Abstract
The hormone insulin plays a central role in the metabolism of carbohydrates, lipids, and proteins. In relation to protein metabolism, insulin stimulates amino acid uptake and activates protein synthesis in responsive cells by modulation of signal transduction pathways, such as associated to Akt/PkB, mTOR, S6Ks, 4E-BP1, and several translation initiation/elongation factors. In this context, there is no information on direct cellular treatment with insulin and effects on eukaryotic translation initiation factor 5A (eIF5A) regulation. The eIF5A protein contains an exclusive amino acid residue denominated hypusine, which is essential for its activity and synthesized by posttranslational modification of a specific lysine residue using spermidine as substrate. The eIF5A protein is involved in cellular proliferation and differentiation processes, as observed for satellite cells derived from rat muscles, revealing that eIF5A has an important role in muscle regeneration. The aim of this study was to determine whether eIF5A expression and hypusination are influenced by direct treatment of insulin on L6 myoblast cells. We observed that insulin increased the content of eIF5A transcripts. This effect occurred in cells treated or depleted of fetal bovine serum, revealing a positive insulin effect independent of other serum components. In addition, it was observed that hypusination follows the maintenance of eIF5A protein content in the serum depleted cells and treated with insulin. These results demonstrate that eIF5A is modulated by insulin, contributing the protein synthesis machinery control, as observed by puromycin incorporation in nascent proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Insulin is a peptide hormone produced in the islets of Langerhans by the pancreatic β-cells, which plays a central role in the metabolism of carbohydrates, lipids, and proteins [1, 2]. In protein metabolism, insulin stimulates the uptake of amino acids and activates the translational machinery through the PI3K/AKT/mTOR signaling pathway in responsive tissues [3]. The mTOR protein forms two distinct complexes, the rapamycin-sensitive mTORC1 complex and the mTORC2 complex which is relatively resistant to this compound [4]. The mTORC1 activates the ribosomal protein S6 kinase (p70S6K) through phosphorylation, which subsequently phosphorylates the ribosomal protein S6 intensifying the protein synthesis and cell proliferation [5]. In addition, mTORC1 is responsible for the phosphorylation of the 4E-BPs, which are protein elF4E binding proteins and act as translation repressors [6, 7]. When phosphorylated by mTOR, 4E-BP1 dissociates from the eIF4E, allowing it to connect to the cap present at the 5′ end of the mRNAs and to the elF4G factor, forming the elF4F complex. As a result, it activates cap-dependent translation [8]. Moreover, insulin modulates other protein synthesis effectors, such as some initiation and elongation translation factors (Table 1), among which the eIF5A protein has not been characterized in this context so far.
Eukaryotic translation initiation factor 5A (eIF5A) is an essential protein of approximately 17 kDa and highly conserved in eukaryotes and among its homologous of Archaea [31,32,33,34]. eIF5A is the only protein known featuring the amino acid residue hypusine, derived from a posttranslational modification (hypusination), involving the polyamine spermidine [35]. It is known that hypusination of eIF5A is essential for cell viability because the cells in which it was blocked became nonviable [32, 34, 36].
The eIF5A protein is involved in the initiation and elongation of translation and is related to the processes of transcription, the transit of macromolecules through the nuclear pore complex, nonsense-mediated mRNA decay and cell proliferation and differentiation [32, 37,38,39,40,41,42,43,44,45,46]. The eIF5A protein has also been linked to inflammatory processes, diabetes, cancer, malaria and viral infections, such as HIV-1 and Ebola [47,48,49,50,51,52,53,54,55,56]. eIF5A is associated with ribosomes actively engaged in translation, strengthening the involvement of this protein in the protein synthesis process [57, 58]. Moreover, it has been shown that eIF5A acts in the translation elongation of proteins containing proline-rich motifs [59].
As previously described, several studies relate insulin to different translational factors; however, there is no evidence relating its action on eIF5A. Thus, this study aimed to evaluate changes in the eIF5A expression and hypusination profile in relation to the action of insulin.
Materials and methods
Cell culture
The myoblast cell line L6, derived from skeletal muscle was acquired from the laboratory of Prof. Dr. Rui Curi (ICB - USP) and it was grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin (Life Technologies, Inc., Carlsbad, CA, USA) at 37 °C in a humidified atmosphere containing 5% CO2.
Real-time RT-PCR
Total RNA was extracted from L6 cells with the TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) and 2 µg was used as a template for the synthesis of cDNA using RevertAid Reverse Transcriptase (Invitrogen). Real-time PCR reactions were performed using 100 ng cDNA, 0.6 µM primers (Exxtend, Paulínia, SP, Brazil), and SYBR® Select Master Mix (Applied Biosystem, Warrington, UK). For the PCR running, it was used the following parameters: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Each set of primers was designed to recognize unique regions of eIF5A cDNA (Forward 5′AATAACTGGCTTCCAGGGTGG3′ and Reverse 5′TGGATAGGAGTCCAGCCAAG3′) and the constitutively expressed endogenous β2M cDNA (Forward 5′TGAATTCACACCCACCGAGA3′ and Reverse 5′TTACATGTCTCGGTCCCAGG3′). The β2M constitutive gene was chosen because it was not significantly altered by the treatments performed. Relative content of mRNAs was determined after normalization with β2M gene using the comparative Ct (cycle threshold) method. This method is presented as the fold over of target gene of control. Primers concentrations were optimized for maximum amplification efficiency by testing combinations of different concentrations of each primer (0.15, 0.3, and 0.6 µM). Different dilutions of the target cDNAs were also assessed to determine the amplification efficiency (400, 80, 16, 3.2, and 0.64 ng).
Antibodies
Mouse monoclonal anti-eIF5A1 antibody (Cat. Number: E1783) was acquired from Sigma-Aldrich, Inc. (Saint Louis, MO, USA). Rabbit polyclonal anti-hypusine antibody (IU-88) was provided by Raghavendra G. Mirmira and Bernhard Maier [60]. Rabbit polyclonal anti-4E-BP1 (Cat. Number: 9452) was acquired from Cell Signaling Technology, Inc. (Danvers, MA, USA), rabbit polyclonal anti-GAPDH antibody (Cat. Number: CSB-PA00025A0Rb) was acquired from Cusabio Technology, Co., Ltd (Wuhan, Hubei, China), and mouse monoclonal anti-puromycin antibody (Cat. Number: MABE343) was acquired from Merck Millipore (Darmstadt, HE, Germany). The secondary antibodies HRP-goat anti-mouse (Cat. Number: 626520) and HRP-goat anti-rabbit (Cat. Number: 656120) were acquired from Invitrogen.
Immunoblotting
L6 cells were washed with phosphate buffered saline (PBS) followed by disruption using lysis buffer (100 mM Tris/HCl, pH 7.4; 10 mM Na2 ethylenediaminetetraacetic acid [EDTA], pH 8.0; 1% Triton X-100; and complete EDTA-free protease inhibitor cocktail [Roche Diagnostics GmbH, Mannheim, Germany]). For the protein separation, it was used 30 µg of total protein extract, which was resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose membranes and blocked with Tris-buffered saline with tween 20 (TBS-T) containing 5% non-fat milk. Membranes were then incubated overnight at 4 °C with the desired primary antibodies diluted in TBS-T containing 5% non-fat milk for anti-eIF5A1 and IU-88, or 5% bovine serum albumin (BSA) for anti-4E-BP1, anti-puromycin, and anti-GAPDH. Membranes were washed with TBS-T and incubated for 1 h at room temperature with the appropriate specific horseradish peroxidase-conjugated anti-immunoglobulin (IgG) (GE Healthcare, Buckinghamshire, UK) diluted in TBS-T containing 5% non-fat milk. Lastly, membranes were washed with TBS-T and immunoreactive proteins were identified using an enhanced chemiluminescence (ECL) reagent (GE Healthcare).
Cell treatment
L6 cells were seeded at a density of 4 × 105 cells per well in six-well plates and grown for 24 h in the regular culture medium. The cells were treated with 50 µM GC7 in regular culture medium for 48 h. The GC7 was protected against the action of amine oxidases in serum using 1 mM aminoguanidine (Sigma-Aldrich), it is used for others studies with GC7 [61, 62]. The GC7-negative control was also treated with 1 mM aminoguanidine. After the treatment period, cells were grown in regular culture medium or serum-free culture medium with or without 430 nM insulin (Sigma-Aldrich) for 24 h. Finally, we used the surface sensing of translation (SUnSET) technique by adding 1 µM puromycin dihydrochloride (Life Technologies) for 30 min before harvesting cells for further analysis [63, 64]. This technique specifically comprises the use of the anti-puromycin antibody for the immunological detection of puromycin-labeled peptides.
Cell counting and viability
Cells were collected by trypsinization from cell culture plates and cell counting and cell viability were performed in triplicates after Trypan Blue staining using Countess® Automated Cell Counter (Invitrogen). For the assay, 10 µL of sample was mixed with 10 µL of trypan blue, and 10 µL pipetted into a Countess chamber slide for cell counting. The procedure followed the manufacturer’s protocol.
Statistical analysis
Three to five independents experiments were performed with cells pertaining to different passages. All values were expressed as mean ± SEM. Different treatments were compared by One-Way ANOVA, followed by post-hoc Tukey test or analyzed by Student’s t-test. For nonparametric data, different treatments were compared by Kruskal–Wallis one-way ANOVA on Ranks test. Statistical analyses were carried out using SigmaStat version 3.5 for Windows (Systat Software, Inc., Point Richmond, CA). Differences were considered statistically significant when probabilities were < 0.05 (P < 0.05). Different letters indicate significant differences.
Results
The Content of hypusinated eIF5A is modulated by insulin
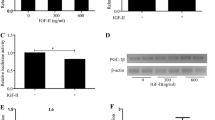
To assess the protein content of total and hypusinated eIF5A, L6 cells were incubated for 24 h in culture medium without fetal bovine serum (FBS) (Fig. 1a). Later, using qRT-PCR analysis, we observed a significant reduction in the content of the transcript of eIF5A. However, when the FBS depleted cells were treated with insulin for 24 h, it was observed an increase of the eIF5A transcript content, not only in relation to the cells submitted to the FBS depletion but also in relation to the cells grown in regular medium (10% FBS) (Fig. 1g).
Analysis of the action of insulin on eIF5A expression and its repercussion on eIF5A hypusination and on general protein synthesis in L6 cells. a Diagram of the experimental procedures used in the treatment of L6 cells. b Immunoblotting test using protein extracts of L6 myoblasts. Representative image of polypeptides marked with puromycin (SUnSET), GAPDH (normalizer), as well as of the profile of phosphorylation of protein 4E-BP1, total eIF5A and hypusinated eIF5A (IU-88), in response to the treatment with insulin and/or total removal of bovine fetal serum for 24 h. c Graphic representation of the puromycin normalized with GAPDH. d Graphic representation of the 4E-BP1 normalized with GAPDH. e Graphic representation of the eIF5A normalized with GAPDH. f Graphic representation of the hypusinated eIF5A (IU-88) normalized with GAPDH. Representative images of 3 independent experiments. g Analysis of the relative content of the eIF5A transcript by real-time RT-PCR. Relative content of mRNAs was determined after normalization with β2M gene using the comparative cycle threshold (Ct) method. This method is presented as the fold over of target gene of control. The results are presented as mean ± SEM of 3–5 independent experiments. Different letters indicate significant differences (P < 0.05)
In relation to the protein content of eIF5A analyzed by immunoblotting, it was found that the FBS depletion promoted decrease of the protein content of eIF5A and of its fraction hypusinated form (IU-88). However, the treatment with insulin caused the maintenance of total and hypusinated protein content, when compared with the regular culture conditions (Fig. 1b, e, f). Similar effects were observed both in the general protein synthesis pattern, by the observation of the polypeptide chains with the incorporation of puromycin, and in the profile of 4E-BP1 phosphorylation (Fig. 1b–d). The FBS depletion promoted a reduction of the general protein synthesis and of the 4E-BP1 phosphorylation, while the treatment with insulin in the absence of serum preserved the expression profile in a similar level to the observed in the regular culture conditions (Fig. 1b–d).
GC7 promotes a persistent inhibitory action on deoxyhypusine synthase (DHS), causing an expressive inhibitory action on protein synthesis profile
In order to assess the general protein synthesis after inhibition of eIF5A hypusination and to determine whether the action of insulin occurs directly on the reaction of hypusination, we used the analogous of spermidine N1-guanyl-1,7-diaminoheptane (GC7), an inhibitor of the DHS enzyme [65]. Treatment with GC7 caused a reduction in cell proliferation (Fig. 2a, b), an effect already described in the literature [61, 66], but caused no effects on cell viability in the conditions assayed (Fig. 2c). Thus, L6 cells with approximately 70% of confluence, grown in regular medium, were first treated with GC7 (50 µM) for 48 h, to reduce the content of hypusinated eIF5A. Then, the GC7 was removed by removing the regular culture medium, which was replaced with medium without FBS in the presence or absence of insulin (430 nM), conditions under which the cells remained for 24 h. At the end of the 24-h period, the cells were treated with puromycin (1 µM) for 30 min, in order to observe the general protein synthesis profile (Fig. 3a).
Assessment of viability and number of cells after treatment with GC7 for 48 h. a Microphotographs of L6 cells, before and 48 h after treatment with GC7 (50 µM) + aminoguanidine (AG) (1 mM) for 48 h. The GC7-negative control was also treated with AG (1 mM) for 48 h. b Assessment of the total number of cells with Countess® Automated Cell Counter (Invitrogen). c Assessment of cell viability by staining with trypan blue. The results are presented as mean ± SEM of 3 independent experiments. Different letters indicate significant differences (P < 0.05)
Analysis of the action of insulin on eIF5A expression and its repercussion on eIF5A hypusination and on general protein synthesis in L6 cells previously treated with an inhibitor of hypusination (GC7). a Diagram of the experimental procedures used in the treatment of L6 cells. b Immunoblotting test using protein extracts of L6 myoblasts. Representative image of polypeptides marked with puromycin (SUnSET), GAPDH (normalizer), as well as of the phosphorylation profile of the protein 4E-BP1, eIF5A and hypusinated eIF5A (IU-88), in response to the treatment with insulin in cells previously treated with GC7 (50 µM) + aminoguanidine (AG) (1 mM) for 48 h and total removal of bovine fetal serum for 24 h. The GC7-negative control was also treated with AG (1 mM). Representative images of 3 independent experiments. c Graphic representation of the puromycin normalized with GAPDH. d Graphic representation of the 4E-BP1 normalized with GAPDH. e Graphic representation of the eIF5A normalized with GAPDH. f Graphic representation of the hypusinated eIF5A (IU-88) normalized with GAPDH. Representative images of 3 independent experiments. g Analysis of the relative content of the eIF5A transcript by real-time RT-PCR. Relative content of mRNAs was determined after normalization with β2M gene using the comparative cycle threshold (Ct) method. This method is presented as the fold over of target gene of control. The results are presented as mean ± SEM of 3–5 independent experiments. Different letters indicate significant differences (P < 0.05)
In relation to the eIF5A transcript, it was observed that the treatment with GC7 promoted an increase in its content in cells grown in regular medium (Fig. 3g). FBS depletion of cells treated with GC7 caused the reduction in the transcript content; however, when compared with the cells grown in regular medium no difference was observed. In the analysis of cells treated with GC7 in the culture medium without FBS and treated with insulin, it was observed an increase in the content of eIF5A compared to the cells grown in regular medium, similarly to the effect observed with GC7 treatment in the presence of FBS. In relation to the eIF5A protein content and of its hypusinated form (IU-88), only the hypusinated form was reduced after treatment with GC7 (Fig. 3b, e, f). In addition, it was observed that cells treated with GC7 presented significant suppression of general protein synthesis (Fig. 3b, c). Addition of either insulin or FBS, despite having promoted the hyperphosphorylation of 4E-BP1, was unable to revert this profile (Fig. 3b, d). This result shows a lasting effect of the inhibition of hypusination promoted by GC7, with clear detriment to the translational machinery of the cells, as observed through the immunoblotting of puromycin.
Discussion
eIF5A is an abundant and highly stable protein, with a half-life of approximately 24 h [67,68,69]. So, the high stability makes it difficult to observe discrete modulatory effects on its protein content in regular growth conditions. In this context, the presence of insulin and growth factors as components of the FBS added in the culture medium impairs the proper determination of the effects of insulin treatment on the protein eIF5A content (data not shown). Therefore, in order to evaluate the isolated action of insulin, the tests were conducted in the absence of FBS. Under these conditions, we observed a reduction of the mRNA contents and protein of total and hypusinated eIF5A. However, when the cells were treated with insulin, in the absence of FBS, there was an increase in the parameters evaluated. This result shows a regulatory effect of insulin on eIF5A. The results obtained suggest different possibilities of action of insulin on the activation of eIF5A. One of the ways would be by modulating the expression of eIF5A transcripts, which may generate a higher availability of the total protein, enabling the increase of its hypusinated form. Another possibility would be by directly favoring the hypusination of eIF5A and its consequent activation. It is important to note that both propositions may occur simultaneously. However, the fact that the treatment with insulin increased the protein content of total eIF5A, similarly to hypusinated eIF5A, makes it impossible to affirm that insulin has a direct action on the hypusination reaction. Thus, even if indirectly, we observed that insulin modulated the activation of eIF5A having clear reflections on cellular protein synthesis. A previous study observed that rat insulinoma INS-1E cells treated with triiodo-l-thyronine (T3) presented an increase in its eIF5A protein content accompanied by insulin increase [70]. Although this aforementioned study presented a similar overview, it was focused on thyroid hormone actions on translation factors. These results in addition to ours corroborate the possibility of an indirect effect of the T3 hormone on eIF5A expression, which could also be a consequence of insulin increase and action in those cells.
Aiming to determine a possible direct action of insulin on the hypusination reaction, we conducted tests using the synthetic compound GC7, the most potent inhibitor of the DHS enzyme [65]. The GC7 acts by blocking the first stage of the eIF5A hypusination process by inhibiting the deoxyhypusine synthase (DHS) enzyme [71]. In this study, we observed decreased cell proliferation of L6 cells treated with GC7, which had no impact on the reduction of cell viability. Furthermore, it has been shown previously that the GC7 caused inhibition of the growth and proliferation of tumor cells by induction of mitochondrial apoptotic pathways and activation of AMPK [72] and also anticancer effects [38].
The treatment of cells with GC7 caused a decrease in the protein content of hypusinated eIF5A, as described previously [73], the same was observed for total eIF5A. However, treatment with GC7 increased the content of the eIF5A transcript. These results suggest that the inhibition of hypusination mediated by GC7 could—through a compensation mechanism—promote increased gene transcription rate of the gene that encodes eIF5A or increased stability of the transcript, in order to restore the content of eIF5A. After removal of GC7 and addition of serum or insulin, we observed that there was no restoration of the protein content of hypusinated eIF5A, showing an expressive and lasting effect of GC7. Moreover, treatment with GC7 promoted a drastic reduction of the general protein synthesis, as observed through the profile of the polypeptides incorporated by puromycin. In addition, the addition of insulin was not capable of stimulating the general protein synthesis in these cells. Recently, it was demonstrated that eIF5A depletion results in a global translation elongation and termination defect, which corroborates to our data and reinforces that hypusinated eIF5A acts on general protein synthesis [74].
Contrarily to the results previously described, we observed an increase in the phosphorylation of 4E-BP1, which would be related to a favoring of protein synthesis. The protein 4E-BP1 is one of the important factors that regulate the initiation of translation, preventing the formation of the eIF4F complex, which is responsible for the activation of the cap-dependent translation of mRNAs [8]. The hyperphosphorylation of 4E-BP1 indicates that its activity is inhibited, which favors protein synthesis. Differently to our results, it was observed in a previous study a decrease in the phosphorylation of 4E-BP1 through the treatment with GC7 [71]. However, in this study, differently from the aforementioned work, after 48 h of treatment with GC7, the cells remained for 24 h in the absence of this protein synthesis inhibitor and in the presence of FBS or insulin. Thus, the increased phosphorylation of 4E-BP1 may be related to the cell’s attempt to resume protein synthesis after the block period.
This study is the first report that demonstrates the modulatory action of insulin on eIF5A in mammalian cell culture, contributing to expanding the knowledge on the regulation of protein synthesis by insulin. Furthermore, it highlights eIF5A as a new target to be incorporated into the list of factors involved in the modulation of translation by the insulin signaling pathway [3, 20, 75, 76]. Therefore, we conclude that it is relevant to intensify the study on eIF5A in other models, aiming to characterize the effects of insulin on metabolism.
References
Best CH (1930) A brief review of certain physiological properties of insulin. Can Med Assoc J 23(2):141–145
Proud CG, Denton RM (1997) Molecular mechanisms for the control of translation by insulin. Biochem J 328(Pt 2):329–341
Proud CG (2006) Regulation of protein synthesis by insulin. Biochem Soc Trans 34(Pt 2):213–216. https://doi.org/10.1042/BST20060213
Foster KG, Fingar DC (2010) Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem 285(19):14071–14077. https://doi.org/10.1074/jbc.R109.094003
Magnuson B, Ekim B, Fingar DC (2012) Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 441(1):1–21. https://doi.org/10.1042/BJ20110892
Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N (2001) Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 15(21):2852–2864. https://doi.org/10.1101/gad.912401
Kim DH, Sabatini DM (2004) Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr Top Microbiol Immunol 279:259–270
Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5(10):827–835. https://doi.org/10.1038/nrm1488
Welsh GI, Stokes CM, Wang X, Sakaue H, Ogawa W, Kasuga M, Proud CG (1997) Activation of translation initiation factor eIF2B by insulin requires phosphatidyl inositol 3-kinase. FEBS Lett 410(2–3):418–422
Shen WH, Boyle DW, Wisniowski P, Bade A, Liechty EA (2005) Insulin and IGF-I stimulate the formation of the eukaryotic initiation factor 4F complex and protein synthesis in C2C12 myotubes independent of availability of external amino acids. J Endocrinol 185(2):275–289. https://doi.org/10.1677/joe.1.06080
Shen W, Mallon D, Boyle DW, Liechty EA (2002) IGF-I and insulin regulate eIF4F formation by different mechanisms in muscle and liver in the ovine fetus. Am J Physiol Endocrinol Metab 283(3):E593–E603. https://doi.org/10.1152/ajpendo.00570.2001
Minich WB, Balasta ML, Goss DJ, Rhoads RE (1994) Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA 91(16):7668–7672
Sullivan JM, Alousi SS, Hikade KR, Bahu NJ, Rafols JA, Krause GS, White BC (1999) Insulin induces dephosphorylation of eukaryotic initiation factor 2alpha and restores protein synthesis in vulnerable hippocampal neurons after transient brain ischemia. J Cereb Blood Flow Metab 19(9):1010–1019. https://doi.org/10.1097/00004647-199909000-00009
Towle CA, Mankin HJ, Avruch J, Treadwell BV (1984) Insulin promoted decrease in the phosphorylation of protein synthesis initiation factor eIF-2. Biochem Biophys Res Commun 121(1):134–140
Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC (2006) mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25(8):1659–1668. https://doi.org/10.1038/sj.emboj.7601047
Brina D, Miluzio A, Ricciardi S, Clarke K, Davidsen PK, Viero G, Tebaldi T, Offenhäuser N, Rozman J, Rathkolb B, Neschen S, Klingenspor M, Wolf E, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, Quattrone A, Falciani F, Biffo S (2015) eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat Commun 6:8261. https://doi.org/10.1038/ncomms9261
Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N (2000) Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J 19(3):434–444. https://doi.org/10.1093/emboj/19.3.434
Vary TC, Lynch CJ (2007) Nutrient signaling components controlling protein synthesis in striated muscle. J Nutr 137(8):1835–1843
Morley SJ, Traugh JA (1990) Differential stimulation of phosphorylation of initiation factors eIF-4F, eIF-4B, eIF-3, and ribosomal protein S6 by insulin and phorbol esters. J Biol Chem 265(18):10611–10616
Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N (2006) The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 25(12):2781–2791. https://doi.org/10.1038/sj.emboj.7601166
Holz MK, Ballif BA, Gygi SP, Blenis J (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123(4):569–580. https://doi.org/10.1016/j.cell.2005.10.024
Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23(1):26–37
Dennis MD, Jefferson LS, Kimball SR (2012) Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J Biol Chem 287(51):42890–42899. https://doi.org/10.1074/jbc.M112.404822
Piazzi M, Bavelloni A, Faenza I, Blalock W, Urbani A, D’Aguanno S, Fiume R, Ramazzotti G, Maraldi NM, Cocco L (2010) eEF1A phosphorylation in the nucleus of insulin-stimulated C2C12 myoblasts: Ser53 is a novel substrate for protein kinase C βI. Mol Cell Proteomics 9(12):2719–2728. https://doi.org/10.1074/mcp.M110.003152
Peters HI, Chang YW, Traugh JA (1995) Phosphorylation of elongation factor 1 (EF-1) by protein kinase C stimulates GDP/GTP-exchange activity. Eur J Biochem 234(2):550–556
Chang YW, Traugh JA (1997) Phosphorylation of elongation factor 1 and ribosomal protein S6 by multipotential S6 kinase and insulin stimulation of translational elongation. J Biol Chem 272(45):28252–28257
Venema RC, Peters HI, Traugh JA (1991) Phosphorylation of elongation factor 1 (EF-1) and valyl-tRNA synthetase by protein kinase C and stimulation of EF-1 activity. J Biol Chem 266(19):12574–12580
Chang YW, Traugh JA (1998) Insulin stimulation of phosphorylation of elongation factor 1 (eEF-1) enhances elongation activity. Eur J Biochem 251(1–2):201–207
Levenson RM, Nairn AC, Blackshear PJ (1989) Insulin rapidly induces the biosynthesis of elongation factor 2. J Biol Chem 264(20):11904–11911
Browne GJ, Proud CG (2004) A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol 24(7):2986–2997
Klier H, Wohl T, Eckerskorn C, Magdolen V, Lottspeich F (1993) Determination and mutational analysis of the phosphorylation site in the hypusine-containing protein Hyp2p. FEBS Lett 334(3):360–364
Park MH, Lee YB, Joe YA (1997) Hypusine is essential for eukaryotic cell proliferation. Biol Signals 6(3):115–123
Park MH, Wolff EC, Folk JE (1993) Hypusine: its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors 4(2):95–104
Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW (1991) Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol 11(6):3105–3114
Park MH (2006) The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J Biochem 139(2):161–169. https://doi.org/10.1093/jb/mvj034
Sasaki K, Abid MR, Miyazaki M (1996) Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett 384(2):151–154
Saini P, Eyler DE, Green R, Dever TE (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459(7243):118–121. https://doi.org/10.1038/nature08034
Jasiulionis MG, Luchessi AD, Moreira AG, Souza PP, Suenaga AP, Correa M, Costa CA, Curi R, Costa-Neto CM (2007) Inhibition of eukaryotic translation initiation factor 5A (eIF5A) hypusination impairs melanoma growth. Cell Biochem Funct 25(1):109–114. https://doi.org/10.1002/cbf.1351
Zuk D, Jacobson A (1998) A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J 17(10):2914–2925. https://doi.org/10.1093/emboj/17.10.2914
Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Gorlich D (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J 19(16):4362–4371. https://doi.org/10.1093/emboj/19.16.4362
Rosorius O, Reichart B, Kratzer F, Heger P, Dabauvalle MC, Hauber J (1999) Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci 112(Pt 14):2369–2380
Luchessi AD, Cambiaghi TD, Hirabara SM, Lambertucci RH, Silveira LR, Baptista IL, Moriscot AS, Costa-Neto CM, Curi R (2009) Involvement of eukaryotic translation initiation factor 5A (eIF5A) in skeletal muscle stem cell differentiation. J Cell Physiol 218(3):480–489. https://doi.org/10.1002/jcp.21619
Parreiras-e-Silva LT, Luchessi AD, Reis RI, Oliver C, Jamur MC, Ramos RG, Oliveira EB, Curi R, Costa-Neto CM (2010) Evidences of a role for eukaryotic translation initiation factor 5A (eIF5A) in mouse embryogenesis and cell differentiation. J Cell Physiol 225(2):500–505. https://doi.org/10.1002/jcp.22229
Huang Y, Higginson DS, Hester L, Park MH, Snyder SH (2007) Neuronal growth and survival mediated by eIF5A, a polyamine-modified translation initiation factor. Proc Natl Acad Sci USA 104(10):4194–4199. https://doi.org/10.1073/pnas.0611609104
Li Y, Fu L, Li JB, Qin Y, Zeng TT, Zhou J, Zeng ZL, Chen J, Cao TT, Ban X, Qian C, Cai Z, Xie D, Huang P, Guan XY (2014) Increased expression of EIF5A2, via hypoxia or gene amplification, contributes to metastasis and angiogenesis of esophageal squamous cell carcinoma. Gastroenterology 146(7):1701–1713.e1709. https://doi.org/10.1053/j.gastro.2014.02.029
Hoque M, Park JY, Chang YJ, Luchessi AD, Cambiaghi TD, Shamanna R, Hanauske-Abel HM, Holland B, Pe’ery T, Tian B, Mathews MB (2017) Regulation of gene expression by translation factor eIF5A: Hypusine-modified eIF5A enhances nonsense-mediated mRNA decay in human cells. Translation 5(2):e1366294. https://doi.org/10.1080/21690731.2017.1366294
de Almeida OP, Toledo TR, Rossi D, Rossetto DeB, Watanabe TF, Galvão FC, Medeiros AI, Zanelli CF, Valentini SR (2014) Hypusine modification of the ribosome-binding protein eIF5A, a target for new anti-inflammatory drugs: understanding the action of the inhibitor GC7 on a murine macrophage cell line. Curr Pharm Des 20(2):284–292
Kaiser A (2012) Translational control of eIF5A in various diseases. Amino Acids 42(2–3):679–684. https://doi.org/10.1007/s00726-011-1042-8
Nakanishi S, Cleveland JL (2016) Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids. https://doi.org/10.1007/s00726-016-2275-3
Imam S, Mirmira RG, Jaume JC (2014) Eukaryotic translation initiation factor 5A inhibition alters physiopathology and immune responses in a “humanized” transgenic mouse model of type 1 diabetes. Am J Physiol Endocrinol Metab 306(7):E791–E798. https://doi.org/10.1152/ajpendo.00537.2013
Olsen ME, Filone CM, Rozelle D, Mire CE, Agans KN, Hensley L, Connor JH (2016) Polyamines and hypusination are required for ebolavirus gene expression and replication. MBio 7(4):e00882-16. https://doi.org/10.1128/mBio.00882-16
Hanauske-Abel HM, Saxena D, Palumbo PE, Hanauske AR, Luchessi AD, Cambiaghi TD, Hoque M, Spino M, D’Alliessi Gandolfi D, Heller DS, Singh S, Park MH, Cracchiolo BM, Tricta F, Connelly J, Popowicz AM, Cone RA, Holland B, Pe’ery T, Mathews MB (2013) Drug-induced reactivation of apoptosis abrogates HIV-1 infection. PLoS ONE 8(9):e74414. https://doi.org/10.1371/journal.pone.0074414
Hoque M, Hanauske-Abel HM, Palumbo P, Saxena D, D’Alliessi Gandolfi D, Park MH, Pe’ery T, Mathews MB (2009) Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology 6:90. https://doi.org/10.1186/1742-4690-6-90
Mathews MB, Hershey JW (2015) The translation factor eIF5A and human cancer. Biochim Biophys Acta 1849(7):836–844. https://doi.org/10.1016/j.bbagrm.2015.05.002
Saxena D, Spino M, Tricta F, Connelly J, Cracchiolo BM, Hanauske AR, D’Alliessi Gandolfi D, Mathews MB, Karn J, Holland B, Park MH, Pe’ery T, Palumbo PE, Hanauske-Abel HM (2016) Drug-based lead discovery: the novel ablative antiretroviral profile of deferiprone in HIV-1-infected cells and in HIV-infected treatment-naive subjects of a double-blind, placebo-controlled, randomized exploratory trial. PLoS ONE 11(5):e0154842. https://doi.org/10.1371/journal.pone.0154842
Mémin E, Hoque M, Jain MR, Heller DS, Li H, Cracchiolo B, Hanauske-Abel HM, Pe’ery T, Mathews MB (2014) Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer Res 74(2):552–562. https://doi.org/10.1158/0008-5472.CAN-13-0474
Jao DL, Chen KY (2006) Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem 97(3):583–598. https://doi.org/10.1002/jcb.20658
Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR (2006) eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun 348(4):1358–1366. https://doi.org/10.1016/j.bbrc.2006.07.195
Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE (2013) eIF5A promotes translation of polyproline motifs. Mol Cell 51(1):35–45. https://doi.org/10.1016/j.molcel.2013.04.021
Nishiki Y, Farb TB, Friedrich J, Bokvist K, Mirmira RG, Maier B (2013) Characterization of a novel polyclonal anti-hypusine antibody. SpringerPlus 2:421. https://doi.org/10.1186/2193-1801-2-421
Park MH, Wolff EC, Lee YB, Folk JE (1994) Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J Biol Chem 269(45):27827–27832
Maier B, Ogihara T, Trace AP, Tersey SA, Robbins RD, Chakrabarti SK, Nunemaker CS, Stull ND, Taylor CA, Thompson JE, Dondero RS, Lewis EC, Dinarello CA, Nadler JL, Mirmira RG (2010) The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J Clin Invest 120(6):2156–2170. https://doi.org/10.1172/JCI38924
Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA (2011) Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 25(3):1028–1039. https://doi.org/10.1096/fj.10-168799
Goodman CA, Hornberger TA (2013) Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exercise Sport Sci Rev 41 (2):107–115. https://doi.org/10.1097/JES.0b013e3182798a95
Jakus J, Wolff EC, Park MH, Folk JE (1993) Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines. J Biol Chem 268(18):13151–13159
Shi XP, Yin KC, Ahern J, Davis LJ, Stern AM, Waxman L (1996) Effects of N1-guanyl-1,7-diaminoheptane, an inhibitor of deoxyhypusine synthase, on the growth of tumorigenic cell lines in culture. Biochim Biophys Acta 1310(1):119–126
Boisvert FM, Ahmad Y, Gierliński M, Charrière F, Lamont D, Scott M, Barton G, Lamond AI (2012) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteom 11(3):M111.011429. https://doi.org/10.1074/mcp.M111.011429
Gerner EW, Mamont PS, Bernhardt A, Siat M (1986) Post-translational modification of the protein-synthesis initiation factor eIF-4D by spermidine in rat hepatoma cells. Biochem J 239(2):379–386
Duncan RF, Hershey JW (1986) Changes in eIF-4D hypusine modification or abundance are not correlated with translational repression in HeLa cells. J Biol Chem 261(27):12903–12906
Goulart-Silva F, Teixeira Sda S, Luchessi AD, Dos Santos LR, Rebelato E, Carpinelli AR, Nunes MT (2012) Potential contribution of translational factors to triiodo-L-thyronine-induced insulin synthesis by pancreatic beta cells. Thyroid 22(6):637–642. https://doi.org/10.1089/thy.2011.0252
Landau G, Bercovich Z, Park MH, Kahana C (2010) The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J Biol Chem 285(17):12474–12481. https://doi.org/10.1074/jbc.M110.106419
Lee SK, Lee J, Lee SI, Bae WJ, Lee YM, Park JS, Park SJ, Min SK, Kim EC (2009) N(1)-guanyl-1,7,-diamineoheptane, an inhibitor of deoxyhypusine synthase, suppresses differentiation and induces apoptosis via mitochondrial and AMPK pathways in immortalized and malignant human oral keratinocytes. J Oral Pathol Med 38(10):792–800. https://doi.org/10.1111/j.1600-0714.2009.00809.x
Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K (2005) Independent roles of eIF5A and polyamines in cell proliferation. Biochem J 385(Pt 3):779–785. https://doi.org/10.1042/BJ20041477
Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R (2017) eIF5A functions globally in translation elongation and termination. Mol Cell 66(2):194–205 e195. https://doi.org/10.1016/j.molcel.2017.03.003
Diggle TA, Moule SK, Avison MB, Flynn A, Foulstone EJ, Proud CG, Denton RM (1996) Both rapamycin-sensitive and -insensitive pathways are involved in the phosphorylation of the initiation factor-4E-binding protein (4E-BP1) in response to insulin in rat epididymal fat-cells. Biochem J 316(Pt 2):447–453
Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136(4):731–745. https://doi.org/10.1016/j.cell.2009.01.042
Acknowledgements
We are grateful to São Paulo Research Foundation (FAPESP) for the financial support. We would like to thank Dr. Rui Curi (USP, Brazil) for L6 cells, Dr. Fernando M. Simabuco (UNICAMP, Brazil) for puromycin and mouse monoclonal anti-puromycin; Dr. Raghavendra G. Mirmira and Dr. Bernhard Maier (IU School of Medicine, USA) for rabbit polyclonal anti-hypusine antibody (IU-88); Dr. Sandro R. Valentini (UNESP, Brazil) for GC7, spermidine and aminoguanidine.
Funding
This work was supported by Grant 2010/18095-0 from the São Paulo Research Foundation (FAPESP). A.R.G.P., K.D.P, L.M, and L.T are recipients of FAPESP scholarships 2013/20504-3, 2013/23620-4, 2014/27154-0 and 2017/21914-1 respectively.
Author information
Authors and Affiliations
Contributions
ADL conceived and supervised the study; ADL and ARGP designed the experiments; ARGP, KDP, LM, and LT performed the experiments; ADL, ARGP, KDP, LM, and LT analyzed the data, interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
de Proença, A.R.G., Pereira, K.D., Meneguello, L. et al. Insulin action on protein synthesis and its association with eIF5A expression and hypusination. Mol Biol Rep 46, 587–596 (2019). https://doi.org/10.1007/s11033-018-4512-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4512-1