Abstract
Insulin resistance may become the most powerful predictor of future development of type 2 diabetes mellitus (T2DM) and a therapeutic target for the treatment of the same. Both Resistin, an adipose derived peptide hormone and Urotensin II a potent vasoconstrictor, are reported to be involved in the development of insulin resistance and T2DM but the results remain contradictory. Therefore, investigations were carried out to study the association of T2DM and single nucleotide polymorphism (SNP) in Resistin (RETN) gene at rs3745367 (+ 299 G > A) and Urotensin II (UTS2) gene at rs228648 (+ 143 G > A) and rs2890565 (+ 3836 C > T) in a North Indian population. Method: The present case–control study, conducted from August 2017 to July 2020, involved 168 T2DM patients and 102 healthy controls. SNPs rs3745367, rs228648 and rs2890565 were amplified from genomic DNA in the studied samples by polymerase chain reaction (PCR) using specific primers. The amplified products were genotyped by restriction fragment length polymorphism (RFLP) using particular restriction endonucleases. Clinical parameters viz. glycosylated haemoglobin (HbA1c), fasting blood glucose (FBG), high density lipoprotein cholesterol (HDL-C), triglycerides (TG), total cholesterol (CHL) and fasting insulin were determined by enzymatic methods. Result and conclusion: A statistically significant association between T2DM and RETN gene at SNP rs3745367 (p = 0.001) and UTS2 gene at SNP rs2890565 (p = 0.001) was observed. In RETN gene SNP rs3745367, insulin and homeostasis model assessment of insulin resistance (HOMA-IR) were found to be higher in GA + AA combined genotype than in GG genotype for T2DM subjects. Regression analysis revealed that SNP rs2890565 and HOMA-IR were independently associated with the risk of development of T2DM when three SNPs were taken as independent variable adjusted for clinical variables. Among four haplotypes, A/T was found associated with increased risk of T2DM as determined for rs228648 and rs2890565 of UTS2 gene. It can be concluded from these results that polymorphism at rs3745367 of RETN gene and at rs2890565 of UTS2 gene are associated with risk of T2DM in North Indian population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Type 2 diabetes mellitus (T2DM) is a chronic disease that develops due to multiple factors and it is characterized by insulin resistance, impaired insulin secretion or both [1]. Globally 415 million people diagnosed with this disease and number is expected to increase more than 640 million by 2040 [2]. T2DM is considered to be a polygenic disease and many genetic markers including cytokines have been reported to be involved in development of T2DM [3,4,5,6]. Resistin is a cysteine rich adipose derived peptide hormone encoded by resistin (RETN) gene [7]. It is secreted in humans by monocytes, macrophages and adipocytes and reported to regulate the glucose, energy and lipid homeostasis [8,9,10]. Resistin treated macrophages and adipocytes showed accumulation of lipids whereas a decreased fatty acid uptake in skeletal muscle cells by resistin treatment leads to lipid toxicity and insulin resistance [11,12,13]. Some studies reported an increased RETN gene expression when there is insulin resistance, T2DM and obesity [14, 15] but others failed to show any change in RETN expression in same conditions [16,17,18]. A genome wide association study showed that polymorphisms of rs3745367 and rs1423096 in RETN gene and haplotypes (A/G and G/G) determined by these SNPs were significantly associated with higher resistin levels and increased T2DM risk [19]. Although, worldwide several association studies were conducted on various genetic variations of RETN gene viz. -420 C > G in promoter region, + 156 C > T and + 299 G > A in intron 2 (SNP rs3745367), + 1084 G > A and + 62 G > A in 3′ untranslated region, however most of them showed negative association with T2DM except SNP + 299 G > A which showed association with T2DM in some ethnicities whereas in other, contradictory results have been observed [20,21,22,23,24]. As association between different SNPs of RETN gene and T2DM except rs3745367 could not be observed in different population studies and information regarding the association of SNP rs3745367 with T2DM in Indian population is lacking, so the current study was planned.
Urotensin 2 (UTS2) gene specify an 11 amino acid cyclic peptide called Urotensin II (U-II), which is most potent mammalian vasoconstrictor identified to date that exerts its biological effects via interaction with a member of a G-protein coupled receptor superfamily, originally termed GPR14 [25,26,27]. Recent evidence suggests that vascular factor dysfunction contributes to insulin resistance [28]. In a study conducted on bovines, it was observed that both UTS2 and UTS2 receptor (UTS2R) genes were significantly associated with regulation of skeletal muscle fat accumulation and fatty acid metabolism [29]. U-II reportedly reduces glucose-induced insulin secretion in the perfused rat pancreas [30] and also the genetic markers present in UTS2 gene were reported to be involved in the prevalence of T2DM [31,32,33]. GGT (605G, 143G and 3836T) haplotype in UTS2 gene are associated with higher plasma level of U-II and insulin, and higher homeostasis model assessment of insulin resistance index (HOMA-IR) and β-cell function (HOMA-β) [33]. The present study was performed to assess possible association between RETN gene polymorphism at SNP rs3745367 and UTS2 gene polymorphisms at SNPs rs228648 and rs2890565 with T2DM in North Indian population as information is lacking in this population.
Materials and methods
Study population
The present study comprised of 270 subjects divided in two groups. First group consisting of 168 T2DM subjects aged 45–85 years and second group had 102 healthy control subjects aged 45–83 years from different parts of Haryana (India). Subjects suspected to have liver, kidney, lungs, coronary artery disease (CAD) and other major chronic or inflammatory diseases were excluded from the study. Diabetes was diagnosed according to the Indian council of Medical Research (ICMR), New Delhi criteria for diabetes which include fasting blood glucose > 126 mg/dl and glycosylated haemoglobin > 6.5% and information about the duration of diabetes was obtained from medical records. Subjects who were 45 or more years of age, no past history of diabetes, normal fasting blood glucose (FBG) and < 5.6% glycosylated haemoglobin (HbA1c) were included in the study as control subjects. The study was approved by the Ethical committee of Kurukshetra University, Kurukshetra and all the subjects who participated in the study had signed the informed consent form.
Body measurements and blood collection
Age, sex, height, weight and personal history were revealed in standardized questionnaire for all the enrolled subjects. Body mass index (BMI) was calculated by dividing the weight in kilogram (Kg)/height in meter square (m2). After 10–12 h fasting, 5 ml venous blood was collected from all the subjects in the morning. Blood was collected in two types of vials, plain vial for serum collection and in ethylene diammine tetra acetate (EDTA) coated vial for FBG, HbA1c evaluation. Fasting insulin concentration was determined by enzyme linked immunosorbent assay (ELISA). HbA1c, high density lipoprotein cholesterol (HDL-C), triacylglycerides (TG), total cholesterol (CHL) were measured using enzymatic methods. Homeostasis model assessment (HOMA-IR) score were calculated by using the formula fasting insulin (IU/ml) × fasting blood glucose (mg/dl)/405.
DNA extraction and genotyping
DNA was extracted from whole blood stored at −20 °C in EDTA coated vials using Qiamp DNA extraction mini kit (QIAGEN, New Delhi, India) according to manufacturer instructions and stored at −20 °C for further use. Quality of DNA was checked on UV-spectrophotometer.
Genotypes were determined by PCR–RFLP method. The PCR amplification was performed in a 25 µl reaction mixture containing master mix 2X (GoTaq® Green Master Mix, Promega, India), 7.5 pmol of each forward and reverse primer (sequence shown in Table 1), 40–60 ng of genomic DNA and best quality water. Assay conditions included an initial denaturation at 95 °C for 5 min, followed by 35–40 cycles of denaturation at 95 °C for 30 s, reannealing at 50–60 °C for 1 min and extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min.
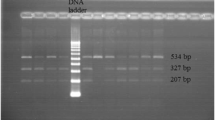
Amplified products of RETN and UTS2 gene containing SNPs of interest were run on 1.5% agarose gels which were stained using ethidium bromide and visualised on UV-transilluminator. A 10 µl aliquot of the PCR products was completely digested with 2–5 units of restriction enzymes and restriction digested PCR products were resolved on 3% agarose gel. G to A transition mutation in resistin gene at SNP rs3745367 generates an AluI restriction enzyme site. Wild type DNA in resistin gene SNP rs3745367 yielded four bands with 243, 55, 52 and 23 bp, while the mutated DNA showed five bands with 158, 85, 55, 52 and 23 bp. Transition mutation from C to T in UTS2 gene polymorphism at SNP rs228648 creates a N1aIII site. Wild type DNA yielded two bands of 109 and 32 bp and mutated DNA showed 3 bands with 57, 52 and 32 bp. Transition mutation from G to A in UTS2 gene at SNP rs2890565 eliminates a CviQI site. The wild type DNA expected to show three bands with 161, 84 and 18 bp whereas mutated DNA yielded two bands with 245 and 18 bp.
Statistical analysis
The Hardy–Weinberg equilibrium for genotypic distributions was examined using the χ2 goodness-of-fit test. Data for the biochemical measurements was analysed by Student’s t-test and data are presented as means ± SD. Differences were considered to be significant at p < 0.05. To evaluate the association between type 2 diabetes as dependent variable and potential clinical parameters, logistic regression was applied. Haplotype analysis was done using SHEsis online software.
Results
Analysis of clinical and demographic profile
Current study is comprised of 168 T2DM patients and 102 healthy individuals as control. Biochemical parameters and demographic profile of both T2DM and control subjects are shown in Table 2. Results showed a significant increase in FBG, HbA1c%, fasting insulin, HOMA-IR, total CHL and low density lipoprotein cholesterol (LDL-C) in T2DM group (Table 2).
Genotyping analysis of genetic markers
Out of three SNPs, two were detected to be highly polymorphic (rs3745367 and rs2890565), whereas SNP (rs228648) showed no significant difference in case and control population. Genotypic differences of SNPs in T2DM and control group are shown in Tables 3 and 4 as accessed by chi square test.
Association of RETN gene polymorphism (rs3745367) with T2DM
AA genotype of RETN gene SNP at rs3745367 was significantly higher in T2DM group as compared to control group (χ2 = 11.29, p = 0.003). Allelic frequency of A allele was found to be increased in T2DM group as compared to control group (G VS A, OR = 1.84, 95% CI: 1.26–2.69, p = 0.001). Significant differences in genotype distribution of polymorphism rs3745367 were seen in three genetic models viz. additive, recessive model and dominant genetic model. (Table 3).
Association of UTS2 gene polymorphisms (rs228648 and rs2890565) with T2DM
For UTS2 gene polymorphism at SNP rs228648, differences in genotypic distribution in two study groups could not be observed (χ2 = 0.396, p = 0.82). Significant differences could not be observed between allele frequencies of T2DM and control subjects (G vs A, OR = 1.09, 95% CI: 0.77–1.56, p = 0.60) (Table 4).
Significant differences were observed in genotypic distribution of UTS2 gene polymorphism at SNP rs2890565 between T2DM and control groups (χ2 = 8.72, p = 0.01). Allele frequency of T allele was found to be significantly higher in T2DM group as compared to control group (12.2 vs 3.9%, C vs T, OR = 3.4, 95% CI: 1.56–7.41, p = 0.001). In addition, three genetic models viz. additive, recessive and dominant genetic model showed significant association (Table 4).
Comparisons of genotypes with clinical profiles of subjects
Clinical comparisons between T2DM and control subjects were performed using dominant model (GG vs GA + AA). There was a statistical significant association in both T2DM and control subjects having GA + AA genotypes with p-value of 0.01 and 0.04 respectively regarding insulin levels for RETN gene SNP rs3745367. There was lowered HOMA-IR in T2DM subjects having GG genotypes with p-value of 0.004 (Table 5). Significant association for clinical parameters with gene polymorphism at SNP rs228648 and rs2890565 of UTS2 gene could not be observed except SBP which was found to be higher in CT + TT genotype as compared to wild type (CC) for SNP rs2890565 in both T2DM and control groups (p-value = 0.04 and 0.01) (Table 6) and HDL-C was found to be lowered in GA + AA genotypes compared to wild type (GG) for SNP rs228648 (p-value = 0.008) (Table 7).
Haplotype analysis
SHEsis online software was used for haplotype analysis of UTS2 gene SNPs (Shi and He 2005). All possible haplotypes were examined from the observed genotypes of studied group for UTS2 gene SNPs rs228648 (G > A) and rs2890565 (C > T) and haplotype (A/T) was found to be significantly associated with the increase risk of developing T2DM (Table 8).
Regression analysis
Multiple logistic regression analysis was done using SPSS software in which T2DM was taken as dependent variable and three polymorphisms were taken as independent variable. It was observed that SNP rs3745367 and rs2890565 were independently associated with T2DM (p = 0.01 and p = 0.009 respectively) and HOMA-IR and SNP rs2890565 were associated with T2DM after adjusted for age, sex and HOMA-IR (p = 0.00 and p = 0.01 respectively) (Table 9).
Discussion
Studies elucidated that resistin protein might impair insulin action, glucose tolerance, decreased glucose uptake in skeletal muscle cells and play a role in obesity [34, 35]. As SNP rs3745367 is present in 2nd intron of RETN gene that has no known regulatory function, however studies have shown that SNPs in the non-coding region, such as the 3-untranslated region can affect gene expression [36]. RETN gene variant at SNP rs3745367 and obesity were reported as potential risk factors for T2DM in a study conducted on Caucasians [37]. This SNP also affects serum resistin concentration which was found to be higher in patients with T2DM [38,39,40]. It might be a risk factor for stroke in patients with T2DM [41] and associated with unfavourable outcomes in T2DM patients [42]. Polymorphism of RETN gene rs3745367 is involved in the development of insulin resistance and T2DM as reported in Iraqi population [22], meta-analysis study [43] and observed in the present study. Contrarily, significant association between RETN gene polymorphism at SNP rs3745367 and risk of T2DM could not be observed in Japanese population [44]. Minor allele A was more frequent in T2DM patients than in the control group (p = 0.001) [23] and GG may be a protective factor against T2DM [43] and as reported in the present study. Serum resistin concentration as well as HOMA-IR was remarkably more in mutant genotype (GA + AA) as compared with wild type (GG) in German [45], Thailand [21] and Egyption [46] T2DM subjects. In contrast, there was no difference in GG and GA + AA genotypes in Han Chinese patients thus neglecting the risk of T2DM incidence [47].
U-II protein may be involved in the pathogenesis of T2DM [48] as it may increase blood glucose level through the mobilization of glucose. Circulating U-II concentrations were significantly higher in T2DM patients with metabolic syndrome compared to those without metabolic syndrome (p < 0.001) but significant correlation could not be observed between U-II concentrations and insulinemia in Belgium population [49]. Levels of U-II were found higher in women with gestational diabetes mellitus (GDM) and its level showed a positive correlation with insulin resistance marker (HOMA-IR) [50] and increased cardiovascular risk as observed in patients with polycystic ovary syndrome [51]. UTS2 gene SNPs at rs228648 and at rs2890565 were found essentially associated with T2DM in Han Chinese and Japanese population respectively [31, 52, 53] and these SNPs were suggested as risk factors in developing diabetic retinopathy (DR) in Caucasians [54]. SNP rs2890565 was found to be significantly associated with T2DM in our study. T Allele frequency was significantly higher in T2DM subjects as compared to control subjects (12.2 vs 3.9%, p = 0.001) so carrier of T allele may be prone to pathogenesis of T2DM but significant association could not be observed between clinical parameters, genotypes and T2DM in our study.
Conclusion
It can be concluded from the present study that individuals with AA genotype of RETN gene SNP rs3745367 are more prone to develop insulin resistance and T2DM as compared to those carrying GG genotype and G allele at this locus may be protective factor against T2DM. T allele in UTS2 gene SNP rs2890565 is more frequent in T2DM subjects than in controls thus may be a predisposing factor in development of T2DM in Indian population.
References
Adeghate E, Schattner P, Dunn E (2006) An update on the etiology and epidemiology of diabetes mellitus. Ann NY Acad Sci 1084(1):1–29
IDF (2018) International Diabetes Federation Diabetes Atlas, 8th edn. IDF
Kubaszek A, Pihlajamäki J, Komarovski V et al (2003) Promoter polymorphisms of the TNF-α (G-308A) and IL-6 (C-174G) genes predict the conversion from impaired glucose tolerance to type 2 diabetes: the Finnish Diabetes Prevention Study. Diabetes 52(7):1872–1876
Solaleh E, Arash HN, Azam N et al (2009) Promoter resistin gene polymorphism in patients with type 2 diabetes and its influence on concerned metabolic phenotypes. J Diabetes Metab Disord 8:17
Arababadi MK, Mirzaei MR, Sajadi SM et al (2012) Interleukin (IL)-10 gene polymorphisms are associated with type 2 diabetes with and without nephropathy: a study of patients from the southeast region of Iran. Inflammation 35(3):797–802
Saxena M, Srivastava N, Banerjee M (2013) Association of IL-6, TNF-α and IL-10 gene polymorphisms with type 2 diabetes mellitus. Mol Bio Rep 40(11):6271–6279
Wang H, Chu WS, Hemphill C et al (2002) Human resistin gene: molecular scanning and evaluation of association with insulin sensitivity and type 2 diabetes in Caucasians. J Clin Endo Meta 87(6):2520–2524
Patel L, Buckels AC, Kinghorn IJ et al (2003) Resistin is expressed in human macrophages and directly regulated by PPARγ activators. Bio Bio Res Comm 300(2):472–476
Jung HS, Park KH, Cho YM et al (2006) Resistin is secreted from macrophages in atheromas and promotes atherosclerosis. Cardio Res 69(1):76–85
Sadashiv ST, Paul BN, Kumar S et al (2012) Over expression of resistin in adipose tissue of the obese induces insulin resistance. World J Diabetes 3(7):135
Lee TS, Lin CY, Tsai JY et al (2009) Resistin increases lipid accumulation by affecting class A scavenger receptor, CD36 and ATP-binding cassette transporter-A1 in macrophages. Life Sci 84(3–4):97–104
Ikeda Y, Tsuchiya H, Hama S et al (2013) Resistin affects lipid metabolism during adipocyte maturation of 3T3‐L1 cells. FEBS J 280(22):5884–5895
Wen F, Zhang H, Bao C et al (2015) Resistin increases ectopic deposition of lipids through miR-696 in C2C12 cells. Biochem Genet 53(4–6):63–71
McTernan PG, Fisher FM, Valsamakis G et al (2003) Resistin and type 2 diabetes: regulation of resistin expression by insulin and rosiglitazone and the effects of recombinant resistin on lipid and glucose metabolism in human differentiated adipocytes. J Clin Endo Meta 88(12):6098–6106
Degawa-Yamauchi M, Bovenkerk JE, Juliar BE et al (2003) Serum resistin (FIZZ3) protein is increased in obese humans. J Clin Endo Meta 88(11):5452–5455
Lee JH, Chan JL, Yiannakouris N et al (2003) Circulating resistin levels are not associated with obesity or insulin resistance in humans and are not regulated by fasting or leptin administration: cross-sectional and interventional studies in normal, insulin-resistant, and diabetic subjects. J Clin Endo Meta 88(10):4848–4856
Utzschneider KM, Carr DB, Tong J et al (2005) Resistin is not associated with insulin sensitivity or the metabolic syndrome in humans. Diabetologia 48(11):2330–2333
Gerber M, Boettner A, Seidel B et al (2005) Serum resistin levels of obese and lean children and adolescents: biochemical analysis and clinical relevance. J Clin Endo Metab 90(8):4503–4509
Chung CM, Lin TH, Chen JW et al (2014) Common quantitative trait locus downstream of RETN gene identified by genome‐wide association study is associated with risk of type 2 diabetes mellitus in Han Chinese: a Mendelian randomization effect. Diabetes Metab Res Rev 30(3): 232–240
Conneely KN, Silander K, Scott LJ et al (2004) Variation in the resistin gene is associated with obesity and insulin-related phenotypes in Finnish subjects. Diabetologia 47(10):1782–1788
Suriyaprom K, Phonrat B, Namjuntra P et al (2009) The+ 299 (G> A) resistin gene polymorphism and susceptibility to type 2 diabetes in Thais. J Clin Bio Nutr 44(1):104–110
Al-Hilali HA, Abduljaleel AK (2015) The role of TNF and Resistin Gene+ 299 (G/A) polymorphism in the development of insulin resistance in non-obese Type 2 Diabetes Mellitus Iraqi patients. Int J Curr Microbiol App Sci 4(10):475–486
Kaur H, Kapoor R, Singh N (2016) Circulating levels of resistin and +299 (G>A) Resistin gene polymorphism in type 2 diabetes mellitus, a study from North India. Int J Curr Res 8(07):34692–34696
Montiel-Tellez BS, Nieva-Vazquez A, Porchia LM et al (2016) + 62G> A and g.-420C> G RETNPolymorphisms and the risk of developing type 2 diabetes and obesity: original research on a Mexican population and meta-analysis. Endocrinol Metab Syndr 5(228). https://doi.org/10.4172/2161-1017.1000228
Ames RS, Sarau HM, Chambers JK et al (1999) Human urotensin-II is a potent vasoconstrictor and agonist for the orphan receptor GPR14. Nature 401(6750):282–286
Douglas SA, Sulpizio AC, Piercy V et al (2000) Differential vasoconstrictor activity of human urotensin‐II in vascular tissue isolated from the rat, mouse, dog, pig, marmoset and cynomolgus monkey. British J Pharma 131(7):1262–1274
Böhm F, Pernow J (2002) Urotensin II evokes potent vasoconstriction in humans in vivo. British J Pharma 135(1):25–27
Baron AD (2002) Insulin resistance and vascular function. J Dia Comp 16(1):92–102
Jiang Z, Michal JJ, Tobey DJ et al (2008) Comparative understanding of UTS2 and UTS2R genes for their involvement in type 2 diabetes mellitus. Int J Biol Sci 4(2):96
Silvestre RA, Rodriguez-Gallardo J, Egido EM et al (2001) Inhibition of insulin release by urotensin II-a study on the perfused rat pancreas. Hormon Meta Res 33(06):379–381
Wenyi Z, Suzuki S, Hirai M et al (2003) Role of urotensin II gene in genetic susceptibility to Type 2 diabetes mellitus in Japanese subjects. Diabetologia 46(7):972–976
Suzuki S, Wenyi Z, Hirai M et al (2004) Genetic variations at urotensin II and urotensin II receptor genes and risk of type 2 diabetes mellitus in Japanese. Peptides 25(10):1803–1808
Ong KL, Wong LY, Man YB et al (2006) Haplotypes in the urotensin II gene and urotensin II receptor gene are associated with insulin resistance and impaired glucose tolerance. Peptides 27(7):1659–1667
McTernan CL, McTernan PG, Harte AL et al (2002) Resistin, central obesity, and type 2 diabetes. Lancet 359(9300):46–47
Rajala MW, Obici S, Scherer PE et al (2003) Adipose-derived resistin and gut-derived resistin-like molecule–β selectively impair insulin action on glucose production. J Clin Invest 111(2):225–230
Pesole G, Mignone F, Gissi C et al (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276(1–2):73–81
Ma X, Warram JH, Trischitta V et al (2002) Genetic variants at the resistin locus and risk of type 2 diabetes in Caucasians. J Clin Endo Meta 87(9):4407–4410
Lau CH, Muniandy S (2011) Adiponectin and resistin gene polymorphisms in association with their respective adipokine levels. Ann Hum Genet 75(3):370–382
Bouchard L, Weisnagel SJ, Engert JC et al (2004) Human resistin gene polymorphism is associated with visceral obesity and fasting and oral glucose stimulated C-peptide in the Quebec Family Study. J Endo Invest 27(11):1003–1009
Osawa H, Onuma H, Ochi M et al (2005) Resistin SNP-420 determines its monocyte mRNA and serum levels inducing type 2 diabetes. Biochem Bio Res Comm 335(2):596–602
Nakashima E, Watarai A, Tsukahara T et al (2010) Association of resistin polymorphism, its serum levels and prevalence of stroke in Japanese type 2 diabetic patients. J Diabetes Invest 1(4):154–158
Kapłon‐Cieślicka A, Tymińska A, Rosiak M et al (2019) Resistin is a prognostic factor for death in type 2 diabetes. Diabetes Meta Res Rev 35(2):e3098
Kumar V, Singh J, Aneja A et al (2019) Association of RETN gene polymorphism at+ 299 G> A with type 2 diabetes mellitus: a meta-analysis. Int J Diabetes Dev Count 40:1–9
Osawa H, Onuma H, Murakami A et al (2002) Systematic search for single nucleotide polymorphisms in the resistin gene: the absence of evidence for the association of three identified single nucleotide polymorphisms with Japanese type 2 diabetes. Diabetes 51(3):863–866
Gouni‐Berthold I, Giannakidou E, Faust M, et al (2005) Resistin gene 3′‐untranslated region+ 62G→ A polymorphism is associated with hypertension but not diabetes mellitus type 2 in a German population. J Intern Med 258(6):518–526
Khalil O, Alnahal A, Ghonium M et al (2014) Does resistin gene polymorphisms+ 299 (G> A) participate in insulin resistance in Egyption non-obese type 2 diabetes? Int J Genet Med 2:117
Jiang B, Liu Y, Liu Y et al (2014) Association of four insulin resistance genes with type 2 diabetes mellitus and hypertension in the Chinese Han population. Mol Bio Rep 41(2):925–933
Ong KL, Wong LY, Cheung BM (2008) The role of urotensin II in the metabolic syndrome. Peptides 29(5):859–867
Gruson D, Rousseau MF, Ketelslegers JM et al (2010) Raised plasma urotensin II in type 2 diabetes patients is associated with the metabolic syndrome phenotype. J Clin Hypertension 12(8):653–660
Calan M, Arkan T, Kume T et al (2019) The relationship between urotensin II and insulin resistance in women with gestational diabetes mellitus. Hormones 18(1):91–97
Yilmaz Ö, Calan O, Kume T et al (2013) The relationship of urotensin II with insulin resistance and hs-CRP in patients having PCOS. Gyneco Endo 29(11):970–973
Sun HX, Du WN, Zuo J et al (2002) The association of two single nucleotide polymorphisms in PRKCZ and UTS2 respectively with type 2 diabetes in Han people of northern China. Zhongguo yi xue ke xue yuan xue bao. Acta Acad Med Sinicae 24(3):223–227
Zhu F, Ji L, Luo B (2002) The role of urotensin II gene in the genetic susceptibility to type 2 diabetes in Chinese population. Zhonghua Yi Xue Za Zhi 82(21):1473–1475
Okumus S, Igci YZ, Taskin T et al (2012) Association between Thr21Met and Ser89Asn polymorphisms of the urotensin-II (UTS2) gene, diabetes mellitus, and diabetic retinopathy. Curr Eye Res 37(10):921–929
Acknowledgement
The authors are thankful to Dr. Ashok Chaudhary and Dr. Sapna Grewal, Bio and Nano Technology Department, Guru Jambheshwar University of Science and Technology (G.J.U), Hisar, Haryana, for providing access to instruments, which have been used in present research work, in their laboratory.
Author information
Authors and Affiliations
Contributions
All authors contributed to this work significantly and all are in agreement with the content of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The research work was approved by the Ethical committee of Kurukshetra University, Kurukshetra and all the subjects who participated in the study had signed the informed consent form.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, V., Singh, J., Bala, K. et al. Association of resistin (rs3745367) and urotensin II (rs228648 and rs2890565) gene polymorphisms with risk of type 2 diabetes mellitus in Indian population. Mol Biol Rep 47, 9489–9497 (2020). https://doi.org/10.1007/s11033-020-05991-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05991-6