. urticae reared on P. vulgaris colonized with the EEPF B. bassiana. Observed survivals and hatchings are presented using curves with markers; circle: Treatment (T. urticae reared on EEPF-colonized P. vulgaris); triangle: control (T. urticae reared on the P. vulgaris)
Expression analysis of genes involved in the growth of B. bassiana
The amplification conditions were optimized for each gene. The melting temperatures were unique for each gene indicating specific amplification for the target genes. The amplification efficacies were evaluated for each primer set and they varied between 98.2 and 102.2% indicating good reaction conditions (Table 1).
Genes encoding for degrading enzymes
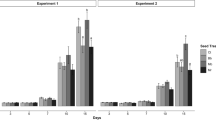
The Cdep1 gene, responsible for encoding a subtilisin protease, was significantly induced at 2- and 7-DPI (F = 1178, df = 2, P < 0.05). A significant increase of 11.1- fold change was recorded at 2 DPI when compared to the calibrator; afterward, an increase of 2.07- fold change was recorded at 7 DPI with no significant difference when compared to the calibrator. The presence of the TSSM showed an effect only at 7 DPI where a significant induction (6.8 fold change) was recorded (F = 82.022, df = 1, P < 0.05) (Fig. 2).
Fold expression levels (± S.E.) of Cdep1, Cyp5337A1, Bbchit1, BBA_08728, Hyd1, Hyd2,BbCHS, BbFKS, Bbbeas, Bbbsls, and Bbhog1 genes of B. bassiana at 0-, 2-, and 7- days post inoculation in P. vulgaris in the presence or absence of T. urticae. Means followed by distinct letters within each gene differ by the Tukey test at the 5% probability level
The Bbchit1 gene, responsible for encoding a chitinase, was significantly induced at 7-DPI (F = 410.005, df = 2, P < 0.05) during the presence of the herbivore (F = 437.270, df = 1, P < 0.05).
The Cyp5337A1 gene, responsible for encoding a fatty acid hydroxylase, was always significantly induced post-inoculation (F = 71.941, df = 2, P < 0.05). At 2 days post-inoculation, the expression was significantly increased (7.2- fold change) when compared to the calibrator. At 7 days post-inoculation, the expression was increased (1.5- fold change) with no significant difference when compared to the calibrator (Fig. 2). Here, the effect of the TSSM was not notable at 2- and 7- DPI. (F = 0, df = 1, P > 0.05).
Genes encoding for hydrophobins
The hydrophobin coding genes Hyd1 (F = 146.749, df = 2, P < 0.05) and Hyd2 (F = 141.408, df = 2, P < 0.05) were only significantly induced at 7- days post-inoculation when compared to the calibrator (Fig. 2). No significant difference was recorded to the Hyd1 (F = 0.259, df = 1, P > 0.05) and Hyd2 (F = 0.123, df = 1, P > 0.05) gene expression after the inoculation of the TSSM (Fig. 2).
Genes encoding for cell wall components
The genes BbCHS (F = 107.407, df = 2, P < 0.05) and BbFKS (F = 350.028, df = 2, P < 0.05) involved in the synthesis of chitin and glucan, respectively were significantly induced at 7- days post inoculation relative to the calibrator (Fig. 2). The presence of the herbivor has no significant effect on the expression of BbCHS (F = 0.001, df = 1, P > 0.05) and BbFKS (F = 0.364, df = 1, P > 0.05).
Genes encoding for secondary metabolites
Significantly different induction levels of the beauvericin coding gene Bbbeas were recorded at different timings of the essay (F = 186.603, df = 2, P < 0.05). The recorded fold changes in the expression were 3.9 and 1.1 at 2- and 7- days post-inoculation respectively. Similarly, significantly induction levels of the bassianolide coding gene Bbbsls were recorded at 2 and 7 days post-inoculation (F = 48.062, df = 1, P < 0.05). The recorded fold changes in the expression were 2.1 and 1.1 at 2- and 7- days post-inoculation respectively. The presence of the herbivore did not show any significant difference on the expression levels of Bbbeas (F = 0, df = 1, P > 0.05) and Bbbsls (F = 0.436, df = 1, P > 0.05) (Fig. 2).
Gene encoding for sugar transporter
The BBA_08728 gene responsible for the expression of the sugar transporters was induced at 2- and 7- days post-inoculation with a significant difference (F = 357.709, df = 2, P < 0.05). An increase of 2.2- fold change was recorded at 2 days post-inoculation; afterward, a significant increase of 4.19 fold-change was recorded at 7 days post-inoculation (Fig. 2). The presence of herbivore resulted in a significant (F = 77.291, df = 1, P < 0.05) induction of the gene (8.95- fold increase) at 7DPI (Fig. 2).
Gene encoding for high osmolarity glycerol
The mitogen-activated protein kinase-encoding gene, Bbhog1, was significantly induced at all times of the experiment (F = 78.952, df = 2, P < 0.05). At 2- and 7- DPI, 8.67- and 4.59- fold increases were recorded, respectively, when compared to the calibrator. The presence of the herbivore did not show any effect on the expression (F = 0.189, df = 1, P > 0.05) (Fig. 2).
Discussion
Several authors reported the possibility of B. bassiana to dwell in different P. vulgaris tissues [55]. A study conducted by Afandhi et al. [26] confirmed that such colonization can enhance the growth of the common bean. In this present study, we have demonstrated the endophytic colonization of B. bassiana in different tissues (leaf, stem, and roots) of P. vulgaris via spraying of infective propagules. As indications, it was relied upon the fungal re-isolation as well as the detection of fungal DNA from surface-sterilized plant tissues (leaf, stem, and root) located farthest from the area of inoculation. Based on both detection methods the localization of (EEPF) is high, moderate, and low at the leaves, stem, and roots respectively. These results could be hypothesized by an vertical colonization of B. bassiana with the tissues of P. vulgaris. A study conducted by Parsa et al. [56] previously demonstrated the systemic vertical growth of B. bassiana in P. vulgaris; however, in this present study, the same was demonstrated, for the first time, using the molecular technique of fungal genomic detection. Several studies suggested that for the endophytic biological control strategy the best option will be to localize the EEPF onto lower parts of the P. vulgaris (e.g. seed soaking, soil/root drench); thus, allowing endophytic growth towards the uppermost of the host [21, 28, 57]. In this study, it was demonstrated that a foliar application of B. bassiana could also result in the internal colonization of the host. This goes in line with the results obtained from Wagner and Lewis [37] who confirmed and described the endophytic growth of B. bassiana within the internal tissues of Zea mays after inoculation with a foliar spray of conidia.
Our findings also revealed that the colonization rates of Beauveria spp. can be relative to the detection method (cultivation-dependant method > cultivation-independent method). Such risk of error has been previously discussed by McKinnon et al. [57]. It is hypothesized that the low concentration of the fungal DNA within the teated plant extracts may lead to amplification failure. In addition, Healey et al. [58] related PCR inhibition to the presence of plant secondary metabolites. Therefore, to ensure accurate results it would be best to adapt several techniques for the detection of fungal endophytism.
It was notable that most of the studies evaluated the insecticidal effect of EEPF with no differentiation of the herbivore life stage. The present study demonstrated that the EEPF activity caused high, moderate, and low mortality rates against adults, nymphs, and eggs of T. urticae respectively. These results are in accordance with those obtained from Al Khoury et al. [59] who observed similar mortality rates within the different developmental stages of T. urticae after inundative application of B. bassiana aerial conidia. However, the endophytic approach of entomopathogenic fungi may have an advantage relative to the conventional inundative application of different types of infective propagules. A study conducted by Tehri et al. [60] revealed that inadequate environmental conditions are considered the principal limitation and concern during biological control with B. bassiana. It is hypothesized that the endophytic fungal growth within the internal tissues of the host plant will minimize their exposure to abiotic factors; thus, enhancing their survival capacity. Moreover, the mycosis percentages obtained in this study are extremely low when compared to those attained after inundative application of B. bassiana [59, 61]. The localization of endophytic B. bassiana within the parenchymal cells of the host has been previously demonstrated [37, 62, 63]. The feeding behavior of the TSSM consists of withdrawing the plant cellular content via their stylet, including the parenchyma-localized fungal cells [64]. Infections caused by conidial ingestion have been previously reported [12, 65]. As far as we are aware, studies demonstrating that the ingestion of B. bassiana colonized plant tissue results in mycosis are not available in the literature. A study conducted by Mannino et al. [66] hypothesized that the host microbiota could inhibit the entomopathogenic fungal growth following per os infection. Taken together, these findings provide additional support to the hypothesis that the endophytic colonization of B. bassiana per se is not responsible for the antagonistic activity; nevertheless, the fungal growth may be indirectly involved in the reduction of herbivore damage.
The knowledge gap regarding the tri-trophic interaction between EEPF-plant–herbivore is notable. Through rt-PCR, a series of genes involved in the development and growth of B. bassiana were analyzed for their expression during endophytic colonization within P. vulgaris in the presence or absence of T. urticae.
The expression of degrading enzyme genes Cdep1 and Cyp5337A1 (excluding Bbchit1) was found to increase at 2 DPI suggesting that B. bassiana utilizes some depolymerases for the penetration of the plant tissues. Thus, it is suggested that EEPF invasion is not limited to the natural opening (e.g. stomata) but it could also occur via degradation holes. These results are in accordance with those from Wagner and Lewis [37], who described the penetration process of Z. mays by B. bassiana. On the contrary to arthropods, the induction of chitinases may not be necessary for the penetration of the plant leaf tissues. This may be hypothesized by the fact that chitin is not found in the plant cell walls [67]. Afterward, the expression of the depolymerases genes was repressed during the development of the fungi (7 DPI). This could be hypothesized by the fact that the endophytic growth of EEPF does not harm the plant host,thus, the former can acquire the needed nutrients for development in a non-conventional approach. Interestingly, after the introduction of the herbivore, the cuticle degrading enzymes were significantly induced at 7 DPI. Therefore, it can be speculated that the EEPF may induce various depolymerases targeting the cuticle of arthropods in order to transfer insect-derived nutrients to their plant host. This assumption ties well with the results from Behie et al. [35] who demonstrated that the endophytism and arthropod parasitism of Metarhizim robertsii are combined so that the fungus functions as a channel to provide insect-derived nutrients to plant hosts.
To further explain the nutrient exchanges between all parties, we have analyzed the expression of the gene encoding sugar transporters. As might have been expected, a limited expression was recorded at 2 DPI (penetration phase) since the fungus may obtain the required nutrients directly from the lysed plant tissue. However, after the internal colonization of the plant tissues, a significant induction of the gene was recorded. This is in complete agreement with results obtained by Behie et al. [68] who demonstrated that the host plant provides photosynthate to EEPF. Remarkably, the presence of the herbivore results in a significant fold increase in the sugar transporter-encoding gene. The correlation between the levels of insect-derived nitrogen and the plant-derived carbon is noteworthy. As it seems, after translocating the nitrogen from the arthropod to the plant, the fungus increased its carbon uptake from the plant host. A "fair" nutrient trade is established during this mutualistic association.
The remarkable observation to emerge is the induction of Hyd1 and Hyd2 expression during different timings of the essay. Despite the fact that Branine et al. [69] discussed the similar mechanisms adapted by EEPF to infect both plants and insect hosts, our results suggest a different post-penetration infection strategy between endophytism and parasitism. A study conducted by Holder et al. [70] showed that once the entomopathogenic fungi reach the hemocoel, blastospores (yeast-like cells) are produced and disseminated throughout the arthropod. Unexpectedly, the analysis did not show any dimorphic transition of the fungi during the post-penetration events during the presence and absence of the herbivore.
The mechanism underlying this novel role of EEPF as plant bodyguards remains unknown. Vidal and Jaber [28] suggested that the antagonistic activity against herbivorous pests may be due to changes in the metabolism of the host plant. It is hypothesized that ligands (chitin and glucans) present on the cell wall of B. bassiana, known as pathogen-associated molecular patterns (PAMPs), can be recognized by the host immune system [13]. A study conducted by Pieterse et al. [71] revealed that such antigens are able to trigger the induced system resistance (ISR) for enhanced defenses. Results in this study showed that synthesis of PMAPs is significantly induced during the post-penetration events,therefore, it may be that the plant pattern recognition is translated into ISR. This result ties well with the previous study of Jaber and Ownley [29] who suggested that the endophytic growth can initiate the ISR and result in the secretion of compounds with insecticidal activities.
Furthermore, contrary to the earlier research that indicates secondary metabolite production by Beauveria spp. after breaching the arthropod surface [72], this study suggests the non-involvement of the secondary metabolites (beauvericin and bassianolide) post-penetration of plant tissues by the EEPF. It may be suggested that host plants initially identify the EEPF as possible pathogens,thus, the former activate their immune system and the latter produce the secondary metabolites to overpower it. Afterward, the beneficial endophytic growth will be established and the EEPF will be recognized as a mutualist by its host. This is in good agreement with previous findings of Barelli et al. [73] indicating that EEPF-plant relation is mediated by diffusible communication molecules, as in other plant-mutualistic fungi, indicating they are not pathogens. This is consistent with what has been found in a review conducted by Zimmermann [74] who argued that the secondary metabolites found in plants and food are mostly produced by Fusarium spp. The mammal toxicity of secondary metabolites such as beauvericin has been previously discussed [75], therefore, the present result may overcome a major hurdle in the consideration of EEPF for potential biocontrol applications. However, it is important to note that our findings are based on the work on P. vulgaris plants grown in sterile conditions, the expression of the Bbbeas and Bbbsls genes may be significantly different under the natural conditions as Beauveria spp. can produce secondary metabolites to prevent microbial competition for limited nutrients.
A further novel finding is the repression of BBhog1 gene implicated in the stress management of B. bassiana in planta (7 DPI) when compared to that obtained in natura (0- and 2- DPI). This finding is directly in line with those from Mascarin and Jaronski [76], who indicated that the endophytic growth of B. bassiana provides protection from abiotic stresses. Knowing that inconvenient environmental conditions are considered the first limitation during the inundative application of B. bassiana [58], it is believed that the internal colonization of EEPF would transcend this major obstacle leading towards a more persistent and cost-effective biocontrol agent.
Conclusion
This study presented the first evidence of the possibility of B. bassiana to grow within P. vulgaris with preferential tissue colonization. The internal growth of EEPF was most favorable within the leaves followed by the stem and roots of P. vulgaris. Furthermore, it is clarified that the report of endophytes is method dependent; therefore, advanced studies should investigate the EEPF-plant relationship using different methodologies (infection, detection methods, etc.). In addition, this is the first analysis of genes involved in the endophytic colonization of B. bassiana in planta. This insight into the endophytic growth of EEPF would enhance the engineering and production of a more efficient endophytic biocontrol agent. A transcriptome analysis would reveal the biological pathways for the mutualism relationship. Nevertheless, this work has allowed us to conclude that EEPF adopt different mechanisms relative to its intended host.
Data availability
All data are available.
References
Smith GS, O'Day MH, Reid W (2019) Pecan pest management: insects and diseases. Department of entomology, Kansas State University. https://extension2.missouri.edu/mp711. Accessed July 2020
Aktar W, Sengupta D, Chowdhury A (2009) Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol 2:1–12
Plant R, Freudenberger D (2005) Changes in global agriculture: a framework for diagnosing ecosystem effects and identifying response options. WWF Macroeconomics Program Office Washington, DC, USA
Faria MR, Wraight SP (2007) Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol Control 43:237–256
Cafarchia C, Immediato D, Iatta R, Ramos RAN, Lia RP, Porretta D, Figueredo LA, Dantas-Torres F, Otranto D (2015) Native strains of Beauveriabassiana for the control of Rhipicephalussanguineus sensu lato. Parasite Vectors 8:80
Chandler D, Davidson G, Jacobson RJ (2005) Laboratory and glasshouse evaluation of entomopathogenic fungi against the two-spotted spider mite, Tetranychusurticae (Acari: Tetranychidae), on tomato, Lycopersicon esculentum. Biocontrol Sci Technol 15:37–54
Immediato D, Camarda A, Iatta R, Puttilli MR, Ramos RAN, Di Paola G, Giangaspero A, Otranto D, Cafarchia C (2015) Laboratory evaluation of a native strain of Beauveriabassiana for controlling Dermanyssusgallinae (De Geer, 1778)(Acari: Dermanyssidae). Vet Parasitol 212:478–482
Kaiser D, Handschin S, Rohr RP, Bacher S, Grabenweger G (2020) Co-formulation of Beauveriabassiana with natural substances to control pollen beetles–synergy between fungal spores and colza oil. Biol Control 140:104106
Morales AD, Castillo A, Cisneros J, Valle JF, Gómez J (2019) Effect of spinosad combined with Beauveriabassiana (Hypocreales: Clavicipitaceae) on Hypothenemushampei (Coleoptera: Curculionidae) under laboratory conditions. J Entomol Sci 54:106–109
Mwamburi LA, Laing MD, Miller RM (2019) External development of the entomopathogenic fungus beauveria bassiana on the housefly (Muscadomestica). Proc Natl Acad Sci India B 89:833–839
Yanar Y, Yanar D, Demir B, Karan YB (2019) Effects of local entomopathogen Beauveriabassiana isolates against Sitophilusgranaries (Coleoptera). Poljoprivreda i Sumarstvo 65:49–55
Feng MG, Poprawski TJ, Khachatourians GG (1994) Production, formulation and application of the entomopathogenic fungus Beauveriabassiana for insect control: current status. Biocontrol Sci Technol 4:3–34
Butt TM, Coates CJ, Dubovskiy IM, Ratcliffe NA (2016) Entomopathogenic fungi: new insights into host–pathogen interactions. Anonymous advances in genetics, vol 94. Elsevier, Amsterdam, pp 307–364
Mondal S, Baksi S, Koris A, Vatai G (2016) Journey of enzymes in entomopathogenic fungi. Pac Sci Rev A 18:85–99
Zhang S, Xia YX, Kim B, Keyhani NO (2011) Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveriabassiana. Mol Microbiol 80:811–826
Tartar A, Shapiro AM, Scharf DW, Boucias DG (2005) Differential expression of chitin synthase (CHS) and glucan synthase (FKS) genes correlates with the formation of a modified, thinner cell wall in in vivo-produced Beauveriabassiana cells. Mycopathologia 160:303–314
Xu Y, Orozco R, Wijeratne EK, Espinosa-Artiles P, Gunatilaka AL, Stock SP, Molnár I (2009) Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveriabassiana. Fungal Genet Biol 46:353–364
Xu Y, Orozco R, Wijeratne EK, Gunatilaka AL, Stock SP, Molnár I (2008) Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveriabassiana. Chem Biol 15:898–907
Xiao G, Ying S, Zheng P, Wang Z, Zhang S, Xie X, Shang Y, Leger RJS, Zhao G, Wang C (2012) Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveriabassiana. Sci Rep 2:483
Zhang Y, Zhao J, Fang W, Zhang J, Luo Z, Zhang M, Fan Y, Pei Y (2009) Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveriabassiana regulates environmental stress responses and virulence to insects. Appl Environ Microbiol 75:3787–3795
Ramakuwela T, Hatting J, Bock C, Vega FE, Wells L, Mbata GN, Shapiro-Ilan D (2020) Establishment of Beauveriabassiana as a fungal endophyte in pecan (Caryaillinoinensis) seedlings and its virulence against pecan insect pests. Biol Control 140:104102
Ownley BH, Griffin MR, Klingeman WE, Gwinn KD, Moulton JK, Pereira RM (2008) Beauveriabassiana: endophytic colonization and plant disease control. J Invertebr Pathol 98:267–270
Russo ML, Scorsetti AC, Vianna MF, Allegrucci N, Ferreri NA, Cabello MN, Pelizza SA (2019) Effects of endophytic Beauveriabassiana (Ascomycota: Hypocreales) on biological, reproductive parameters and food preference of the soybean pest Helicoverpagelotopoeon. J King Saud Univ Sci 31:1077–1082
Vega FE (2018) The use of fungal entomopathogens as endophytes in biological control: a review. Mycologia 110:4–30
Barra-Bucarei L, France Iglesias A, Gerding González M, Silva Aguayo G, Carrasco-Fernández J, Castro JF, Ortiz Campos J (2020) Antifungal activity of Beauveriabassiana endophyte against botrytis cinerea in two solanaceae crops. Microorganisms 8:65
Afandhi A, Widjayanti T, Emi AAL, Tarno H, Afiyanti M, Handoko RNS (2019) Endophytic fungi Beauveriabassiana Balsamo accelerates growth of common bean (Phaeseolusvulgaris L.). Chem Biol 6:11
Gange AC, Koricheva J, Currie AF, Jaber LR, Vidal S (2019) Meta-analysis of the role of entomopathogenic and unspecialized fungal endophytes as plant bodyguards. New Phytol 223:2002–2010
Vidal S, Jaber LR (2015) Entomopathogenic fungi as endophytes: plant–endophyte–herbivore interactions and prospects for use in biological control. Curr Sci 109:46–54
Jaber LR, Ownley BH (2018) Can we use entomopathogenic fungi as endophytes for dual biological control of insect pests and plant pathogens? Biol Control 116:36–45
Quesada-Moraga E (2020) Entomopathogenic fungi as endophytes: their broader contribution to IPM and crop production. Biocontrol Sci Technol. https://doi.org/10.1080/09583157.2020.1771279
Quesada-Moraga E, Yousef-Naef M, Garrido-Jurado I (2020) Advances in the use of entomopathogenic fungi as biopesticides in suppressing crop insect pests. In: Nick Birch N, Glare T (eds) Biopesticides for sustainable agriculture. Burleigh Dodds Science Publishing, Cambridge, pp 63–98
González-Mas N, Sánchez-Ortiz A, Valverde-García P, Quesada-Moraga E (2019) Effects of endophytic entomopathogenic ascomycetes on Aphisgossypii glover life-history traits and interaction with melon plants. Insects 10:165
Gurulingappa P, Sword GA, Murdoch G, McGee PA (2010) Colonization of crop plants by fungal entomopathogens and their effects on two insect pests when in planta. Biol Control 55:34–41
Akutse KS, Maniania NK, Fiaboe K, Van den Berg J, Ekesi S (2013) Endophytic colonization of Viciafaba and Phaseolusvulgaris (Fabaceae) by fungal pathogens and their effects on the life-history parameters of Liriomyzahuidobrensis (Diptera: Agromyzidae). Fungal Ecol 6:293–301
Behie SW, Zelisko PM, Bidochka MJ (2012) Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science 336:1576–1577
Behie SW, Jones SJ, Bidochka MJ (2015) Plant tissue localization of the endophytic insect pathogenic fungi Metarhizium and Beauveria. Fungal Ecol 13:112–119
Wagner BL, Lewis LC (2000) Colonization of corn, Zeamays, by the entomopathogenic fungus Beauveriabassiana. Appl Environ Microbiol 66:3468–3473
Myers JR, Kmiecik K (2017) Common bean: economic importance and relevance to biological science research. Anonymous the common bean genome. Springer, Cham, pp 1–20
Capinera L (2008) Twospotted spider mite, Tetranychusurticae Koch (Acari: Tetranychidae). In: Capinera JL (ed) Encyclopedia of entomology. Springer, Dordrecht
Boubou A, Migeon A, Roderick GK, Navajas M (2011) Recent emergence and worldwide spread of the red tomato spider mite, Tetranychusevansi: genetic variation and multiple cryptic invasions. Biol Invasions 13:81–92
Dagli F, Tunc I (2001) Dicofol resistance in Tetranychus cinnabarinus: resistance and stability of resistance in populations from Antalya, Turkey. Pest Manag Sci 57:609–614
Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L (2010) Acaricide resistance mechanisms in the two-spotted spider mite Tetranychusurticae and other important Acari: a review. Insect Biochem Mol 40:563–572
Goettel MS, Inglis GD (1997) Fungi: hyphomycetes. Manual of techniques in insect pathology. Elsevier, Amsterdam, pp 213–249
Posada F, Aime MC, Peterson SW, Rehner SA, Vega FE (2007) Inoculation of coffee plants with the fungal entomopathogen Beauveriabassiana (Ascomycota: Hypocreales). Mycol Res 111:748–757
Rehner SA, Minnis AM, Sung G, Luangsa-ard JJ, Devotto L, Humber RA (2011) Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103:1055–1073
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 18:315–322
Landa BB, López-Díaz C, Jiménez-Fernández D, Montes-Borrego M, Muñoz-Ledesma FJ, Ortiz-Urquiza A, Quesada-Moraga E (2013) In-planta detection and monitorization of endophytic colonization by a Beauveriabassiana strain using a new-developed nested and quantitative PCR-based assay and confocal laser scanning microscopy. J Invertebr Pathol 114:128–138
Batta YA (2018) Efficacy of two species of entomopathogenic fungi against the stored-grain pest, Sitophilusgranarius L. (Curculionidae: Coleoptera), via oral ingestion. Egypt J Biol Pest Control 28:44–52
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408
Zhou Y, Zhang Y, Luo Z, Fan Y, Tang G, Liu L, Pei Y (2012) Selection of optimal reference genes for expression analysis in the entomopathogenic fungus Beauveriabassiana during development, under changing nutrient conditions, and after exposure to abiotic stresses. Appl Microbiol Biotechnol 93:679–685
Fang W, Feng J, Fan Y, Zhang Y, Bidochka MJ, Leger RJS, Pei Y (2009) Expressing a fusion protein with protease and chitinase activities increases the virulence of the insect pathogen Beauveriabassiana. J Invertebr Pathol 102:155–159
Fang W, Leng B, Xiao Y, Jin K, Ma J, Fan Y, Feng J, Yang X, Zhang Y, Pei Y (2005) Cloning of Beauveriabassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71:363–370
Abdo C, Nemer N, Nemer G, Jawdah YA, Atamian H, Kawar NS (2008) Isolation of Beauveria species from Lebanon and evaluation of its efficacy against the cedar web-spinning sawfly, Cephalciatannourinensis. Biocontrol 53:341–352
Parsa S, García-Lemos AM, Castillo K, Ortiz V, López-Lavalle LAB, Braun J, Vega FE (2016) Fungal endophytes in germinated seeds of the common bean, Phaseolusvulgaris. Fungal Biol 120:783–790
Parsa S, Ortiz V, Vega FE (2013) Establishing fungal entomopathogens as endophytes: towards endophytic biological control. J Vis Exp. https://doi.org/10.3791/50360
McKinnon AC, Saari S, Moran-Diez ME, Meyling NV, Raad M, Glare TR (2017) Beauveriabassiana as an endophyte: a critical review on associated methodology and biocontrol potential. Biocontrol 62:1–17
Healey A, Furtado A, Cooper T, Henry RJ (2014) Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10:21
Al Khoury C, Guillot J, Nemer N (2020) Susceptibility and development of resistance of the mite Tetranychusurticae to aerial conidia and blastospores of the entomopathogenic fungus Beauveriabassiana. Syst App Acarol 25:429–443
Tehri K, Gulati R, Geroh M, Dhankhar SK (2015) Dry weather: a crucial constraint in the field efficacy of entomopathogenic fungus BeauveriaBassiana against Tetranychusurticae Koch (Acari: Tetranychidae). J Entomol 3:287–291
Ortucu S, Algur OF (2017) A laboratory assessment of two local strains of the Beauveriabassiana (Bals.) Vuill. against the Tetranychusurticae (acari: Tetranychidae) and their potential as a mycopesticide. J Pathog 2107:1–7
Gómez-Vidal S, Lopez-Llorca LV, Jansson H, Salinas J (2006) Endophytic colonization of date palm (Phoenixdactylifera L.) leaves by entomopathogenic fungi. Micron 37:624–632
Quesada-Moraga E, Ruiz-García A, Santiago-Alvarez C (2006) Laboratory evaluation of entomopathogenic fungi Beauveriabassiana and Metarhiziumanisopliae against puparia and adults of Ceratitiscapitata (Diptera: Tephritidae). J Econ Entomol 99:1955–1966
Bensoussan N, Zhurov V, Yamakawa S, O’Neil CH, Suzuki T, Grbić M, Grbić V (2018) The digestive system of the two-spotted spider mite, Tetranychusurticae Koch, in the context of the mite-plant interaction. Front Plant Sci 9:1206
Charnley AK, Leger RS (1991) The role of cuticle-degrading enzymes in fungal pathogenesis in insects. In: Ann R (ed) The fungal spore and disease initiation in plants and animals. Springer, Boston, p 267
Mannino MC, Huarte-Bonnet C, Davyt-Colo B, Pedrini N (2019) Is the insect cuticle the only entry gate for fungal infection? insights into alternative modes of action of entomopathogenic fungi. J Fungi 5:33
Wan J, Zhang X, Neece D, Ramonell KM, Clough S, Kim S, Stacey MG, Stacey G (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20:471–481
Behie SW, Moreira CC, Sementchoukova I, Barelli L, Zelisko PM, Bidochka MJ (2017) Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat Commun 8:1–5
Branine M, Bazzicalupo A, Branco S (2019) Biology and applications of endophytic insect-pathogenic fungi. PLoS Pathog 15:e1007831
Holder DJ, Kirkland BH, Lewis MW, Keyhani NO (2007) Surface characteristics of the entomopathogenic fungus Beauveria (Cordyceps) bassiana. Microbiology 153:3448–3457
Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA (2014) Induced systemic resistance by beneficial microbes. Ann Rev Phytopathol 52:347–375
Wu Q, Patocka J, Nepovimova E, Kuca K (2018) A review on the synthesis and bioactivity aspects of beauvericin, a fusarium mycotoxin. Front Pharmacol 9:1338
Barelli L, Moonjely S, Behie SW, Bidochka MJ (2016) Fungi with multifunctional lifestyles: endophytic insect pathogenic fungi. Plant Mol Biol 90:657–664
Zimmermann G (2007) Review on safety of the entomopathogenic fungi Beauveriabassiana and Beauveriabrongniartii. Biocontrol Sci Technol 17:553–596
Strasser H, Vey A, Butt TM (2000) Are there any risks in using entomopathogenic fungi for pest control, with particular reference to the bioactive metabolites of Metarhizium, Tolypocladium and Beauveria species? Biocontrol Sci Technol 10:717–735
Mascarin GM, Jaronski ST (2016) The production and uses of Beauveriabassiana as a microbial insecticide. World J Microbiol Biotechnol 32:177
Funding
This work was funded by the Lebanese National Council for Scientific Research (CNRS), grant name (CNRS-L/USEK) and “Coopération pour l'évaluation et le développement de la recherche” CEDRE grant number 37349SA.
Author information
Authors and Affiliations
Contributions
The study conception and design, material preparation, data collection and analysis were performed by Charbel Al Khoury.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Al Khoury, C. Molecular insight into the endophytic growth of Beauveria bassiana within Phaseolus vulgaris in the presence or absence of Tetranychus urticae. Mol Biol Rep 48, 2485–2496 (2021). https://doi.org/10.1007/s11033-021-06283-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06283-3