Abstract
This study was conducted to perform an epidemiological survey of carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates for nosocomial pneumonia (NP) and visit their Gram-negative bacteria neighbors (GNNs) from the respiratory tracts. Pulsed-field gel electrophoresis and multi locus sequence typing were performed to evaluate the clonal relationships of these isolates. Statistical methods were then used to understand their possible relationship between CRKP and their GNNs. Among the 63 CRKP isolates, 84.1% produced a KPC-2 carbapenemase gene, followed by blaNDM-1 and blaIMP-38 like. 13 different STs and 29 clones were identified. Less heterogeneous clone backgrounds were observed in 53 KPC-2 K. pneumoniae isolates, and 6 STs have been found to contain KPC-2, of which the predominant ST11 harbored 47 KPC-2-producing K. pneumoniae isolates. Particularly, the two infrequent isolates co-possessing blaKPC-2 and blaIMP-38 like shared a different clonal relationship. 87 GNNs isolates of CRKP were established, the analysis present that different strains showed significant or no difference with CRKP upon antibiotics susceptibility. Conclusion, ST11 harboring KPC-2-producing K. pneumoniae isolates were dominant for NP, and both clonal spread and horizontal transfer contributed to the dissemination of CRKP. However, additional studies are needed to explore the biological relationship with their GNNs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nosocomial pneumonia (NP) is one of the most common hospital-acquired infections and is associated with considerable morbidity and mortality [1]. Gram-negative pathogens (including enterobacteriaceae and nonfermenters) which are responsible for the respiratory infections in hospital patients that are difficult to treat with antimicrobial therapy, particularly carbapenem-resistant Gram-negative bacteria (CR-GNB) [2]. Of greater concerns are the populations at risk of acquiring respiratory infections for carbapenem-resistant K. pneumoniae (CRKP) which is regarded as the key pathogen in NP [3, 4]. A 7 years retrospective study on carbapenem-resistant enterobacteriaceae infections in three tertiary care hospitals of China showed that the patients of 68.8% diagnosed with hospital-acquired bacterial pneumonia were identified for CRKP infection, of which 25% died within 28 days [5]. Qureshi et al. [6] described that 10 of 41 patients with pneumonia from hospital or health care caused by CRKP for death. Currently, the prevalence and the high mortality associated with CRKP in NP also have been confirmed in other literature [7, 8]. In addition, K. pneumoniae is also considered as one of the most challenging multiple drug resistance organisms, especially when they are isolated with other co-pathogens from polymicrobial respiratory [9]. Acquired carbapenemases in intra-species and inter-species facilitate their spread among bacterial pathogens. Encoding carbapenemase genes including serine-type enzymes (KPC, OXA-48) and metallo β-lactamases (NDM, IMP, VIM) in K. pneumoniae have been typically reported [10].
The epidemiology of CRKP strains were described worldwidely. The major clone of ST258 K. pneumoniae undeniably was reported in worldwide, while the high-risk clone of K. pneumoniae ST11 is frequently reported as a nosocomial pathogen in numerous geographic regions, including China [11]. Nevertheless, data on the molecular characterization of these CRKP isolates collected from NP is still limited, and the inter-reaction between the CRKP and the other Gram-negative pathogens, regarding as the neighbors of CRKP isolates from the same specimen category, scarcely been reported.
Gram-negative bacteria (GNB) consist of multiple species in commensal or symbiotic coexistence, and several studies had been recorded to monitor the effects of commensal species on the growth or survival of the target species [12, 13]. With antimicrobial resistance becoming a common development of microbial, GNB are often researched as multi-drug resistance (MDR), which has been recorded for several articles [14, 15], resulting in increased mortality for the limitation of treatment. Whether the species of microbial communities are co-infection or colonization, the sources of specimens are more focused on the gastrointestinal flora [16,17,18], few took insight into the respiratory tract.
In this report, we sought to investigate an molecular epidemiology of CRKP isolates, targeting from NP, received at a China academic tertiary hospital from the year of 2015 to 2016. Besides, we payed a visit to the GNNs of CRKP isolates from the respiratory tract, to understand their distribution, antimicrobial resistance and the possibility of interspecies relevance with CRKP upon the antimicrobial susceptibility.
Materials and methods
Study settings and ethical statement
The study was designed as a retrospective analysis that included CRKP isolates acquired from NP in a tertiary hospital with 2800 beds. Antibiotic resistance data for relevant respiratory isolates in NP, during January 2015 to December 2016, was retrieved from electronic medical records. And the present study was approved by the Ethics Committee of the first affiliated hospital of Anhui Medical University.
Bacterial isolates in NP
Eligible isolates were included that not only showed resistance to carbapenems but also acquired from NP participants, including VAP (ventilator-associated pneumonia). Hospital-acquired pneumonia was defined as occurring at least 48 h after admission to hospital, which based on clinical manifestation to diagnose, including a new or progressive and persistent infiltrate on chest radiographs, also at least one systemic sign and two respiratory symptoms [1, 19]. For this study not only respiratory samples such as sputum, tracheobronchial secretions and bronchoalveolar lavage were eligible for inclusion. In addition, blood samples were as well selected upon bacteremia, when the clinical manifestations of NP were also produced, but no respiratory specimens were detected.
Antibiotic susceptibility testing was performed using a Vitek 2 automated system. Resistance to imipenem and meropenem was evaluated using agar dilution or disc diffusion method and interpreted according to CLSI guidlines [20]. All CRKP isolates in NP were screened for the presence of carbapenemase genes (blaKPC, blaIMP, blaVIM, blaNDM and blaOXA-48) by polymerase chain reaction (PCR).
To evaluate and characterize the clonal relationships of the K. pneumoniae isolates, pulsed-field gel electrophoresis (PFGE) of the XbaI-digested genomic DNA was conducted using a CHEF-DRIII system (Bio-Rad, United Kingdom) and a unique PFGE profile was created for each clonal strain. Concomitantly, multi locus sequence typing (MLST) was performed for CRKP isolates according to the Institute Pasteur MLST website. And sequence types (STs) were further defined by eBURST.
Concurrent interspecies of CRKP neighbours
In this study, CRKP isolates were labeled as the host GNB in NP, furthermore, we payed a visit to their neighbours that defined as concurrent interspecies which were isolated simultaneously or successively from the same specimen source from the single patient during hospitalization and also screened in GNB including enterobacteriaceae and nonfermenters.
Sampling collection of bacterial species, specimen source, MDR, carbapenem-resistant MDR (CR-MDR) and drug susceptibility to 12 selected antibiotics were included in our study. By the way, the definition for MDR made reference to Magiorakos et al. [21]. Strains with MDR showing resistance to imipenem or/and meropenem, namely CR-MDR GNB, become a global healthcare concern. Among MDR isolates, natural resistant strains were only chose several antibiotics for drug-sensitive tests. The above materials were retrieved from electronic medical records.
Statistical analysis
Descriptive statistical data were presented in frequency and percentage. In addition, a comparison of drug susceptibility adopted by CRKP isolates and their GNNs was conducted through independent-samples t test. All statistical analyses were performed using SPSS 21 software (IBM Corp., Armonk, NY).
Results
Of all the diagnosed cases of NP, a total of 63 CRKP isolates (only one strain was moderately resistance to imipenem) were isolated from 63 patients. The distribution of samples in NP was as follows: sputum samples (n = 44, 69.8%), blood samples (n = 10, 15.9%), tracheobronchial secretion samples (n = 3, 4.8%), bronchoalveolar lavage samples (n = 3, 4.8%) and the samples from VAP (n = 3, 4.8%). Among the 63 CRKP isolates diagnosed with NP, 25 samples had been isolated from ICU (25/63, 37.9%), 26 samples from medical wards including respiratory ward (n = 11, 17.5%), neurology ward (n = 6, 9.5%), haematology (n = 4, 6.3%), gastroenterology (n = 3, 4.8%), cardiology and oncology (n = 1, 1.6%, respectively). The others contained burn unit, general surgery and plastic surgery of surgical wards (n = 3, 4.8%) and emergency wards (n = 9, 14.3%).
Of these 63 CRKP strains, they showed resistant rates of 100% to ampicillin/sulbactam, ceftriaxone and meropenem, 98.4% to ceftazidime and imipenem, more than 92% to ceftazidime, aztreonam, ciprofloxacin and piperacillin/tazobactam. Amikacin, gentamicin and levofloxacin showed activities against CRKP with susceptibility rates of 76.2%, 79.4% and 88.9%, respectively. And all isolates were susceptible to Tigecycline (Table 1). Among the 63 studied isolates, 53 strains (53/63, 84.1%) produced a KPC-2 carbapenemase. The other carbapenemases genes were of blaNDM-1 (n = 5) and blaIMP-38 like (n = 2). Of those, one isolate co-harbored blaKPC-2 and blaNDM-1, while two isolates co-possessed blaKPC-2 and blaIMP-38 like. Results of screening for VIM and OXA-48 carbapenemases genes were negative. And 6 isolates were not found correlating to the aforementioned genes.
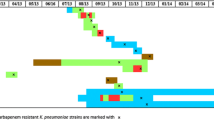
The distribution of the STs among the CRKP isolates for NP showed a similarity and diversity. 13 different STs were identified including the dominating type ST11 (n = 48, 76.2%), ST23 (n = 3, 4.8%), ST15 (n = 2, 3.2%), and ST17, ST35, ST36, ST307, ST340, ST414, ST528, ST661, ST1334 and ST1688 obtaining one strain respectively. Surprisingly, less heterogeneous clone backgrounds were observed in 53 KPC-2 K. pneumoniae isolates in this study, and 6 STs have been found to contain KPC-2, of which the predominant ST11 harbored 47 KPC-2-producing K. pneumoniae isolates (47/48, 97.9%), only one strain identified ST11 was excluded for negative carbapenemases gene. The distribution of STs in the isolated place was as follow: 7 different STs were identified in ICU (18 ST11, 72%; 2 ST23, 8%; 1 ST17/ST340/ST35/ST528/ST661, respectively, 4%), 6 STs in medical wards (21 ST11, 80.8%; 1 ST1334/ST1668/ST307/ST36/ST414, respectively, 3.8%), 2 STs in surgical wards (2 ST15, 66.7%; 1 ST23, 33.4%) and 1 ST in emergency wards (9 ST11, 100%) (Fig. 1).
The genetic relatedness of carbapenem-resistant K. pneumoniae clones in nosocomial pneumonia during the years 2015 and 2016. Isolates were clustered into 29 different clusters (29 PT, PFGE types). And carbapenemase gene alleles, sequence types (STs), time, ward, sample and the date of isolation were presented on the right. (sp sputum, bl blood, trs tracheobronchial secretion, bll bronchoalveolar lavage, vap the samples from ventilator-associated pneumonia (VAP), icu intensive care unit, res respiratory, neu neurology, hem haematology, car cardiology, onc oncology, gas gastroenterology, eme emergency, bur burn unit, gsd general surgery department, pla plastic surgery)
Clonal relationship has been further analyzed using PFGE which showed a moderation upon the 2 year study. 29 clones (PFGE type, PT01–PT29) were identified out of the 62 strains, because one K. pneumoniae isolate showed nucleic acid degradation whose sample from VAP of neurology ward belonging to ST1688 with negative carbapenemases gene. 48 CRKP isolates from clones PT03 to PT21 corresponded to the same type of ST11, of those, the high percentage clones were named PT12 (n = 13), PT07 (n = 7), PT04 (n = 4), PT08 (n = 4) and PT10 (n = 3). The other 14 strains contained 10 types of PT. From the isolated 2 years, the same types of ST11 and ST23 as well as the same PT types such as PT04, PT10 and PT12 were discovered, of which the ST11 and the PT10/PT12 were all isolated from the departments the ICU, medical ward and emergency ward. Besides theses, PT07 corresponding to ST11 or ST23 only appeared in 2016, but existed in most departments of the hospital including the wards of ICU, medical, surgical and emergency. In addition, the three CRKP strains co-harbored two carbapenemase genes were all isolated in 2016. Of which, one isolate co-harboring blaKPC-2 and blaNDM-1 was determined for ST11 possessing clone PT21, and the two strains isolated co-possessing blaKPC-2 and blaIMP-38 like shared a different clonal relationship, one strain was associated to PT07 corresponding to ST11, while the other PT25 corresponding to ST1334 (Fig. 1).
Among the 63 patients diagnosed with NP for CRKP infection, 32 patients had been screened out for concurrent interspecies which were all recovered from respiratory specimens during hospitalization. A total of 120 isolates were recovered, of which 33 were CRKP isolates as mentioned above (n = 32) for a patient owning two CRKP isolates (Table S1). And the 87 remaining isolates of GNB contained nonfermenters (n = 64) and enterobacteriaceae (n = 23). Of the 64 nonfermenters isolates, the predominant species was Acinetobacter baumannii (31/87, 35.6%), followed by Pseudomonas aeruginosa (17/87, 19.5%), Acinetobacter spp., (10/87, 11.5%), Stenotrophomonas maltophilia (3/87, 3.4%), and Acinetobacter haemolyticus, Burkholderia cepacia and Flavobacterium indologenes (1/87, 1.1%, respectively). Among enterobacteriaceae, isolated strains of Klebsiella oxytoca (7/87, 8%), Escherichia coli (5/87, 5.7%), Enterobacter aerogenes (3/87, 3.4%), Enterobacter cloacae (3/87, 3.4%), Serratia marcescens (3/87, 3.4%) and Proteus mirabilis (2/87, 2.3%) were collected from respiratory specimens in our study (Table 2).
Susceptibility testing was performed for 12 antibiotics (ceftazidime, ceftriaxone, cefepime, cefoperazone-sulbactam, aztreonam, amikacin, gentamicin, ciprofloxacin, levofloxacin, piperacillin-tazobactam, imipenem and meropenem) on these respiratory pathogens (Table S1). Excluded the 33 CRKP isolates and 5 strains of intrinsic resistance for nonfermenters, the remainder 82 strains, of which 64 isolates were considered as multidrug-resistant bacteria, species of Acinetobacter spp., K. oxytoca, E. coli and P. mirabilis all represented 100%, A. baumannii represented 96.8% (30/31), while P. aeruginosa showed 35.3% (6/17). Among these, 40 CR-MDR GNB isolates were recovered from the four dominant species including 2 nonfermenters isolates (A. baumannii and P. aeruginosa) and 2 enterobacteriaceae isolates (K. oxytoca and E. coli) and the predominant species was A. baumannii (n = 29, 29/30), followed by K. oxytoca (n = 6, 6/7), P. aeruginosa (n = 5, 5/6) and E. coli (n = 0, 0/5) (Table 1). Due to understand more about the relationship between concurrent interspecies, we used CRKP as a host isolate, which compared to the above four dominant species upon the results of 12 antibiotics susceptibilities. Univariate analysis showed that P. aeruginosa and E. coli were significant difference with CRKP for antibiotics susceptibility, while A. baumannii and K. oxytoca showed no difference with it (Table 3).
Discussion
NP is accepted as the most common of all healthcare-associated infections [22] and is associated with the particular morbidity and mortality. GNB are often known as the primary cause of NP, with the main pathogens isolated being Klebsiella spp., P. aeruginosa, Enterobacter spp., and Acinetobacter spp. [23, 24], which almost showed the commonly antimicrobial resistance. In this study, we described the molecular epidemiology of K. pneumoniae in NP that present the resistance to carbapenem. By the way, the distribution, MDR and the possible relationship of interspecies were carried out among GNB from the same respiratory specimen category.
The epidemiology survey on CRKP isolates associated with NP were not restricted to specific wards in this single-center study. Conversely, all patients with NP were included from ICU (37.9%), medical (17.5% respiratory ward, 9.5% neurology, 6.3% haematology, 4.8% gastroenterology, 1.6% oncology and cardiology, respectively), surgical (4.8%) and emergence department (14.3%) of this tertiary hospital, which allowed the surveillance of the data conclusion on all patients at risk of infection with NP. And obviously, ICU still play a major role in acquisition of carbapenem resistant isolates [25]. In our study, the top type of STs that accounted for CRKP in ICU was ST11 (72%), which also occupied the most important position in the wards of medical and emergency. The same type of ST11 was not only found in the different departments, but also appeared across the 2 years. Therefore, such clonal dissemination may be explained that nosocomial transmission was quite frequent in our hospital. Our medical workers should be aware of the issue and intervene the prevalence.
In this study, the most frequent carbapemases were KPC-2, followed by NDM-1 and IMP-38 like. And no carbapenemase VIM and OXA-48 producing K. pneumoniae was detected. Carbapenem resistance of six isolates was not found the aforementioned carbapemases genes that may due to the combination of AmpC or ESBL with porin defects [10]. We found these carbapemases-producing K. pneumoniae isolates were highly resistant to most antibiotics examined, even if amikacin and gentamicin had some susceptibility to these isolates. However, Tigecycline is remain the useful alternative in patients with serious infections caused by CR-GNB [26].
K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae strains of the hyperepidemic clonal complex 258 (CC258) are detected worldwide as hospital-acquired pathogens. In our study, the frequency of KPC-2 enzymes approximated to 80% among carbapenemase producers, which was higher than the previous report [27]. While ST258 is the predominant clone observed in European countries, ST11, belonging to CC258 (closely related to ST258), is the most frequently detected KPC-producing K. pneumoniae clone in China [28, 29]. Surprisingly, less heterogeneous were observed in 53 KPC-2 K. pneumoniae isolates in our study, and 6 STs have been found to contain KPC-2, of which the predominant ST11 harbored 47 KPC-2-producing K. pneumoniae isolates. High identity of the epidemic KPC-2 K. pneumoniae ST11 lineage in our study is mainly resulted from the clone spread, the others has been discovered in different K. pneumoniae strains, presumably due to the Tn4401-containing plasmid capable of mobilization through transposition [30].
The carbapenemases of NDM-1 and IMP-type belonging to metallo-β-lactamases (MBLs) were frequently identified in Enterobacteriaceae [31]. NDM-1-producing K. pneumoniae was first detected in a patient from India and then the sporadic spread has been reported in China [32, 33]. However, NDM-1 has been detected mostly in Acinetobacter spp., but the previous data [32] showed a proportional NDM-1-producing CRE isolates, including K. pneumoniae, suggesting the possible transmission of blaNDM−1-containing sequences from Acinetobacter spp. to Enterobacteriaceae. IMP was first found in S. marcescens in 1991, following IMP-producing K. pneumoniae have spread in Europe and Asia, including China [33, 34]. IMP-38, maybe a point mutation different from the common IMP-4, was rarely reported [35]. In our present study, IMP-38-like was found in two K. pneumoniae isolates, as well co-harbored blaKPC-2, which was first reported as far as we all know. Contrary to all expectations we carried of the result, the two IMP-38-like K. pneumoniae isolates belonged to the different clone, of which associated to ST11 and ST1334, respectively, and further been documented by PFGE.
Isolates from the different STs displayed a scientific moderation PFGE profiles, further demonstrating their clonal dissemination. However, it’s not just a single inheritance, as well present mutation. 29 clones (PT01–PT29) were identified out of the 62 strains, among these CRKP clones, 48 from clones PT03 to PT21 corresponded to the same type of ST11. PT12 as the dominant PFGE type that appeared across the year of 2015 and 2016, while the other tufted bands (i.e., PT07) that only appeared in 2016 but generally existed in each department of the hospital. Our study indicated that which contributed to the dissemination of CRKP in the tertiary hospital, both clonal spread and horizontal transfer should be considered, as well that seem meet with the rule of natural development.
Not only insight into the knowledge of K. pneumoniae, there is a need to understand the relationship with co-pathogens or commensals, for the ongoing emergence of antimicrobial resistance. In this study, species of K. pneumoniae were targeted as the host strains isolating from the respiratory tract which coexisting with other gram-negative bacteria, of which A. baumannii was the most frequently isolated species, followed by P. aeruginosa in the nonfermenters isolates, and the highest in Enterobacteriaceae was K. pneumoniae and E. coli. This distribution is similar to the list of priority pathogens for NP in previous articles [23, 24]. Surprisingly, they all showed a shocking MDR, except P. aeruginosa showed 35.3%, the rest three species almost 100%. Additionally, MDR-A. baumannii, -P. aeruginosa and -E. coli almost all showed resistance to carbapenems, but none of the 5 MDR-E. coli isolates was CR-MDR strain. Except the species of E. coli, the pathogens of carbapenem-resistant A. baumannii, carbapenem-resistant P. aeruginosa, carbapenem- and 3rd generation cephalosporin-resistant Enterobacteriaceae including K. pneumoniae were grouped into the critical priority tiers according to the release of World Health Organization in 2017 [36]. To further understand whether there is an association between the multidrug resistant strains and the host strain, we just could take a univariate analysis upon the results of 12 antibiotics susceptibilities. The results of which may show that K. pneumoniae was close to A. baumannii and K. oxytoca, and a significant difference from P. aeruginosa and E. coli under the antibiotic resistance. Possibly, in polymicrobial communities, the former shared the same metabolite to promote each other in the context of antibiotic resistance [37, 38], and thus commensal organisms in lung may carry the antibiotic resistance elements that could be transferred to opportunistic bacteria or respiratory pathogens [39]. Oppositely, the latter implied competition among bacterial species. Hafza et al. [40] suggested that competition between ESBL-production genes in E. coli and K. pneumoniae confer a fitness cost may provide us a new perspective. And also, the differences in the main type of carbapenemases production among resistant bacteria should be considered. The most prevalent carbapenemases production of P. aeruginosa was MBLs [41, 42], particularly IMP, which had significantly higher resistance to most antibiotics [41]. Due to evidence of cooperation or competition between bacteria was so few, our understanding about how relationships between different bacterial species occur still remains confusion. Therefore, plenty of studies are need to explore in the future research.
The key limitations of this study were as follow: It is a retrospective analysis that does not allow to draw profound conclusions on the relationships between CRKP isolates and their neighbours for unable to perform analyses of resistance mechanisms. In addition, we did not perform plasmid detection and conjugation analysis on CRKP isolates, particularly co-harbored blaKPC-2 and blaNDM-1 and blaKPC-2 and blaIMP-38 like, which would be explored in a subsequent study.
In conclusion, this is the first study on the molecular assessments of CRKP isolates with NP from the tertiary hospital of China. A significant finding from such study is that the different STs displayed a scientific moderation PFGE profiles and both clonal spread and horizontal transfer contributed to the dissemination of CRKP. ST11 harboring KPC-2-producing K. pneumoniae isolates were dominant for NP. Notably, as far as we all know, the two CRKP isolates co-harbored blaKPC-2 and blaIMP-38 like was first reported, and more unexpected both belonged to a different clone. In addition, our understanding about how relationships between the host CRKP isolates and their GNNs occur still remain obscure. Therefore, additional studies are needed to explore their biological relevance.
References
Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O’Grady NP, Bartlett JG (2016) Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 63:e61–e111
Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308
Guitor AK, Wright GD (2018) Antimicrobial resistance and respiratory infections. Chest 154:1202–1212
O’Donnell JN, Rhodes NJ, Lopez J, Jett R, Scheetz MH (2018) Carbapenems vs. alternative β-lactams for the treatment of nosocomial pneumonia: a systematic review and meta-analysis. Int J Antimicrob Agents 52:451-458.5
Pang F, Jia XQ, Zhao QG, Zhang Y (2018) Factors associated to prevalence and treatment of carbapenem-resistant Enterobacteriaceae infections: a seven years retrospective study in three tertiary care hospitals. Ann Clin Microbiol Antimicrob 17:13
Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y (2012) Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.06268-11
Ahmed SMA, Abdelrahman SS, Saad DM, Osman IS, Osman MG, Khalil EAG (2018) Etiological trends and patterns of antimicrobial resistance in respiratory infections. Open Microbiol J 12:34–40
de Maio Carrilho CM, de Oliveira LM, Gaudereto J, Perozin JS, Urbano MR, Camargo CH, Grion CM, Levin AS, Costa SF (2016) A prospective study of treatment of carbapenem-resistant Enterobacteriaceae infections and risk factors associated with outcome. BMC Infect Dis 16:629
Dickson RP, Huffnagle GB (2015) The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog 11:e1004923
Ruppé É, Woerther P-L, Barbier F (2015) Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care 5:21
Andrade LN, Vitali L, Gaspar GG, Bellissimo-Rodrigues F, Martinez R, Darini AL (2014) Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol 52:2530–2535
Mammina C, Bonura C, Vivoli AR, Di Bernardo F, Sodano C, Saporito MA, Verde MS, Saporito L, Cracchiolo AN (2013) Co-colonization with carbapenem-resistant Klebsiella pneumoniae and Acinetobacter baumannii in intensive care unit patients. Scand J Infect Dis 45:629–634
Marchaim D, Perez F, Lee J, Bheemreddy S, Hujer AM, Rudin S, Hayakawa K, Lephart PR, Blunden C (2012) “Swimming in resistance”: co-colonization with carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii or Pseudomonas aeruginosa. Am J Infect Control 40:830–835
Poirel L, Aires-de-Sousa M, Kubyda P, Kieffer N, Nordmann P (2018) Screening and characterization of multidrug-resistant Gram-negative bacteria from a remote African area, São Tomé and Príncipe. Antimicrob Agents Chemother 62:e01021–18
Yang J, Ye L, Wang W, Luo Y, Zhang Y, Han L (2011) Diverse prevalence of 16S rRNA methylase genes armA and rmtB amongst clinical multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolates. Int J Antimicrob Agents 38:348–351
Venturelli OS, Carr AC, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, Arkin AP (2018) Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol 14:e8157
Mohammed WS, Ziganshina EE, Shagimardanova EI, Gogoleva NE, Ziganshin AM (2018) Comparison of intestinal bacterial and fungal communities across various xylophagous beetle larvae (Coleoptera: Cerambycidae). Sci Rep 8:10073
Kiddee A, Assawatheptawee K, Na-Udom A, Treebupachatsakul P, Wangteeraprasert A, Walsh TR, Niumsup PR (2018) Risk Factors for gastrointestinal colonization and acquisition of carbapenem-resistant gram-negative bacteria among patients in intensive care units in Thailand. Antimicrob Agents Chemother 62:e00341–e00318
Torres A, Zhong N, Pachl J, Timsit JF, Kollef M, Chen Z, Song J, Taylor D, Laud PJ, Stone GG, Chow JW (2018) Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect Dis 18:285–295
Clinical and Laboratory Standards Institute (2016) Performance standards for antimicrobial susceptibility testing: twenty-sixth informational supplement M100-S26. CLSI, Wayne
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281
Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L (2014) Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208
Peleg AY, Hooper DC (2010) Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 362:1804–1813
Jones RN (2010) Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator associated bacterial pneumonia. Clin Infect Dis 51(Suppl 1):S81–S87
Katchanov J, Asar L, Klupp EM, Both A, Rothe C, König C, Rohde H, Kluge S, Maurer FP (2018) Carbapenem-resistant Gram-negative pathogens in a German university medical center: prevalence, clinical implications and the role of novel β-lactam/β-lactamase inhibitor combinations. PLoS ONE 13:e0195757
Pontikis K, Karaiskos I, Bastani S, Dimopoulos G, Kalogirou M, Katsiari M, Oikonomou A, Poulakou G, Roilides E, Giamarellou H (2014) Outcomes of critically ill intensive care unit patients treated with fosfomyc in for infections due to pandrug-resistant and extensively drug-resistant carbapenemase-producing Gram-negative bacteria. Int J Antimicrob Agents 43:52–59
Li H, Zhang J, Liu Y, Zheng R, Chen H, Wang X, Wang Z, Cao B, Wang H (2014) Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis 78:63–65
Pitout JD, Nordmann P, Poirel L (2015) Carbapenemase-producing Klebsiella pneumoniae: a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884
Liu J, Yu J, Chen F, Yu J, Simner P, Tamma P, Liu Y, Shen L (2018) Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur J Clin Microbiol Infect Dis 37:293–299
Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Canton R, Darini AL, Coque TM (2011) Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55:3579–3583
Jeon JH, Lee JH, Lee JJ, Park KS, Karim AM, Lee CR, Jeong BC, Lee SH (2015) Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci 16:9654–9692
Qin S, Fu Y, Zhang Q, Qi H, Wen JG, Xu H, Xu L, Zeng L, Tian H, Rong L, Li Y, Shan L, Xu H, Yu Y, Feng X, Liu HM (2014) High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother 58:4275–4282
Liu Y, Wan LG, Deng Q, Cao XW, Yu Y, Xu QF (2015) First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol Infect 143:376–384
Hawkey PM, Xiong J, Ye H, Li H, M’Zali FH (2001) Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People’s Republic of China. FEMS Microbiol Lett 194:53–57
Jian Z, Li Y, Liu W, Li H, Zhang Y, Li Y, Gu X, Peng W (2014) Detection of the novel IMP-38 among carbapenemase-producing Enterobacteriaceae in a University Hospital, China. J Infect Dev Ctries 8:1044–1048
World Health Organization (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva
Morris BE, Henneberger R, Huber H, Moissl-Eichinger C (2013) Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37:384–406
Yurtsev EA, Conwill A, Gore J (2016) Oscillatory dynamics in a bacterial cross-protection mutualism. Proc Natl Acad Sci USA 113:6236–6241
Perry JA, Westman EL, Wright GD (2014) The antibiotic resistome: what’s new? Curr Opin Microbiol 21:45–50
Hafza N, Challita C, Dandachi I, Bousaab M, Dahdouh E, Daoud Z (2018) Competition assays between ESBL-producing E. coil and K. pneumoniae isolates collected from Lebanese elder: an additional cost on fitness. J Infect Public Health 11:393–397
Osawa K, Shigemura K, Kitagawa K, Fukuda T, Takasaka A, Wakabayashi S, Sato K, Yamamichi F, Shirakawa T, Fujisawa M (2018) Molecular characteristics of carbapenem-resistant Pseudomonas aeruginosaisolated from urine in Hyogo, Japan. Int J Urol. https://doi.org/10.1111/iju.13818
Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K (2015) Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa. Infect Chemother 47:81–97
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 81673242).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, D., Zuo, Y., Wang, Z. et al. Characterize carbapenem-resistant Klebsiella pneumoniae isolates for nosocomial pneumonia and their Gram-negative bacteria neighbors in the respiratory tract. Mol Biol Rep 46, 609–616 (2019). https://doi.org/10.1007/s11033-018-4515-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4515-y